New Bisabolane-Type Sesquiterpenoids from Curcuma longa and Their Anti-Atherosclerotic Activity
Abstract
1. Introduction
2. Results
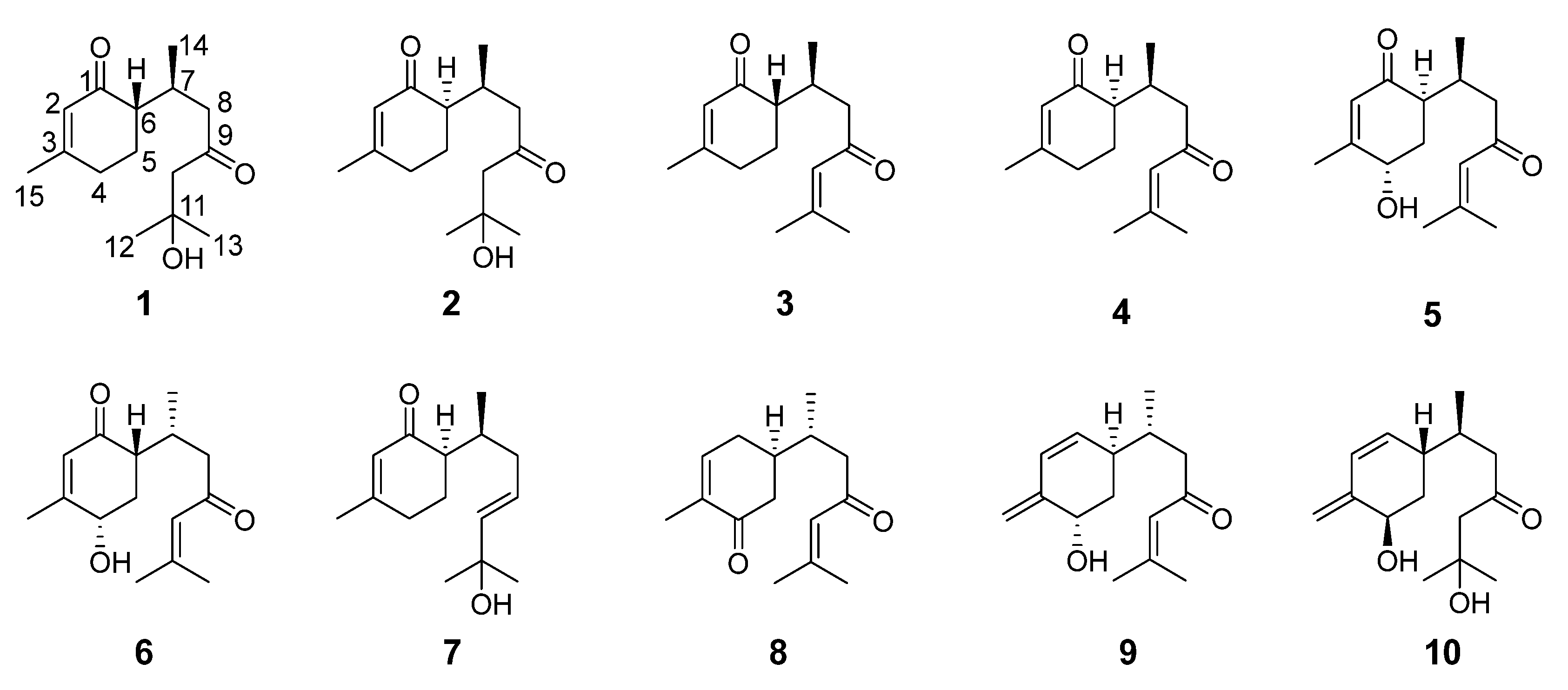
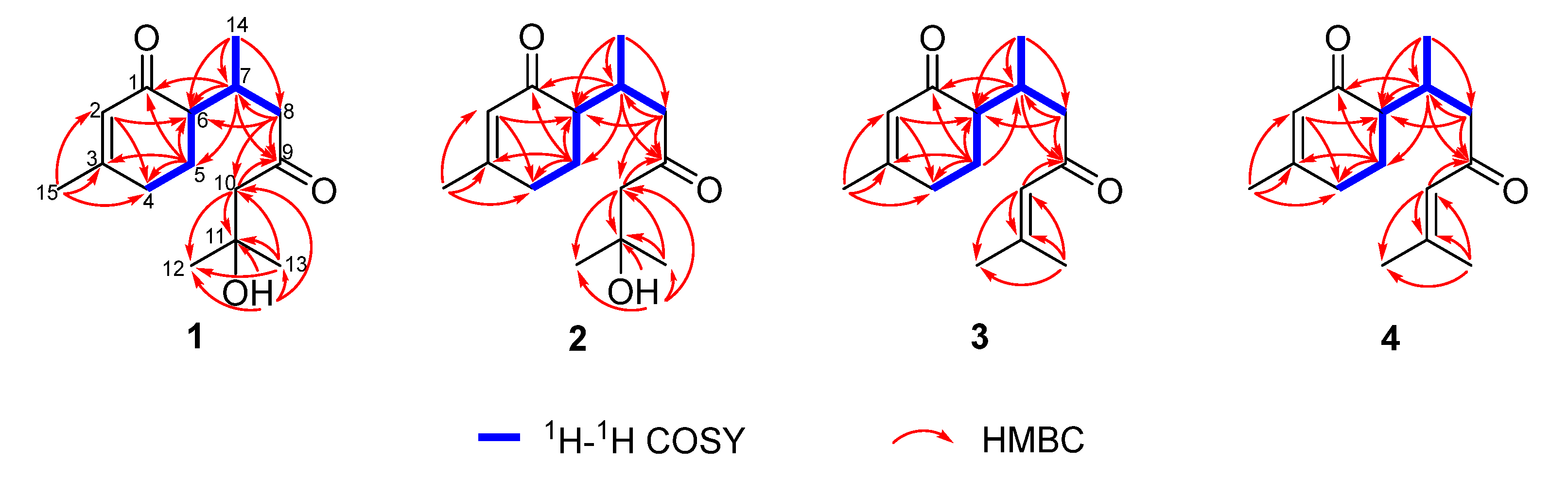
