Beta-Blocker Separation on Phosphodiester Stationary Phases—The Application of Intelligent Peak Deconvolution Analysis
Abstract
1. Introduction
2. Results and Discussion
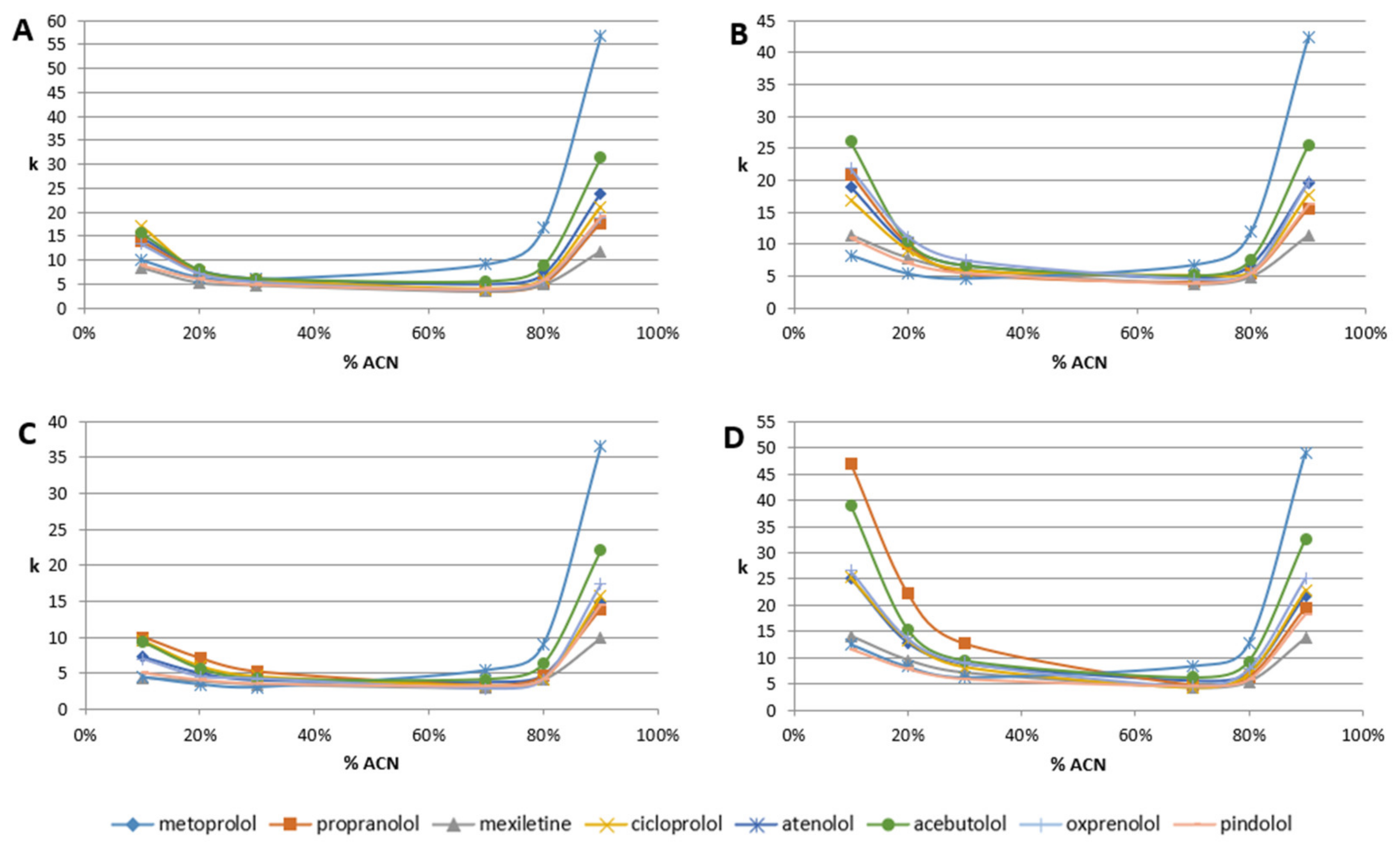
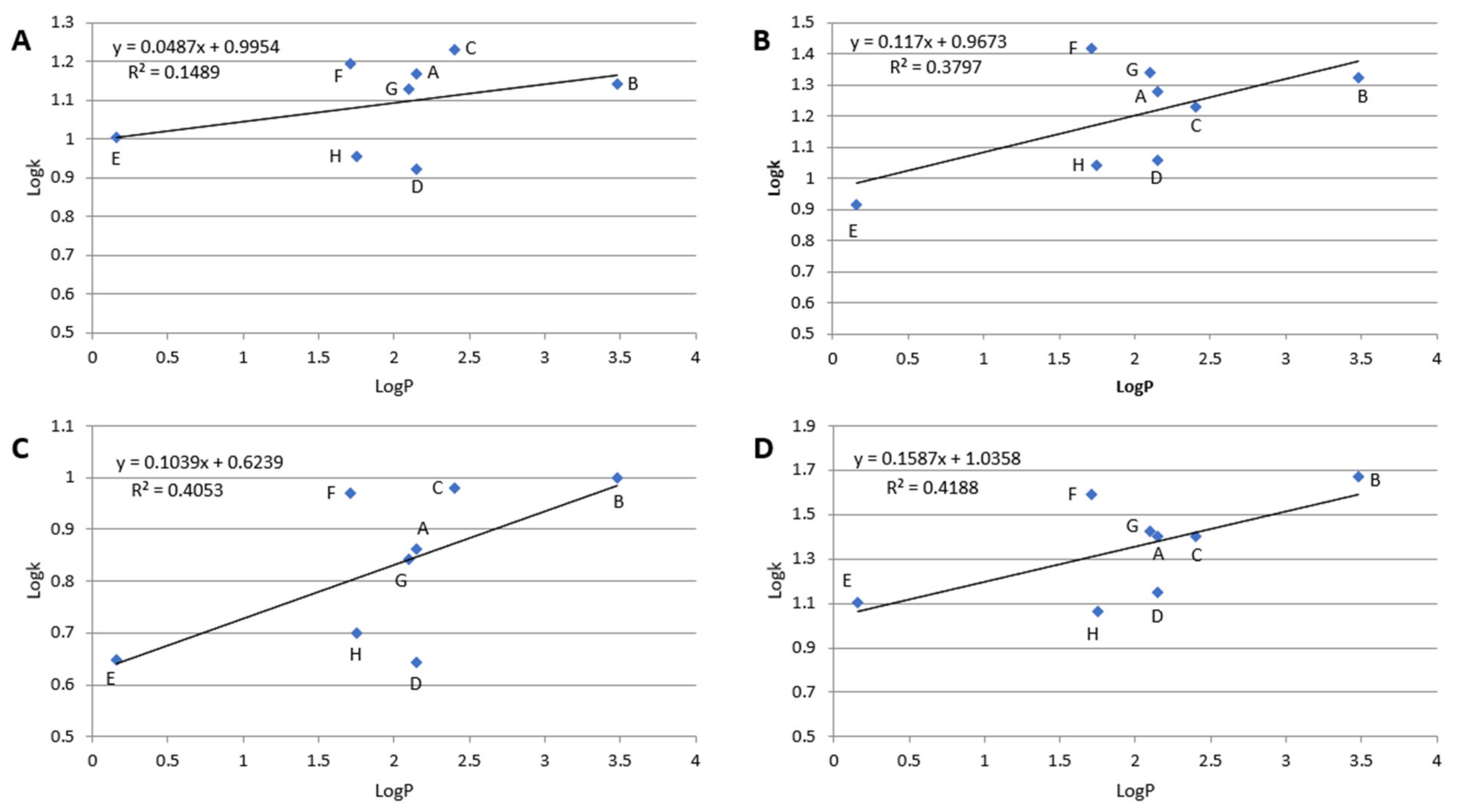
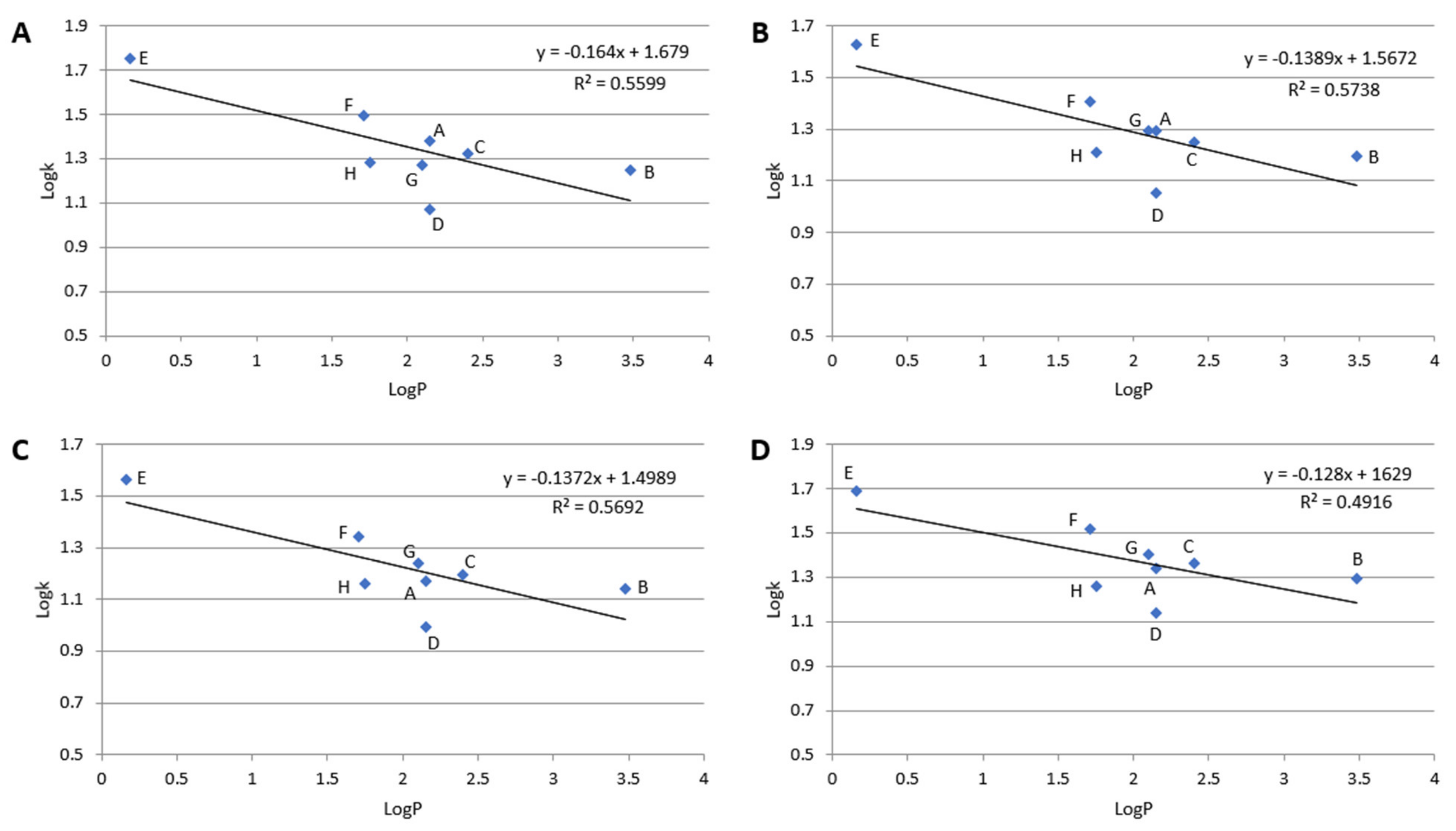
2.1. Retention Investigation
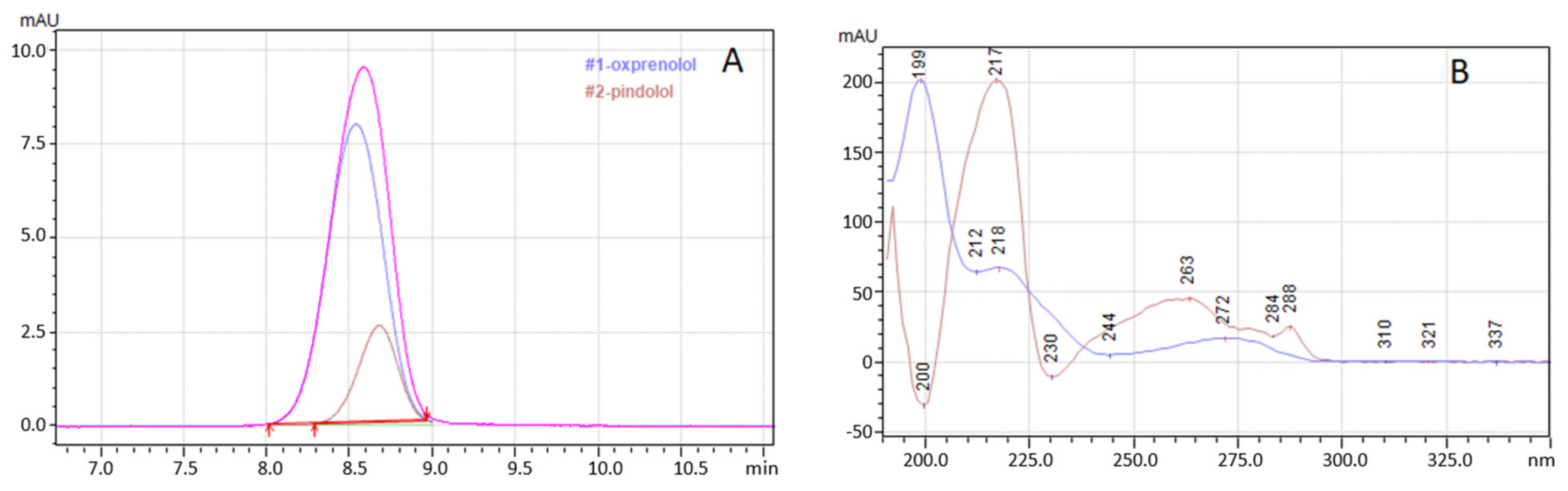
2.2. Separation
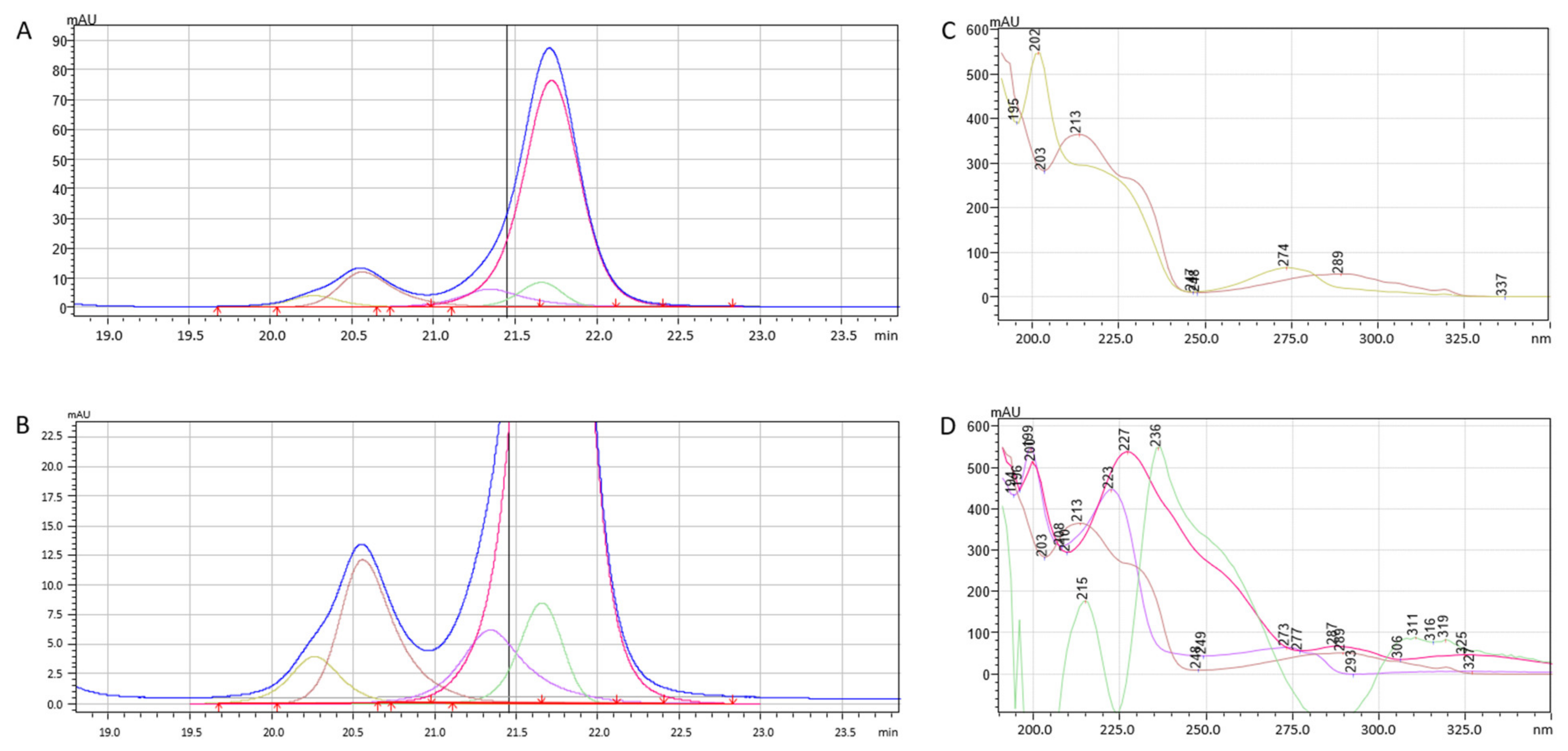
2.3. Intelligent Peak Deconvolution Analysis
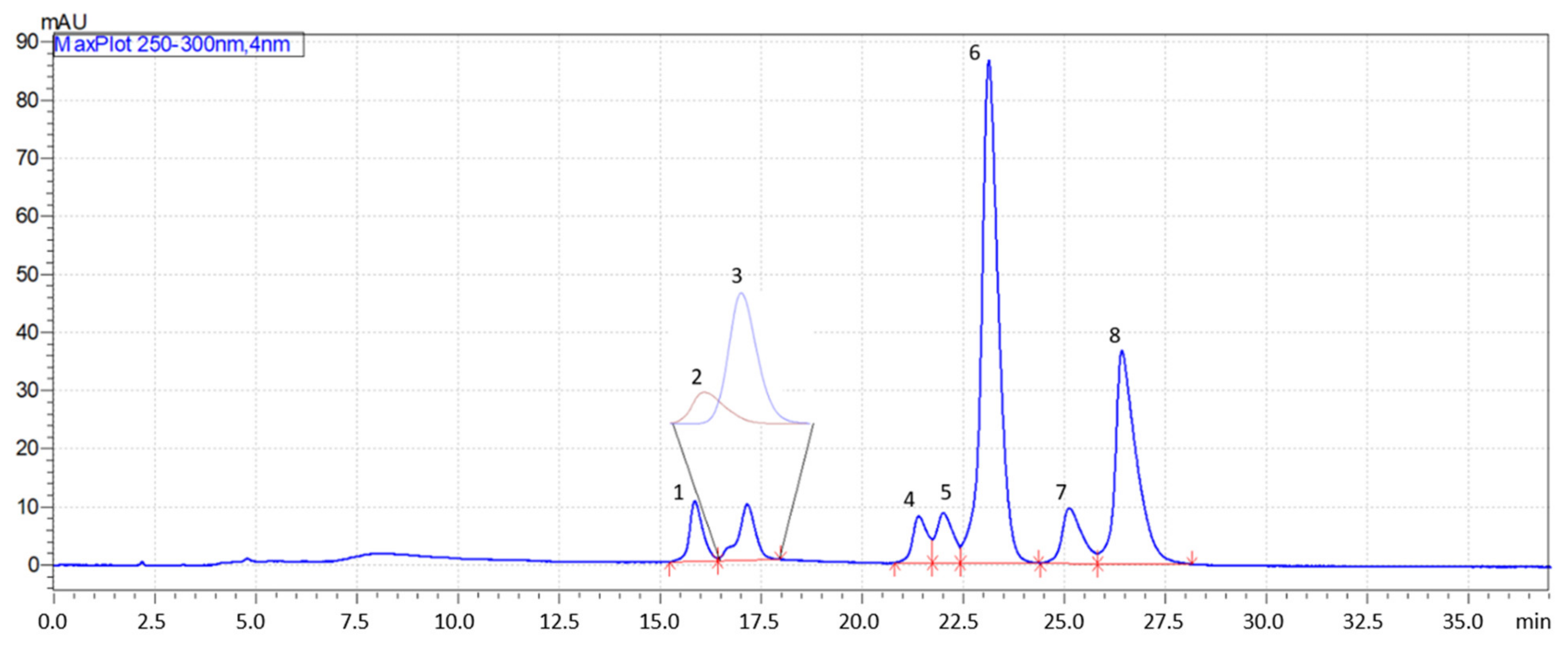
2.4. Gradient Optimization
3. Materials and Methods
3.1. Materials and Reagents
3.2. Compounds
3.3. Instruments
3.4. Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Saleem, K.; Ali, I.; Kulsum, U.; Aboul-Enein, H.Y. Recent developments in HPLC analysis of β-blockers in biological samples. J. Chromatogr. Sci. 2013, 51, 807–818. [Google Scholar] [CrossRef] [PubMed]
- James, P.A.; Oparil, S.; Carter, B.L.; Cushman, W.C.; Dennison-Himmelfarb, C.; Handler, J.; Lackland, D.T.; LeFevre, M.L.; MacKenzie, T.D.; Ogedegbe, O.; et al. 2014 Evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014, 311, 507–520. [Google Scholar] [CrossRef]
- Freemantle, N.; Cleland, J.; Young, P.; Mason, J.; Harrison, J. β blockade after myocardial infarction: Systematic review and meta regression analysis. Br. Med. J. 1999, 318, 1730–1737. [Google Scholar] [CrossRef]
