Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 α-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on β→α Loop 5
Abstract
1. Introduction
2. Results
2.1. Kinetic Analysis of the Reactions of BspAG13_31A Asn258 Mutants with p-Nitrophenyl α-d-Glucopyranoside and G2
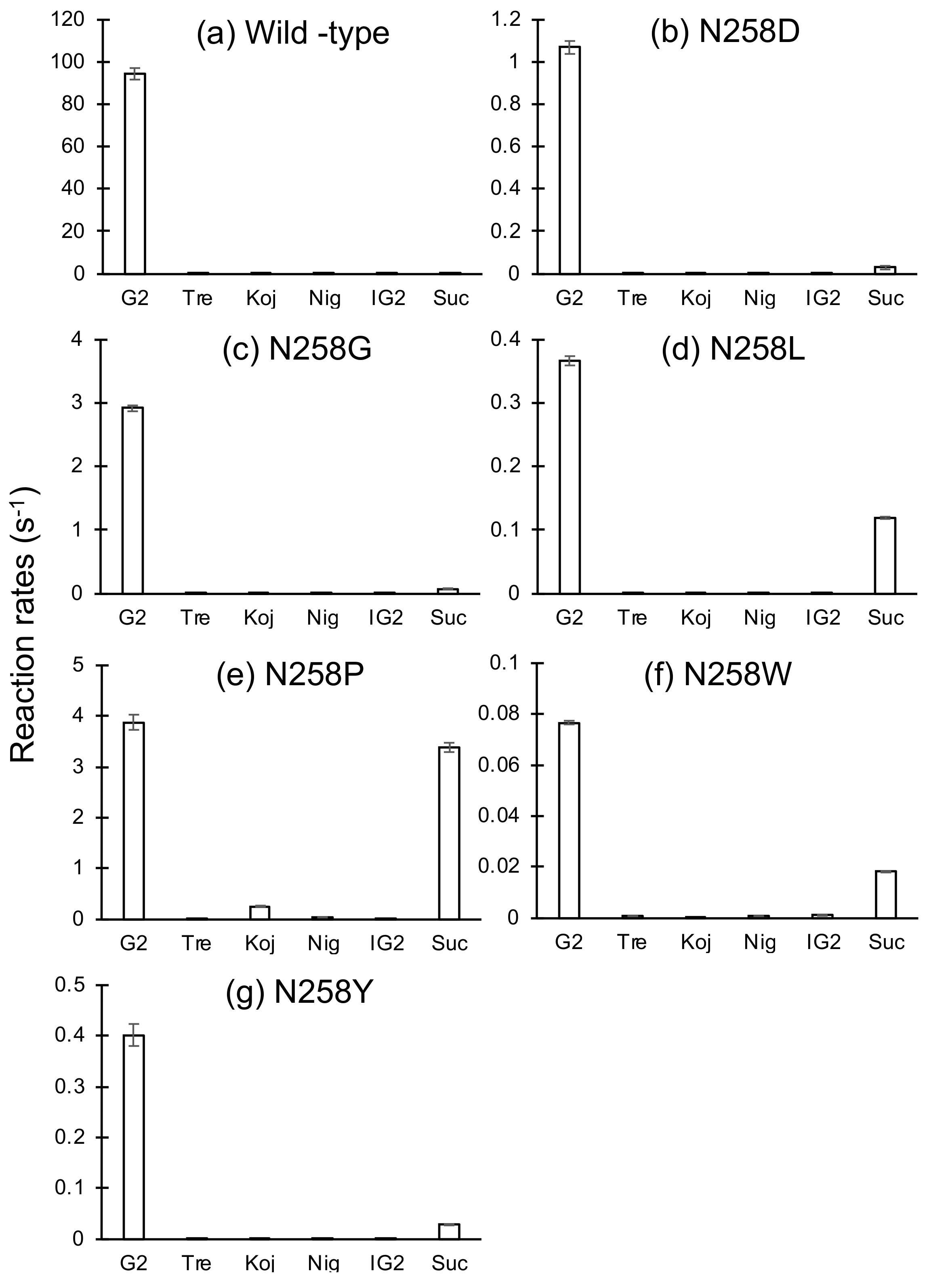
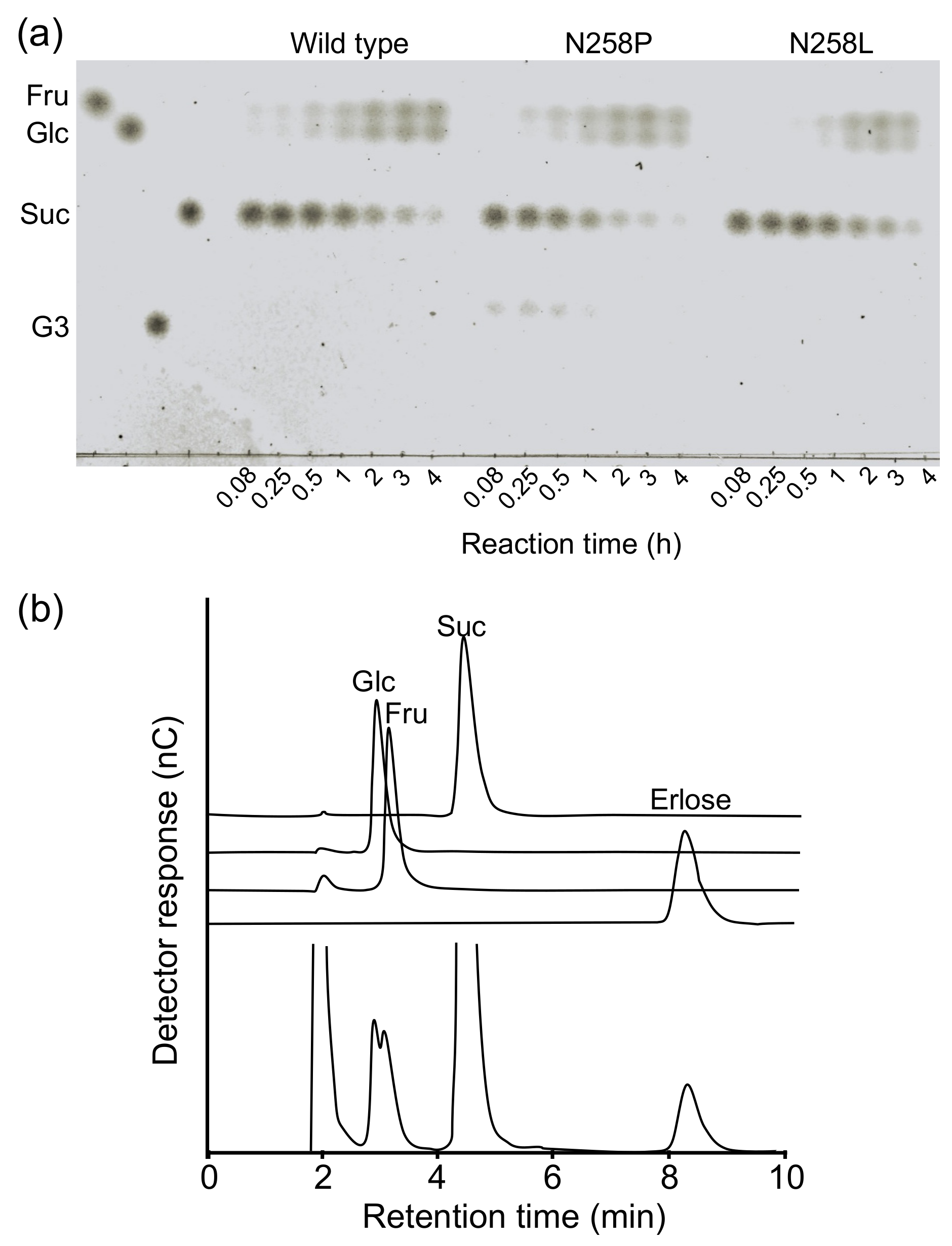
2.2. Reactions of BspAG13_31A Asn258 Mutants with Various Disaccharides
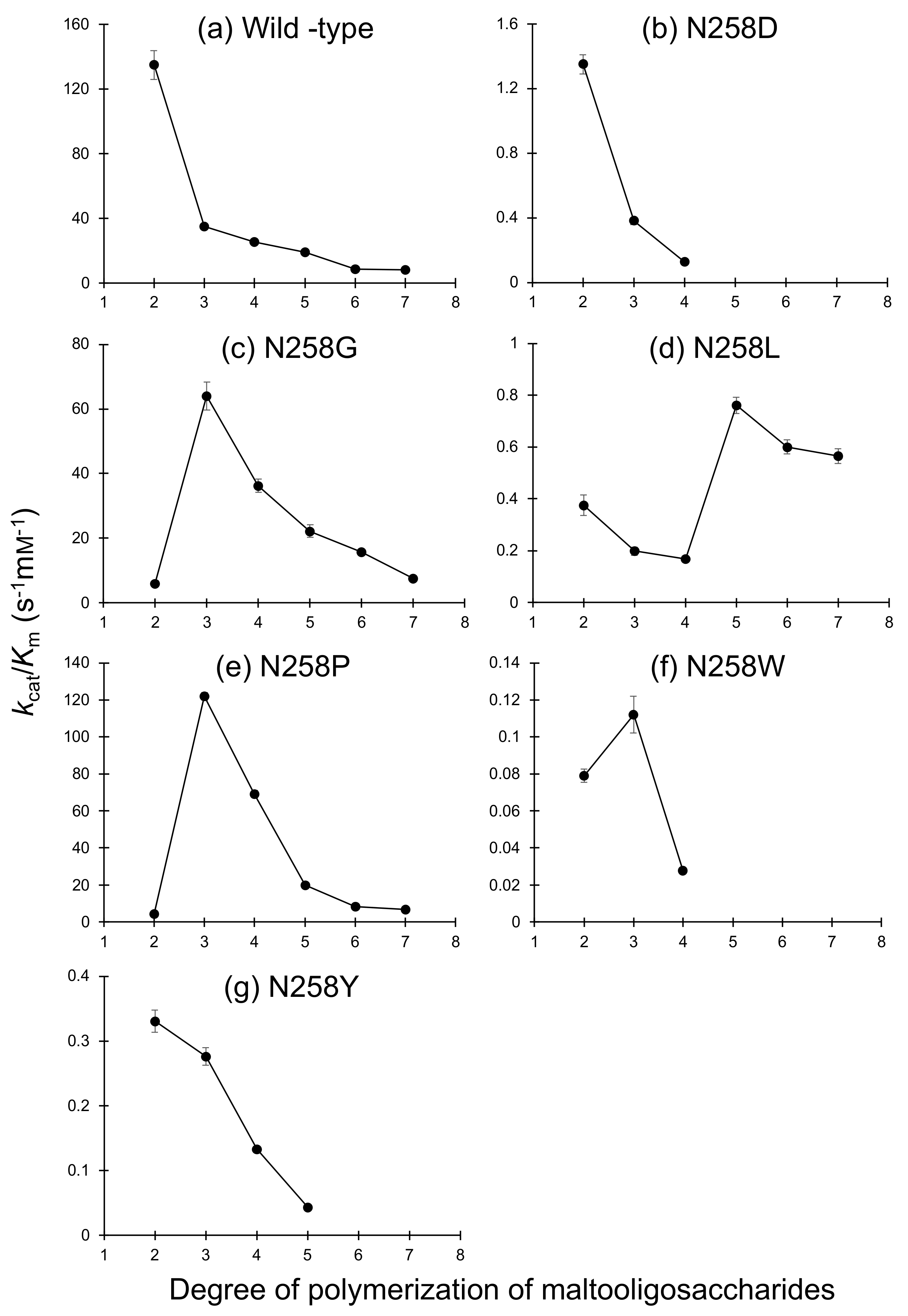
2.3. Substrate Chain-Length Specificity of BspAG13_31A Asn258 Mutants
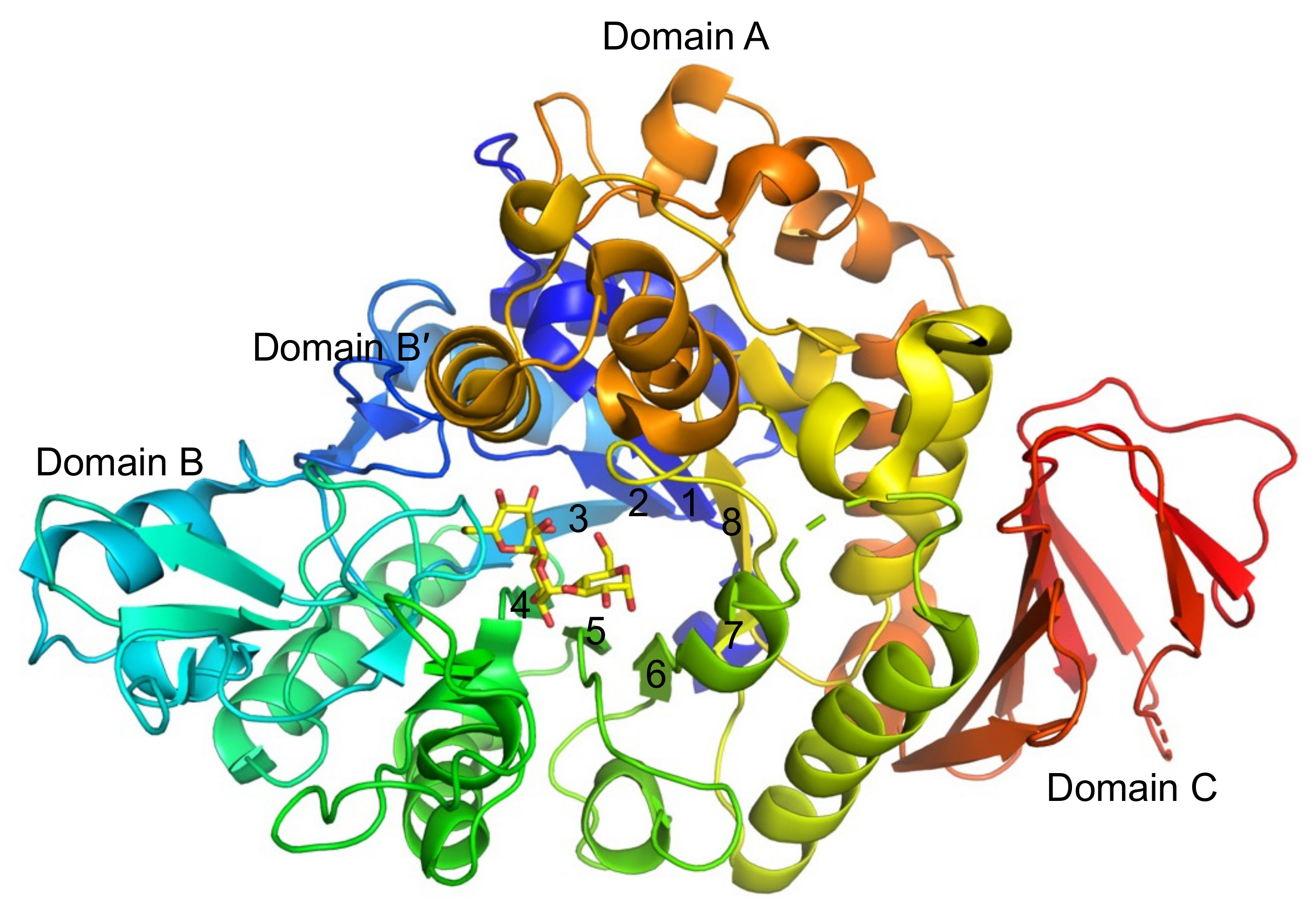
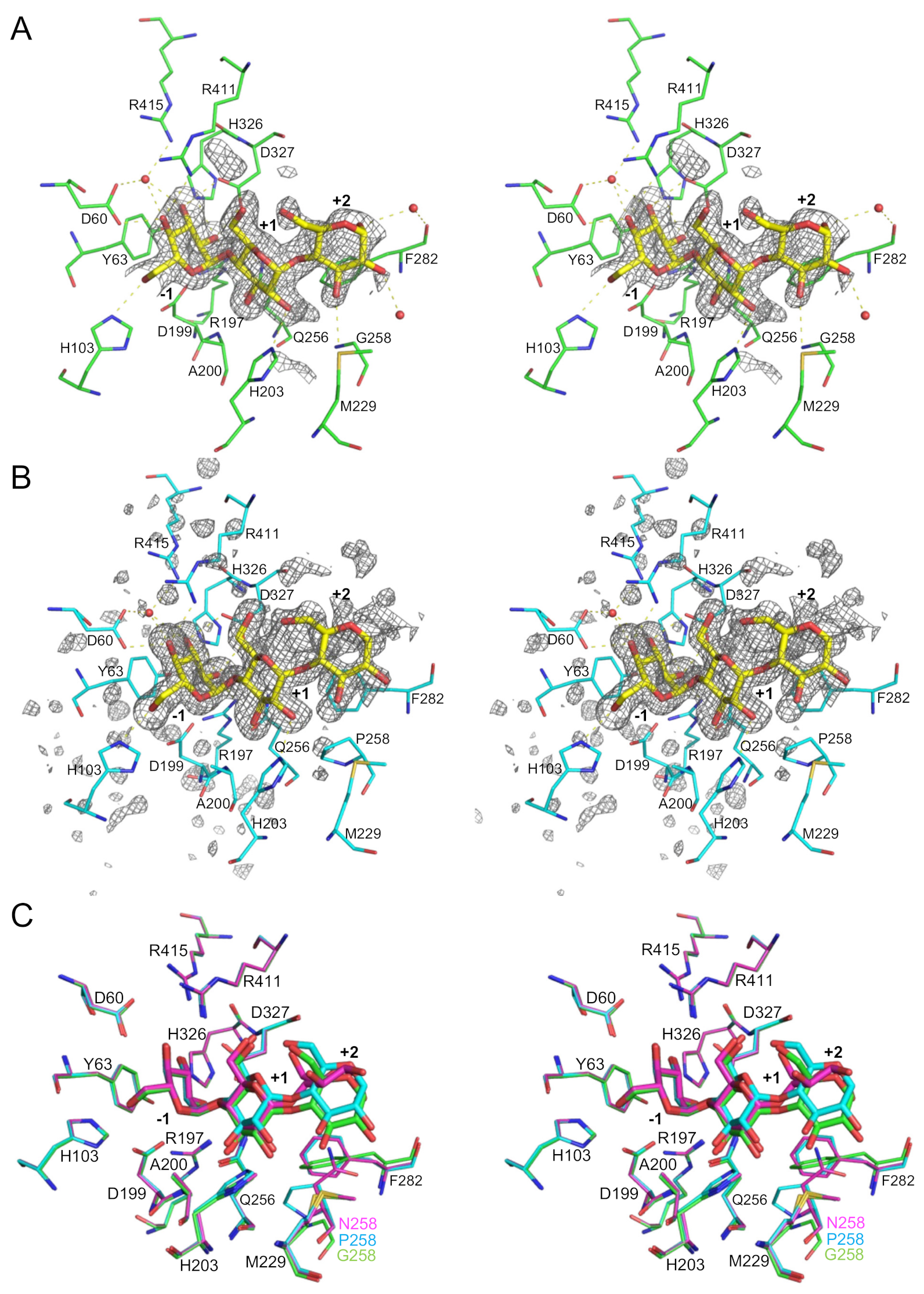
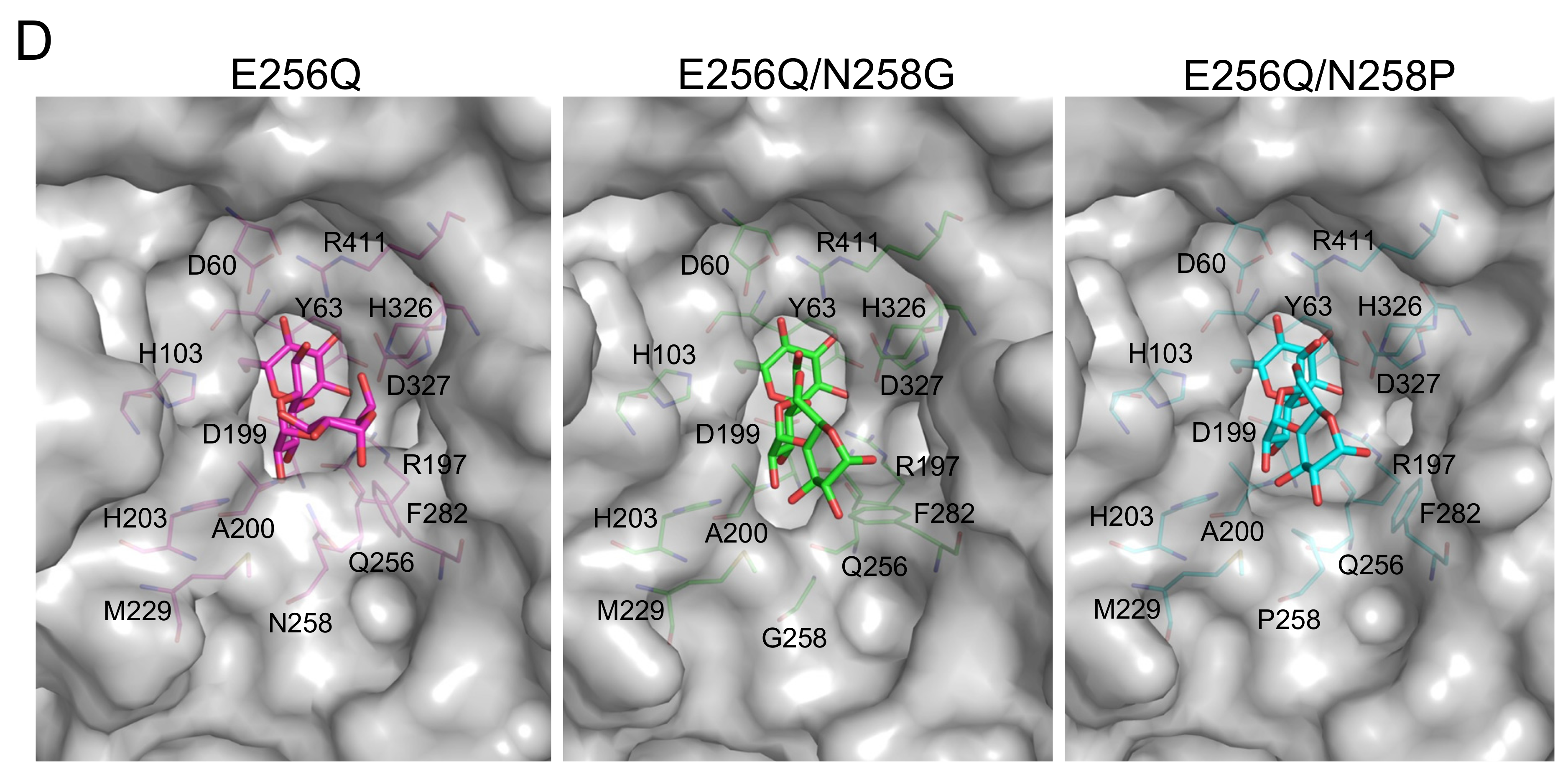
2.4. Structural Analysis of BspAG13_31A E256Q/N258G and E256Q/N258P
3. Discussion
4. Materials and Methods
4.1. Preparation of BspAG13_31A Mutants
4.2. Kinetic Analysis of Reactions with Various Substrates
4.3. Analysis of the Reaction Product from Sucrose
4.4. Crystal Structure Analysis of BspAG13_31A Mutants
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Okuyama, M.; Saburi, W.; Mori, H.; Kimura, A. α-Glucosidases and α-1,4-glucan lyases: Structures, functions, and physiological actions. Cell. Mol. Life Sci. 2016, 73, 2727–2751. [Google Scholar] [CrossRef] [PubMed]
- Takaku, H. Manufacture of oligosaccharides. In Handbook of Amylases and Related Enzymes, The Amylase Research Society of Japan ed.; Pergamon: Oxford, UK, 1988; pp. 212–222. [Google Scholar]
- Konishi, Y.; Shindo, K. Production of nigerose, nigerosyl glucose, and negerosyl maltose by Acremonium sp. S4G13. Biosci. Biotechnol. Biochem. 1997, 61, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Ojima, T.; Aizawa, K.; Saburi, W.; Yamamoto, T. α-Glucosylated 6-gingerol: Chemoenzymatic synthesis using α-glucosidase from Halomonas sp. H11, and its physical properties. Carbohydr. Res. 2012, 354, 59–64. [Google Scholar] [CrossRef]