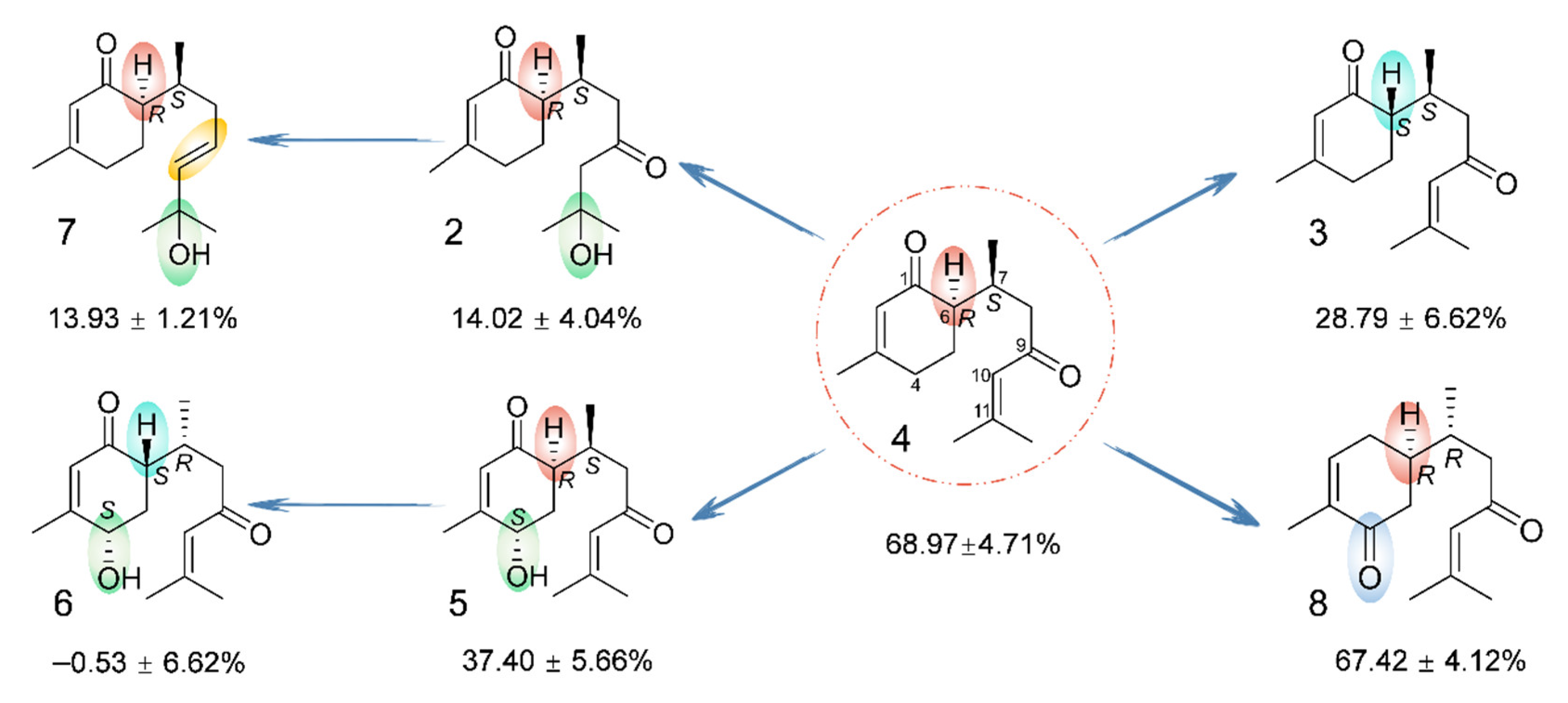
2.1. Chemical Structure Elucidation
2.2. Physicochemical Properties and Spectroscopic Data of Compounds 1–4
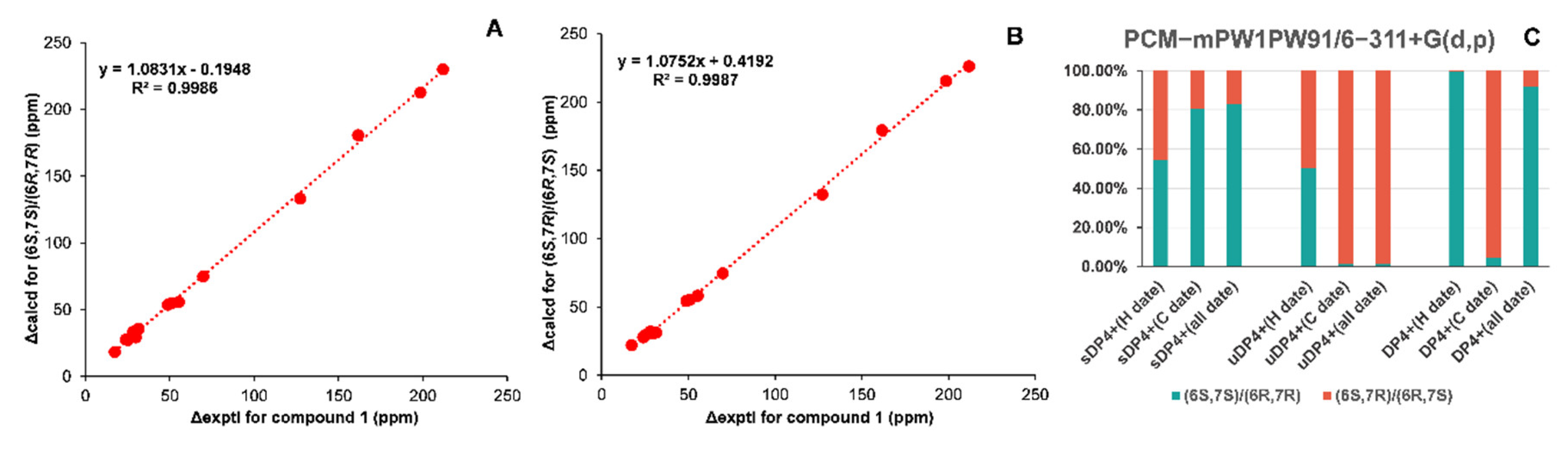
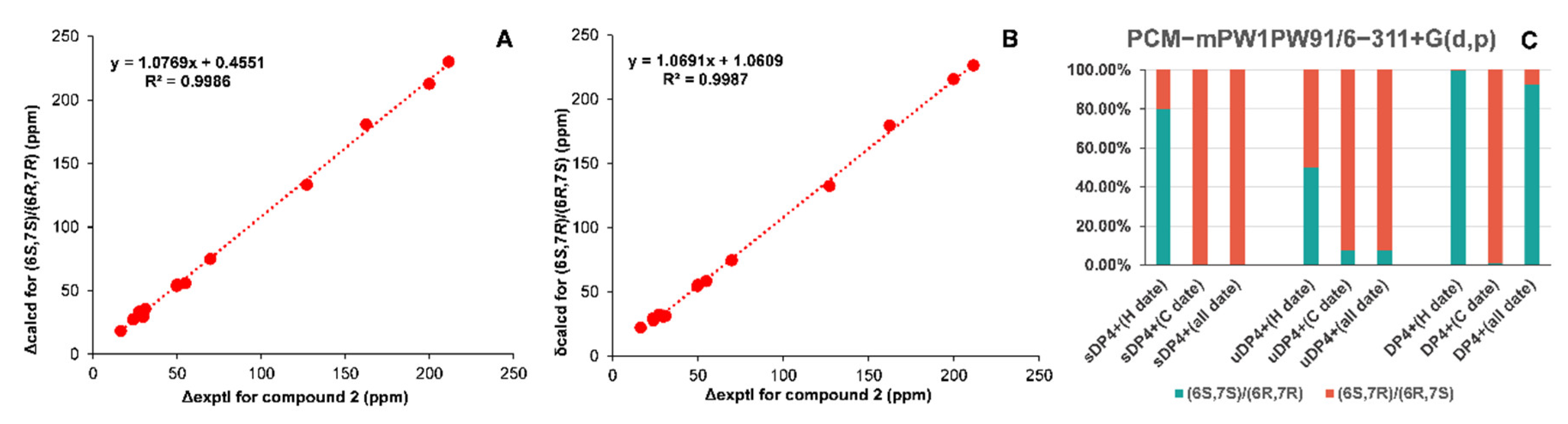