- Tang, X.; Cao, Y.; Yu, J.; Shi, R.; Huang, Y.; Wu, J.; Hu, Y. Development and Validation of HPLC Methods for the Determination of Propranolol Hydrochloride and Hydrochlorothiazide Related Substances in Combination Tablets. Int. J. Drug Dev. Res. Res. 2017, 9, 24–29. [Google Scholar]
- Hakiem, A.F.A.; Hamdy, A.K.; Ali, H.R.H.; Gomaa, M.; Aboraia, A.S. In depth investigation of the retention behavior of structurally related β-blockers on RP-HPLC column: Quality by design and quantitative structure-property relationship complementary approaches for optimization and validation. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1166, 122549. [Google Scholar] [CrossRef]
- Gu, H.W.; Wu, H.L.; Yin, X.L.; Li, Y.; Liu, Y.J.; Xia, H.; Zhang, S.R.; Jin, Y.F.; Sun, X.D.; Yu, R.Q.; et al. Multi-targeted interference-free determination of ten β-blockers in human urine and plasma samples by alternating trilinear decomposition algorithm-assisted liquid chromatography-mass spectrometry in full scan mode: Comparison with multiple reaction monit. Anal. Chim. Acta 2014, 848, 10–24. [Google Scholar] [CrossRef]
- Hashem, H.; Jira, T. Retention behaviour of beta-blockers in HPLC using a monolithic column. J. Sep. Sci. 2006, 29, 986–994. [Google Scholar] [CrossRef]