- Kohmoto, T.; Fukui, F.; Takaku, H.; Machida, Y.; Arai, M.; Mitsuoka, T. Effect of isomalto-oligosaccharides on human fecal flora. Bifidobact. Microflora 1988, 7, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Kohmoto, T.; Fukui, F.; Takaku, H.; Mitsuoka, T. Dose-response test of isomaltooligosaccharides for increasing fecal bifidobacterial. Agric. Biol. Chem. 1991, 55, 2157–2159. [Google Scholar]
- Murosaki, S.; Muroyama, K.; Yamamoto, Y.; Kusaka, H.; Liu, T.; Yoshikai, Y. Immunopotentiating activity of nigerooligosaccharides for the T helper 1-like immune response in mice. Biosci. Biotechnol. Biochem. 1999, 63, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Minamiura, N. α-Glucosidases. In Handbook of Amylases and Related Enzymes, The Amylase Research Society of Japan ed.; Pergamon: Oxford, UK, 1988; pp. 104–116. [Google Scholar]
- Auiewiriyanukul, W.; Saburi, W.; Kato, K.; Yao, M.; Mori, H. Function and structure of GH13_31 α-glucosidase with high α-(1→4)-glucosidic linkage specificity and transglucosylation activity. FEBS Lett. 2018, 592, 2268–2281. [Google Scholar] [CrossRef]
- Suzuki, Y.; Shinji, M.; Eto, N. Assignment of a p-nitrophenyl α-D-glucopyranosidase of Bacillus stearothermophilus ATCC 12016 to a novel exo-α-1,4-glucosidase active for oligomaltosaccharides and α-glucans. Biochim. Biophys. Acta 1984, 787, 281–289. [Google Scholar] [CrossRef]