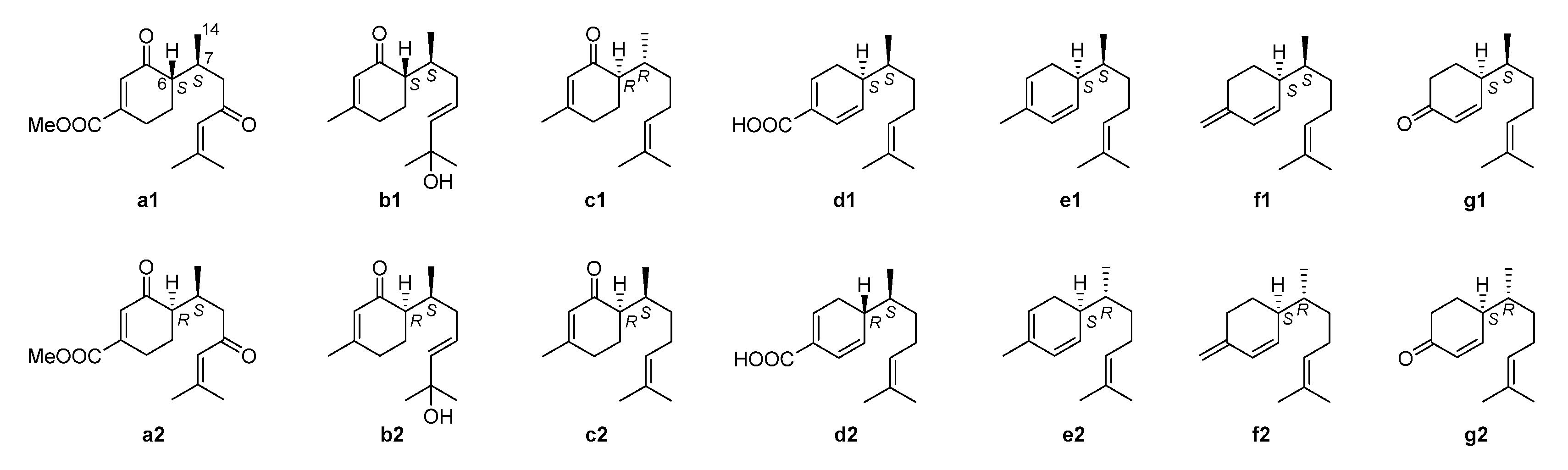
2.3. NMR Data and ECD Calculation
2.4. Cell Viability
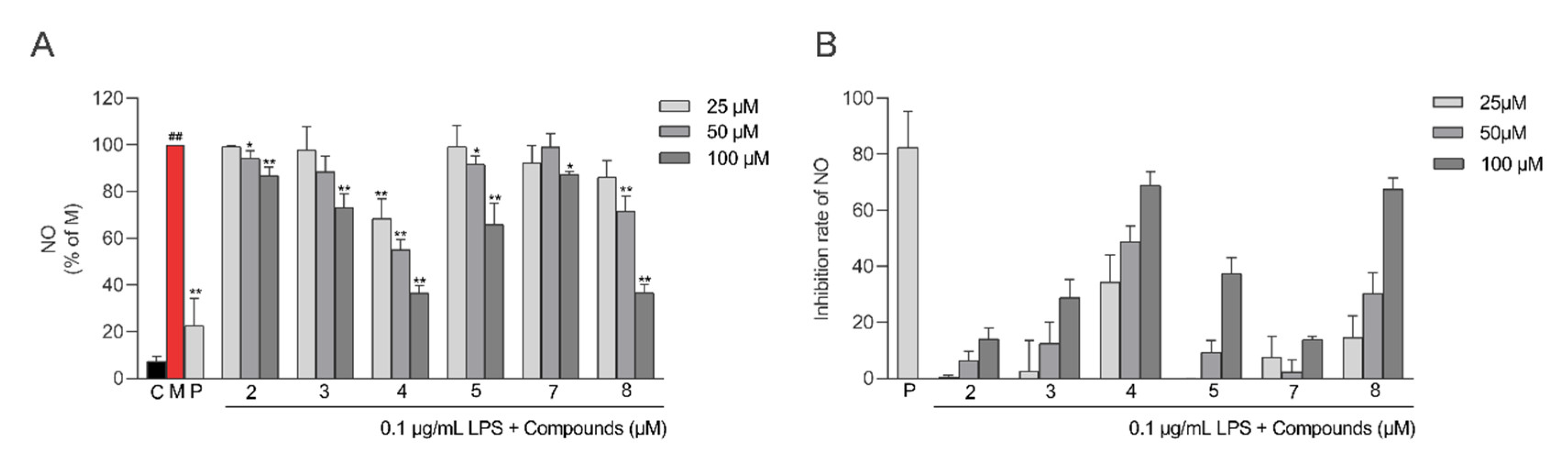
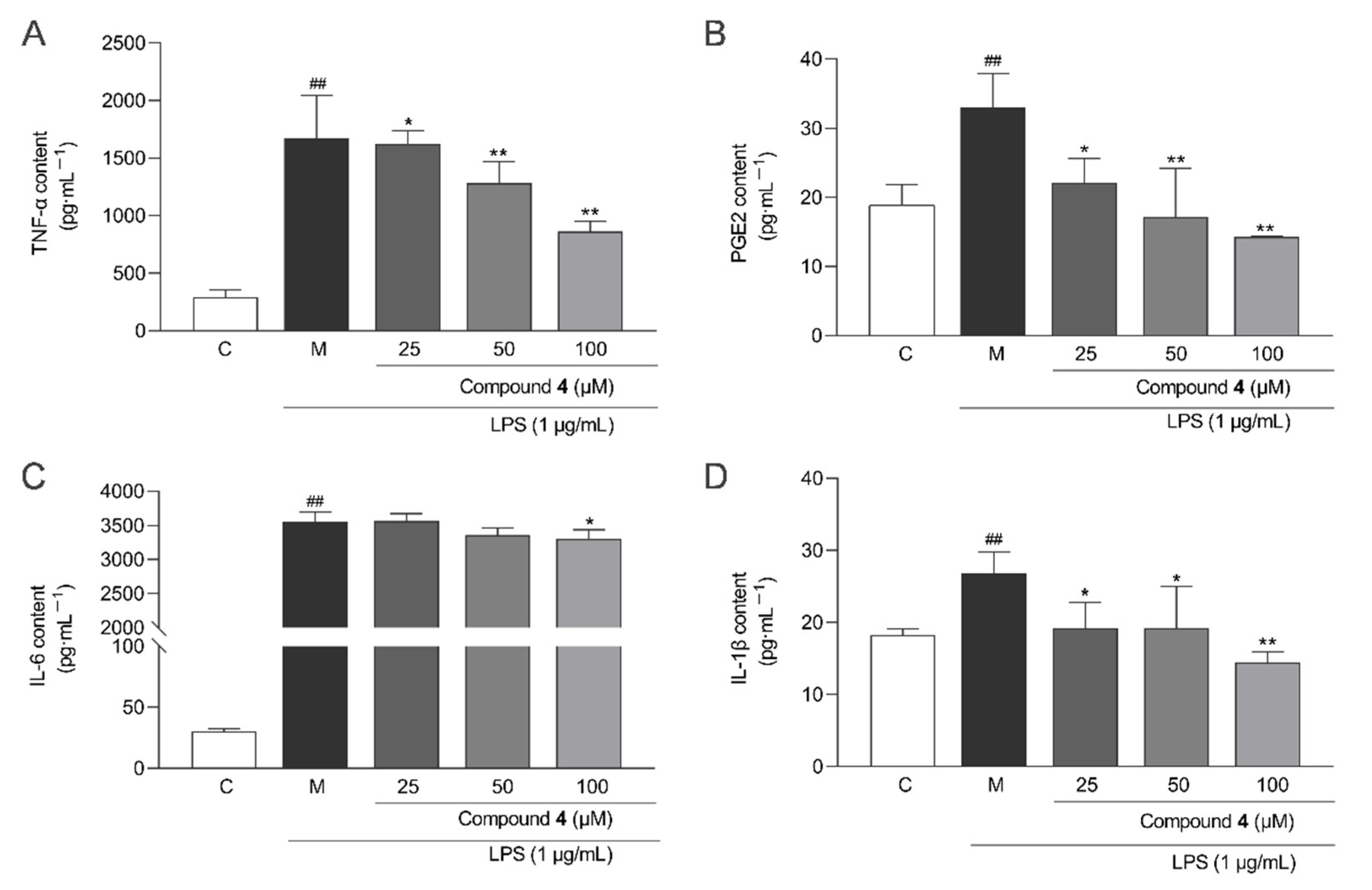
2.5. Anti-Inflammatory Activity
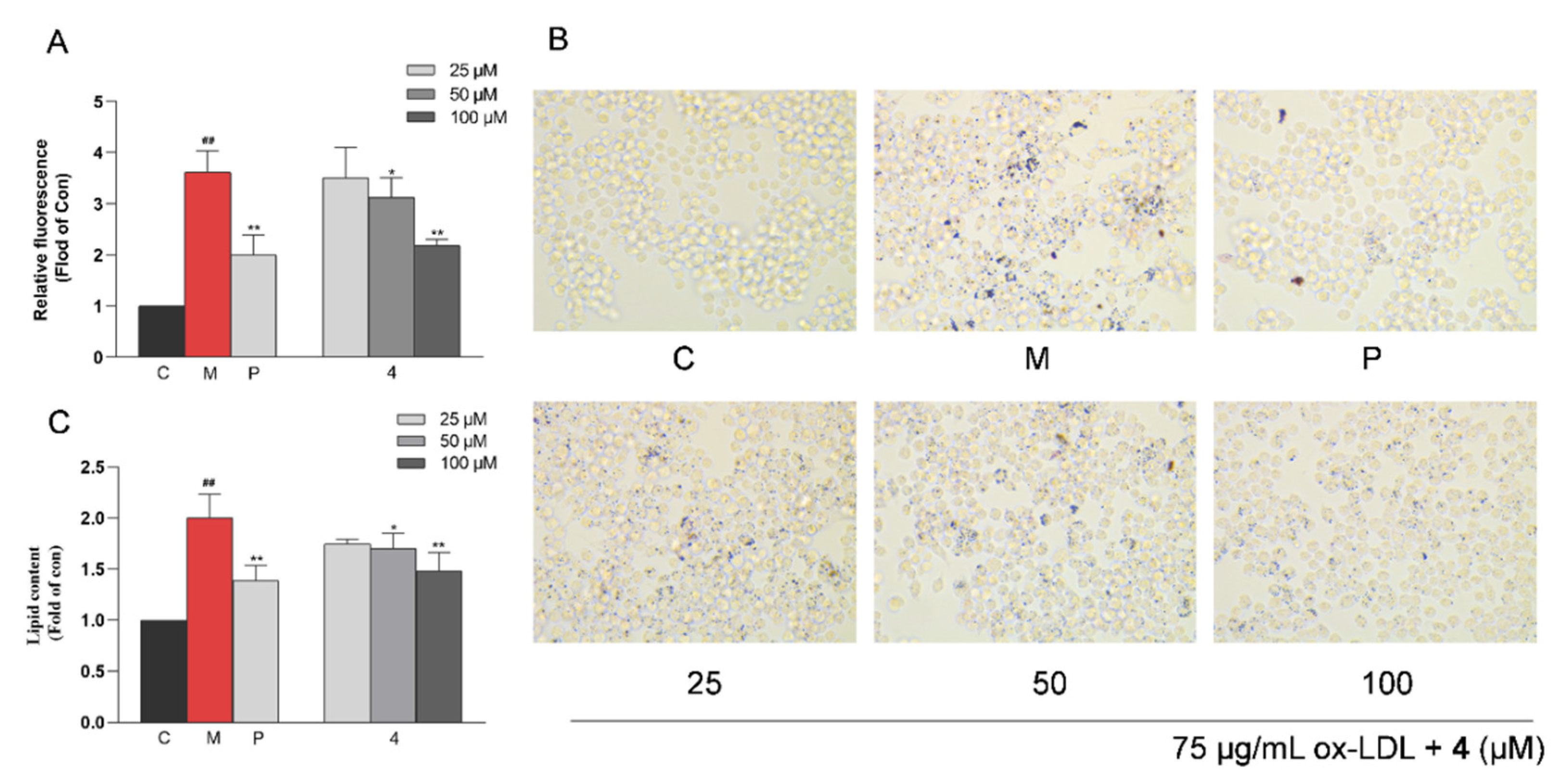