- Canfield, D.V.; Dubowski, K.M.; Whinnery, J.M.; Forster, E.M. Pilot-Reported Beta-Blockers Identified by Forensic Toxicology Analysis of Postmortem Specimens. J. Anal. Toxicol. 2018, 42, 1–5. [Google Scholar] [CrossRef]
- Magiera, S.; Kolanowska, A.; Baranowski, J. Salting-out assisted extraction method coupled with hydrophilic interaction liquid chromatography for determination of selected β-blockers and their metabolites in human urine. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1022, 93–101. [Google Scholar] [CrossRef]
- Mahmoudi, A.; Rajabi, M. Selective determination of some beta-blockers in urine and plasma samples using continuous flow membrane microextraction coupled with high performance liquid chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1128, 121768. [Google Scholar] [CrossRef]
- Cheng, J.Q.; Liu, T.; Nie, X.M.; Chen, F.M.; Wang, C.S.; Zhang, F. Analysis of 27 β-blockers and metabolites in milk powder by high performance liquid chromatography coupled to quadrupole Orbitrap high-resolution mass spectrometry. Molecules 2019, 24, 820. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.J.; Danaceau, J.P. Simultaneous extraction and screening of diuretics, beta-blockers, selected stimulants and steroids in human urine by HPLC-MS/MS and UPLC-MS/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 3857–3864. [Google Scholar] [CrossRef] [PubMed]