- Hung, V.S.; Hatada, Y.; Goda, S.; Lu, J.; Hidaka, Y.; Li, Z.; Akita, M.; Ohta, Y.; Watanabe, K.; Matsui, H.; et al. α-Glucosidase from a strain of deep-sea Geobacillus: A potential enzyme for the biosynthesis of complex carbohydrates. Appl. Microbiol. Biotechnol. 2005, 68, 757–765. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Janeček, Š.; Svensson, B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 2001, 1546, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Svensson, B. Amylolytic glycoside hydrolases. Cell. Mol. Life Sci. 2016, 73, 2601–2602. [Google Scholar] [CrossRef] [PubMed]
- Stam, M.R.; Danchin, E.G.J.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Mareček, F.; Janeček, Š. A novel subfamily GH13_46 of the α-amylase family GH13 represented by the cyclomaltodextrinase from Flavobaceterium sp. No. 92. Molecules 2022, 27, 8735. [Google Scholar] [CrossRef]
- Watanabe, K.; Hata, Y.; Kizaki, H.; Katsube, Y.; Suzuki, Y. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: Structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 1997, 269, 142–153. [Google Scholar] [CrossRef]
- Shirai, T.; Hung, V.S.; Morinaka, K.; Kobayashi, T.; Ito, S. Crystal structure of GH13 α-glucosidase GSJ from one of the deepest sea bacteria. Proteins 2008, 73, 126–133. [Google Scholar] [CrossRef]
- Hondoh, H.; Saburi, W.; Mori, H.; Okuyama, M.; Nakada, T.; Matsuura, Y.; Kimura, A. Substrate recognition mechanism of α-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans. J. Mol. Biol. 2008, 378, 913–922. [Google Scholar] [CrossRef]