- Tomková, J.; Ondra, P.; Kocianová, E.; Václavík, J. Fast and sensitive analysis of beta blockers by ultra-high-performance liquid chromatography coupled with ultra-high-resolution TOF mass spectrometry. Biomed. Chromatogr. 2017, 31, e3911. [Google Scholar] [CrossRef] [PubMed]
- Van Nuijs, A.L.N.; Tarcomnicu, I.; Simons, W.; Bervoets, L.; Blust, R.; Jorens, P.G.; Neels, H.; Covaci, A. Optimization and validation of a hydrophilic interaction liquid chromatography-tandem mass spectrometry method for the determination of 13 top-prescribed pharmaceuticals in influent wastewater. Anal. Bioanal. Chem. 2010, 398, 2211–2222. [Google Scholar] [CrossRef]
- Layne, J. Characterization and comparison of the chromatographic performance of conventional, polar-embedded, and polar-endcapped reversed-phase liquid chromatography stationary phases. J. Chromatogr. A 2002, 957, 149–164. [Google Scholar] [CrossRef]
- Žuvela, P.; Skoczylas, M.; Jay Liu, J.; Baczek, T.; Kaliszan, R.; Wong, M.W.; Buszewski, B. Column Characterization and Selection Systems in Reversed-Phase High-Performance Liquid Chromatography. Chem. Rev. 2019, 119, 3674–3729. [Google Scholar] [CrossRef]
- Aral, H.; Çelik, K.S.; Altındağ, R.; Aral, T. Synthesis, characterization, and application of a novel multifunctional stationary phase for hydrophilic interaction/reversed phase mixed-mode chromatography. Talanta 2017, 174, 703–714. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Jin, J.; Sun, X.; Wang, L.; Chen, J. New reversed-phase/anion-exchange/hydrophilic interaction mixed-mode stationary phase based on dendritic polymer-modified porous silica. J. Chromatogr. A 2014, 1337, 133–139. [Google Scholar] [CrossRef]
- Coym, J.W. Comparison of retention on traditional alkyl, Polar endcapped, and Polar embedded group stationary phases. J. Sep. Sci. 2008, 31, 1712–1718. [Google Scholar] [CrossRef]
- Krzemińska, K.; Bocian, S. The versatility of N, O -dialkylphosphoramidate stationary phase-separations in HILIC, highly aqueous RP LC conditions and purely aqueous mobile phase. Analyst 2018, 143, 1217–1223. [Google Scholar] [CrossRef]
- Vecchietti, D.; Nishio, A.; Fujita, Y.; Yoshida, T.; Yanagisawa, T.; Kou, D. Liquid chromatography coupled with photodiode array and a multivariate curve resolution—Alternating least square algorithm for identification and quantification of co-eluting impurities in pharmaceutical analysis. J. Chromatogr. A 2022, 1678, 463364. [Google Scholar] [CrossRef] [PubMed]
- Domanski, B. Unlocking the Power of 3D Absorbance Data. LC-GC North Am. 2021, 39, 494–495. [Google Scholar]
- Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 25 March 2022).