- Møller, M.S.; Fredslund, F.; Majumder, A.; Nakai, H.; Poulsen, J.C.N.; Lo Leggio, L.; Svensson, B.; Abou Hachem, M. Enzymology and structure of the GH13_31 glucan 1,6-α-glucosidase that confers isomaltooligosaccharide utilization in the probiotic Lactobacillus acidophilus NCFM. J. Bacteriol. 2012, 194, 4249–4259. [Google Scholar] [CrossRef] [PubMed]
- Saburi, W.; Mori, H.; Saito, S.; Okuyama, M.; Kimura, A. Structural elements in dextran glucosidase responsible for high specificity to long chain substrate. Biochim. Biophys. Acta 2006, 1764, 688–698. [Google Scholar] [CrossRef]
- Shen, X.; Saburi, W.; Gai, Z.; Kato, K.; Ojima-Kato, T.; Yu, J.; Komoda, K.; Kido, Y.; Matsui, H.; Mori, H.; et al. Structural analysis of the α-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1382–1391. [Google Scholar] [CrossRef]
- Gessler, K.; Usón, I.; Takaha, T.; Krauss, N.; Smith, S.M.; Okada, S.; Sheldrick, G.M.; Saenger, W. V-Amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose). Proc. Natl. Acad. Sci. USA 1999, 96, 4246–4251. [Google Scholar] [CrossRef] [PubMed]
- Dowd, M.K.; Zeng, J.; French, A.D.; Reilly, P.J. Conformational analysis of the anomeric forms of kojibiose, nigerose, and maltose using MM3. Carbohydr. Res. 1992, 230, 223–244. [Google Scholar] [CrossRef]
- Fujimoto, Z.; Takase, K.; Doui, N.; Momma, M.; Matsumoto, T.; Mizuno, H. Crystal structure of a catalytic-site mutant α-amylase from Bacillus subtilis complexed with maltopentaose. J. Mol. Biol. 1998, 277, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Caner, S.; Kwan, E.; Li, C.; Brayer, G.D.; Withers, S.G. Evaluation of the significance of starch surface binding sites on human pancreatic α-amylase. Biochemistry 2016, 55, 6000–6009. [Google Scholar] [CrossRef] [PubMed]