- Arase, S.; Horie, K.; Kato, T.; Noda, A.; Mito, Y.; Takahashi, M.; Yanagisawa, T. Intelligent peak deconvolution through in-depth study of the data matrix from liquid chromatography coupled with a photodiode array detector applied to pharmaceutical analysis. J. Chromatogr. A 2016, 1469, 35–47. [Google Scholar] [CrossRef]
- Dembek, M.; Bocian, S.; Buszewski, B. Solvent Influence on Zeta Potential of Stationary Phase—Mobile Phase Interface. Molecules 2022, 27, 968. [Google Scholar] [CrossRef] [PubMed]
- Dembek, M.; Szumski, M.; Bocian, S.; Buszewski, B. Optimization of the packing process of microcolumns with the embedded phosphodiester stationary phases. J. Sep. Sci. 2022, 45, 3310–3318. [Google Scholar] [CrossRef] [PubMed]
- Galushko, S.V.; Shishkina, I.; Urtans, E.; Rotkaja, O. ChromSword ®: Software for Method Development in Liquid Chromatography. In Software-Assisted Method Development in High Performance Liquid Chromatography; World Scientific Publishing: Singapore, 2018; pp. 53–94. [Google Scholar] [CrossRef]
- Hewitt, E.F.; Lukulay, P.; Galushko, S. Implementation of a rapid and automated high performance liquid chromatography method development strategy for pharmaceutical drug candidates. J. Chromatogr. A 2006, 1107, 79–87. [Google Scholar] [CrossRef]
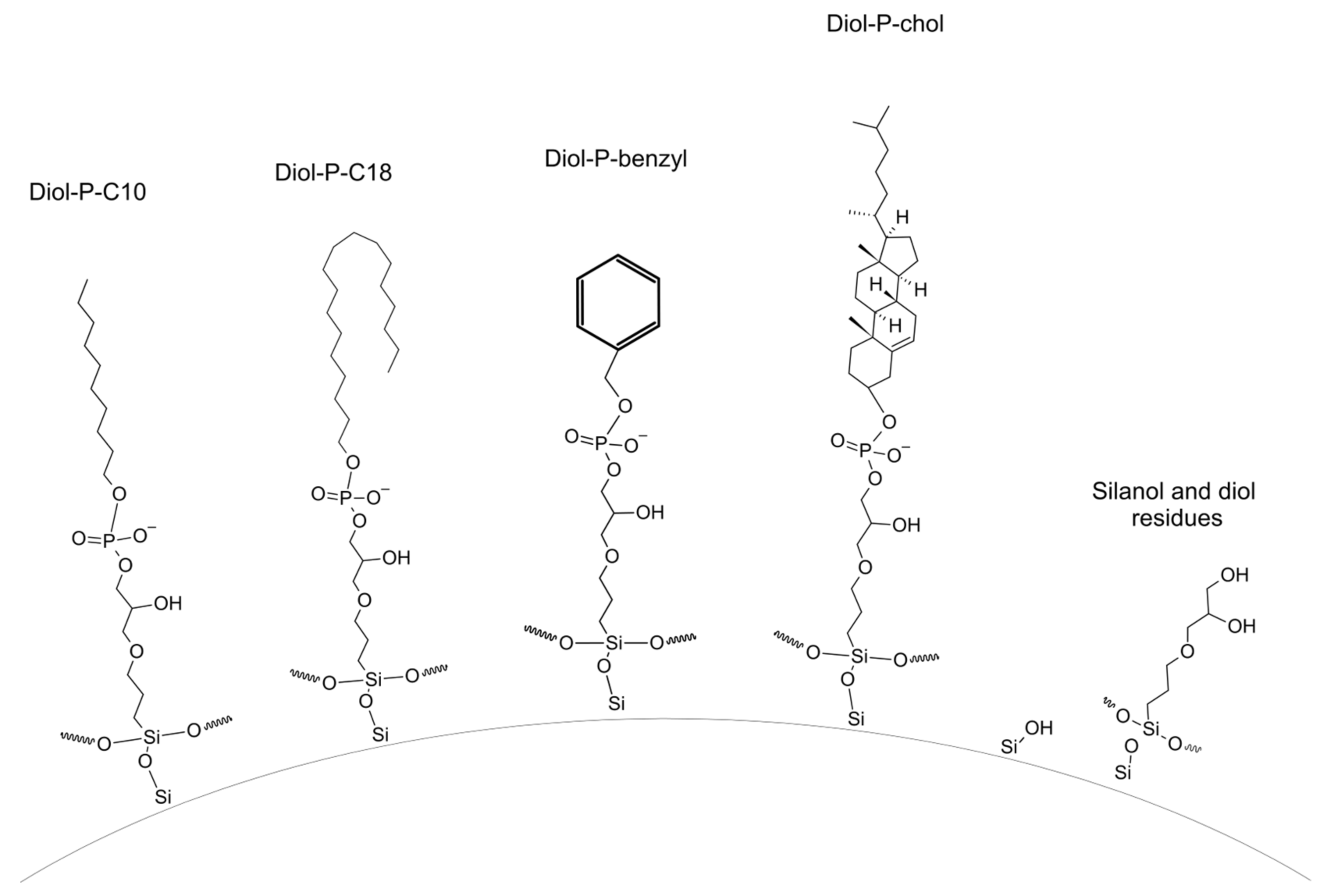
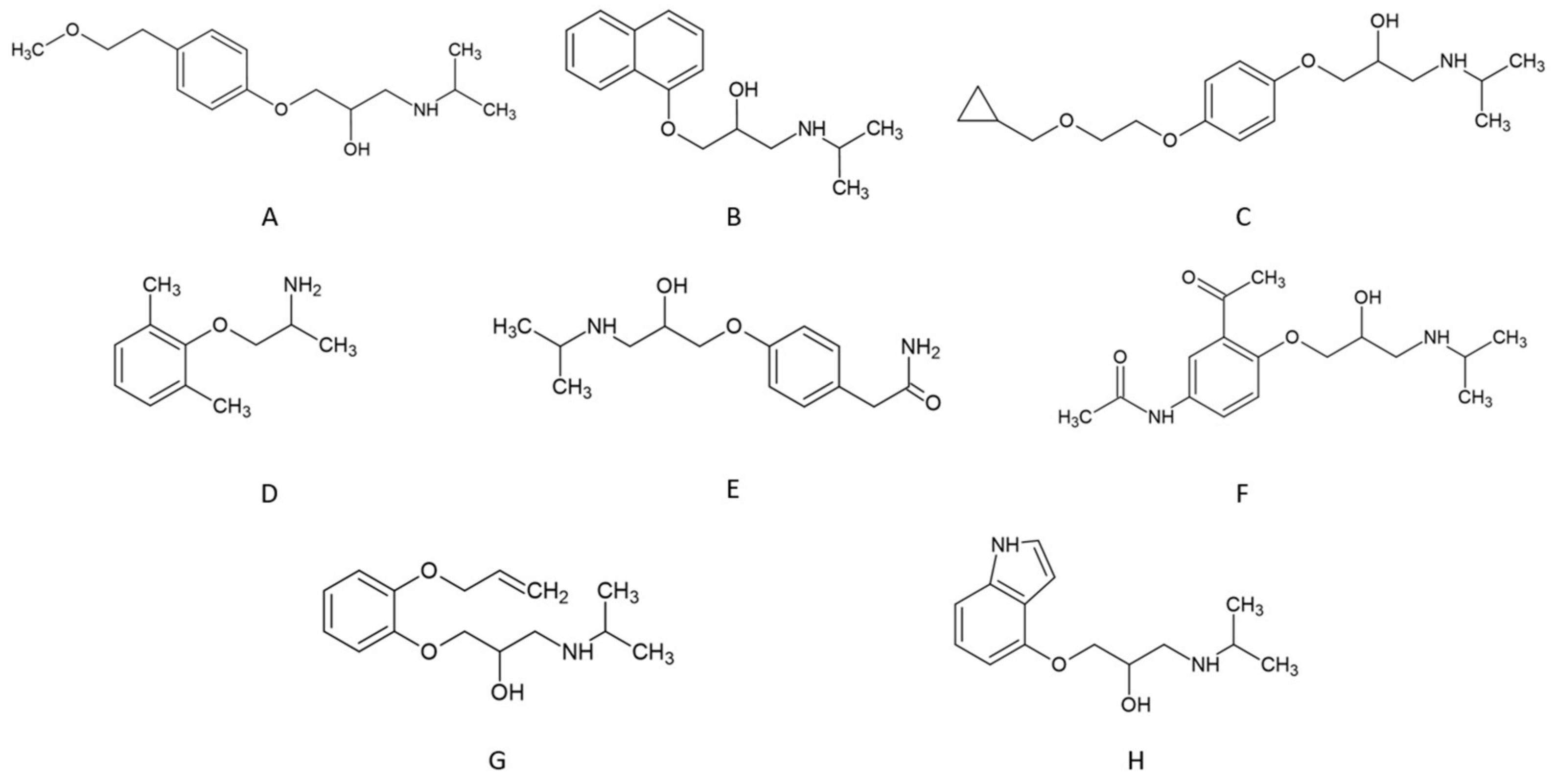
| Abbreviation | Beta-Blocker | Number of Hydrogen Bonds Donor | Number of Hydrogen Bonds Acceptor | Log P * | pKa * |
|---|---|---|---|---|---|
| A | metoprolol | 2 | 4 | 2.15 | 9.56–9.70 |
| B | propranolol | 2 | 3 | 3.48 | 9.53–9.45 |
| C | cicloprolol | 2 | 5 | 2.40 | 9.2 |
| D | mexiletine | 1 | 2 | 2.15 | 9.14–9.15 |
| E | atenolol | 3 | 5 | 0.16 | 9.54–9.60 |
| F | acebutolol | 3 | 6 | 1.71 | 9.52–9.67 |
| G | oxprenolol | 2 | 4 | 2.10 | 9.57 |
| H | pindolol | 3 | 4 | 1.75 | 9.25–9.54 |
| Stationary Phase | Carbon Load [%] | Coverage Density [µmol/m2] |
|---|---|---|
| Diol-P-C10 | 3.43 | 0.56 |
| Diol-P-C18 | 4.18 | 0.42 |
| Diol-P-Benzyl | 2.86 | 0.56 |
| Diol-P-Chol | 9.31 | 0.87 |
| Column | Pair | 10% ACN | Pair | 90% ACN |
|---|---|---|---|---|
| Diol-P-C10 | mexiletine/atenolol | 1.01 | mexiletine/propranolol | 1.41 |
| atenolol/pindolol | 1.13 | propranolol/pindolol | 1.04 | |
| pindolol/metoprolol | 1.39 | pindolol/metoprolol | 1.02 | |
| metoprolol/cicloprolol | 1.05 | metoprolol/cicloprolol | 1.06 | |
| cicloprolol/oxprenolol | 1.28 | cicloprolol/oxprenolol | 1.11 | |
| oxprenolol/acebutolol | 1.02 | oxprenolol/acebutolol | 1.27 | |
| acebutolol/atenolol | 1.04 | acebutolol/atenolol | 1.65 | |
| Diol-P-C18 | atenolol/pindolol | 1.34 | mexiletine/propranolol | 1.38 |
| pindolol/mexiletine | 1.04 | propranolol/pindolol | 1.03 | |
| mexiletine/cicloprolol | 1.48 | pindolol/cicloprolol | 1.10 | |
| cicloprolol/metoprolol | 1.12 | cicloprolol/metoprolol | 1.11 | |
| metoprolol/propranolol | 1.10 | metoprolol/oxprenolol | 1.00 | |
| propranolol/oxprenolol | 1.05 | oxprenolol/acebutolol | 1.29 | |
| oxprenolol/acebutolol | 1.19 | acebutolol/atenolol | 1.66 | |
| Diol-P-Benzyl | mexiletine/pindolol | 1.07 | mexiletine/propranolol | 1.51 |
| pindolol/atenolol | 1.12 | propranolol/oxprenolol | 1.06 | |
| atenolol/oxprenolol | 1.33 | oxprenolol/pindolol | 1.01 | |
| oxprenolol/propranolol | 1.04 | pindolol/cicloprolol | 1.11 | |
| propranolol/metoprolol | 1.06 | cicloprolol/metoprolol | 1.13 | |