- Knegtel, R.M.A.; Strokopytov, B.; Penninga, D.; Faber, O.G.; Rozeboom, H.J.; Kalk, K.H.; Dijkhuizen, L.; Dijkstra, B.W. Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products. J. Biol. Chem. 1995, 270, 29256–29264. [Google Scholar] [CrossRef] [PubMed]
- Hondoh, H.; Kuriki, T.; Matsuura, Y. Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J. Mol. Biol. 2003, 326, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Skov, L.K.; Mirza, O.; Sprogøe, D.; Dar, I.; Remaud-Simeon, M.; Albenne, C.; Monsan, P.; Gajhede, M. Oligosaccharide and sucrose complexes of amylosucrase. Structural implications for the polymerase activity. J. Biol. Chem. 2002, 277, 47741–47747. [Google Scholar] [CrossRef]
- Fersht, A.R.; Shi, J.P.; Knill-Jones, J.; Lowe, D.M.; Wilkinson, A.J.; Blow, D.M.; Brick, P.; Carter, P.; Waye, M.M.Y.; Winter, G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature 1985, 314, 235–238. [Google Scholar] [CrossRef]
- Hiromi, K.; Nitta, Y.; Numata, C.; Ono, S. Subsite affinities of glucoamylase: Examination of the validity of the subsite theory. Biochim. Biophys. Acta 1973, 302, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Hondoh, H.; Mori, H.; Saburi, W.; Okuyama, M.; Kimura, A. Calcium ion-dependent increase in thermostability of dextran glucosidase from Streptococcus mutans. Biosci. Biotechnol. Biochem. 2011, 75, 1557–1563. [Google Scholar] [CrossRef]
- Saburi, W.; Kobayashi, M.; Mori, H.; Okuyama, M.; Kimura, A. Replacement of the catalytic nucleophile aspartyl residue of dextran glucosidase by cysteine sulfinate enhances transglycosylation activity. J. Biol. Chem. 2013, 288, 31670–31677. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]