| metoprolol/acebutolol | 1.06 | metoprolol/acebutolol | 1.31 | |
| acebutolol/cicloprolol | 1.09 | acebutolol/atenolol | 1.81 | |
| Diol-P-Chol | pindolol/atenolol | 1.09 | mexiletine/pindolol | 1.33 |
| atenolol/mexiletine | 1.12 | pindolol/propranolol | 1.07 | |
| mexiletine/metoprolol | 1.78 | propranolol/metoprolol | 1.12 | |
| metoprolol/cicloprolol | 1.00 | metoprolol/cicloprolol | 1.05 | |
| cicloprolol/oxprenolol | 1.05 | cicloprolol/oxprenolol | 1.10 | |
| oxprenolol/acebutolol | 1.47 | oxprenolol/acebutolol | 1.29 | |
| acebutolol/atenolol | 1.20 | acebutolol/atenolol | 1.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalisz, O.; Dembek, M.; Studzińska, S.; Bocian, S. Beta-Blocker Separation on Phosphodiester Stationary Phases—The Application of Intelligent Peak Deconvolution Analysis. Molecules 2023, 28, 3249. https://doi.org/10.3390/molecules28073249
Kalisz O, Dembek M, Studzińska S, Bocian S. Beta-Blocker Separation on Phosphodiester Stationary Phases—The Application of Intelligent Peak Deconvolution Analysis. Molecules. 2023; 28(7):3249. https://doi.org/10.3390/molecules28073249
Chicago/Turabian StyleKalisz, Oktawia, Mikołaj Dembek, Sylwia Studzińska, and Szymon Bocian. 2023. "Beta-Blocker Separation on Phosphodiester Stationary Phases—The Application of Intelligent Peak Deconvolution Analysis" Molecules 28, no. 7: 3249. https://doi.org/10.3390/molecules28073249
APA StyleKalisz, O., Dembek, M., Studzińska, S., & Bocian, S. (2023). Beta-Blocker Separation on Phosphodiester Stationary Phases—The Application of Intelligent Peak Deconvolution Analysis. Molecules, 28(7), 3249. https://doi.org/10.3390/molecules28073249