| Enzyme | kcat1 (s−1) | kcat2 (s−1) | Km1 (mm) | Km2 (mm) | kcat1/Km1 (s−1mm−1) | KTG (mm) |
|---|---|---|---|---|---|---|
| Wild type 1 | 4.72 | 13.9 | 2.05 | 4.71 | 2.30 | 2.48 |
| N258D | 9.88 ± 0.23 | 16.3 ± 1.7 | 1.92 ± 0.03 | 26.9 ± 3.5 | 5.15 | 16.6 |
| N258G | 1.95 ± 0.28 | 2.29 ± 0.09 | 0.194 ± 0.060 | 0.971 ± 0.211 | 10.0 | 0.792 |
| N258L | 5.61 ± 0.30 | 9.70 ± 0.70 | 1.51 ± 0.15 | 7.66 ± 0.71 | 3.72 | 4.43 |
| N258P | 81.3 ± 7.0 | 125 ± 18 | 0.674 ± 0.108 | 10.5 ± 2.8 | 121 | 6.81 |
| N258W | 1.48 ± 0.05 | 2.39 ± 0.22 | 0.193 ± 0.019 | 12.2 ± 1.4 | 7.67 | 7.58 |
| N258Y | 2.50 ± 0.33 | 8.27 ± 0.66 | 0.293 ± 0.006 | 5.17 ± 1.28 | 8.53 | 1.64 |
| Enzyme | vh (s−1) | vtg (s−1) | rtg (%) |
|---|---|---|---|
| Wild type 1 | 193 | 82.9 | 30.1 |
| N258D | 6.72 ± 0.387 | 0.284 ± 0.010 | 4.05 |
| N258G | 2.03 ± 0.76 | 17.4 ± 2.3 | 89.6 |
| N258L | 2.02 ± 0.40 | 0.0975 ± 0.0289 | 4.60 |
| N258P | 11.3 ± 2.4 | 6.86 ± 1.26 | 37.8 |
| N258W | 0.367 ± 0.028 | 0.0137 ± 0.0030 | 3.61 |
| N258Y | 0.967 ± 0.050 | 0.0389 ± 0.0120 | 3.87 |
| Enzyme | kcat1 (s−1) | kcat2 (s−1) | Km1 (mm) | Km2 (mm) | kcat1/Km1 (s−1mm−1) | KTG (mm) |
|---|---|---|---|---|---|---|
| Wild type 1 | 205 | 1890 | 1.52 | 201 | 135 | 21.6 |
| N258D 2 | 13.6 ± 0.2 | N.A. | 10.2 ± 0.6 | N.A. | 1.33 | N.A. |
| N258G | 3.20 ± 0.20 | 190 ± 7 | 0.534 ± 0.089 | 66.9 ± 3.3 | 5.99 | 1.13 |
| N258L 2 | 4.27 ± 0.09 | N.A. | 11.5 ± 1.4 | N.A. | 0.371 | N.A. |
| N258P | 46.8 ± 3.9 | 379 ± 86 | 10.5 ± 1.5 | 219 ± 89 | 4.46 | 27.0 |
| N258W 2 | 0.918 ± 0.087 | N.A. | 10.8 ± 0.6 | N.A. | 0.0850 | N.A. |
| N258Y 2 | 2.06 ± 0.01 | N.A. | 6.25 ± 0.62 | N.A. | 0.330 | N.A. |
| Enzyme | kcat (s−1) | Km (mm) | kcat/Km (s−1mm−1) |
|---|---|---|---|
| Wild type | 4.58 ± 0.1 | 34.2 ± 2.6 | 0.134 |
| N258L | 1.94 ± 0.07 | 18.1 ± 0.5 | 0.107 |
| N258P | 39.2 ± 0.6 | 10.1 ± 0.4 | 3.90 |
| E256Q/N258G-G3 | E256Q/N258P-G3 | |
|---|---|---|
| PDB ID | 8IBK | 8IDS |
| Data collection | ||
| Beamline | Spring-8 BL41XU | Spring-8 BL41XU |
| Space group | P212121 | P212121 |
| Unit cell parameters a, b, c, (Å) | 57.0, 90.0, 128.4 | 56.5, 89.2, 128.3 |
| Wavelength (Å) | 1.00 | 1.00 |
| Resolution range (Å) | 48.0–1.70 (1.76–1.70) | 50.0–1.50 (1.59–1.50) |
| Total No. of reflections | 964,831 (152,139) | 1,374,196 (220,850) |
| No. of unique reflections | 72,935 (11,510) | 104,555 (16,645) |
| Rmeas (%) * | 11.2 (98.1) | 8.8 (83.5) |
| <I/σ(I)> | 17.2 (1.99) | 19.7 (2.76) |
| CC1/2 | 0.999 (0.864) | 0.999 (0.835) |
| Completeness (%) | 99.6 (98.4) | 99.9 (99.6) |
| Redundancy | 13.2 (13.2) | 13.1 (13.3) |
| Refinement | ||
| Rwork/Rfree (%) ** | 18.2/20.9 | 16.5/18.6 |
| No. of atoms | ||
| Macromolecules | 4501 | 4472 |
| Ligand/ion | 36 | 36 |
| Water | 564 | 545 |
| B-factors (Å2) | ||
| Macromolecules | 23.8 | 19.6 |
| Ligand/ion | 25.9 | 27.8 |
| Water | 31.5 | 28.8 |
| RMSD from ideal | ||
| Bond lengths (Å) | 0.006 | 0.016 |
| Bond angles (°) | 0.86 | 1.37 |
| Ramachandran | ||
| Favored (%) Allowed (%) Outliers (%) | 96.85 3.15 0 | 97.21 2.79 0 |
| Residues in Subsites −1 and +1 | Residues in Subsites +1 and +2 | |||||
|---|---|---|---|---|---|---|
| Enzyme | Origin | Ligand | Φ | Ψ | Φ | Ψ |
| BspAG13_31A | Bacillus sp. AHU2216 | |||||
| E256Q 1 | G3 | 42.8° | −151° | 64.6° | −168° | |
| E256QN258G | G3 | 31.6° | −153° | 105° | −113° | |
| E256QN258P | G3 | 43.9° | −156° | 106° | −109° | |
| α-Amylase E208Q 2 | Bacillus subtilis | G5 | 28.6° | −148° | 109° | −125° |
| α-Amylase D300N 3 | Homo sapiens (pancreatic) | G6 | 34.6° | −148° | 112° | −117° |
| CGTase wild type 4 | Bacillus circulans | G4 | 25.2° | −154° | 110° | −125° |
| Neopullulanase E357Q 5 | Geobacillus stearothermophilus | G4 | 33.8° | −144° | 92° | −112° |
| Amylosucrase E328Q 6 | Neisseria polysaccharea | G7 | 33.8° | −155° | 67.1° | −148° |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Auiewiriyanukul, W.; Saburi, W.; Ota, T.; Yu, J.; Kato, K.; Yao, M.; Mori, H. Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 α-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on β→α Loop 5. Molecules 2023, 28, 3109. https://doi.org/10.3390/molecules28073109
Auiewiriyanukul W, Saburi W, Ota T, Yu J, Kato K, Yao M, Mori H. Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 α-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on β→α Loop 5. Molecules. 2023; 28(7):3109. https://doi.org/10.3390/molecules28073109
Chicago/Turabian StyleAuiewiriyanukul, Waraporn, Wataru Saburi, Tomoya Ota, Jian Yu, Koji Kato, Min Yao, and Haruhide Mori. 2023. "Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 α-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on β→α Loop 5" Molecules 28, no. 7: 3109. https://doi.org/10.3390/molecules28073109
APA StyleAuiewiriyanukul, W., Saburi, W., Ota, T., Yu, J., Kato, K., Yao, M., & Mori, H. (2023). Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 α-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on β→α Loop 5. Molecules, 28(7), 3109. https://doi.org/10.3390/molecules28073109