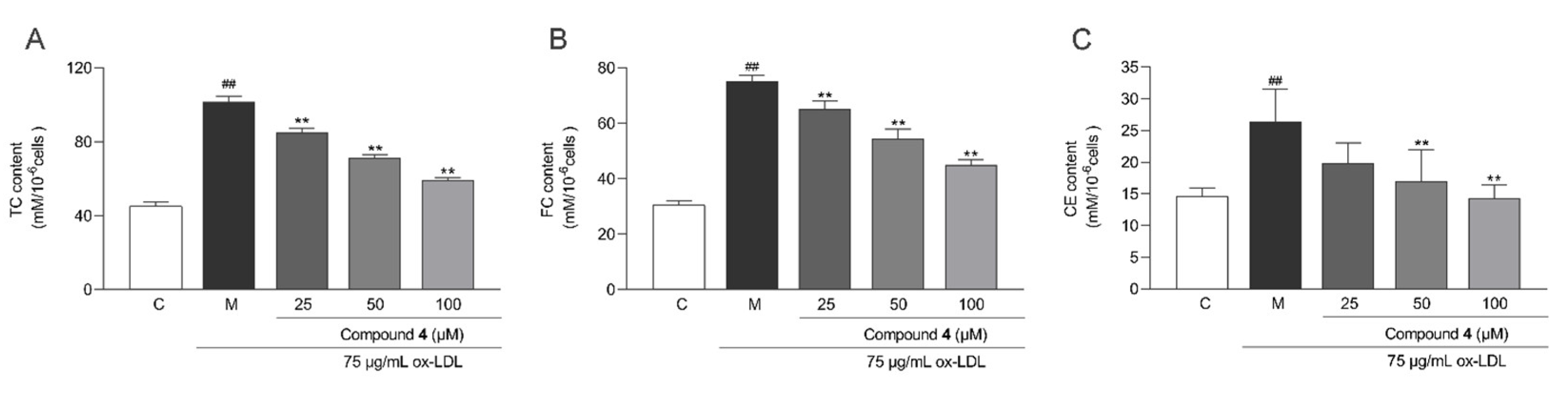
2.6. Anti-Macrophage Foaming Activity
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
4.3. Extraction and Isolation
4.4. Cell Lines and Culture
4.5. Cell Counting Kit-8 Assay
4.6. Inflammatory Factor Assay
4.7. Inhibition of Macrophage Foaming
4.7.1. Fluorometric Determination of Lipid Accumulation by Nile Red
4.7.2. Oil Red O Staining
4.7.3. Intracellular Lipid Content Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Fredman, G.; MacNamara, K.C. Atherosclerosis is a major human killer and non-resolving inflammation is a prime suspect. Cardiovasc. Res. 2021, 117, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.J.; Blaha, M.J.; Chiuve, S.E.; Cushman, M.; Das, S.R.; Deo, R.; de Ferranti, S.D.; Floyd, J.; Fornage, M.; Gillespie, C.; et al. Heart disease and stroke statistics-2017 update: A report from the American heart association. Circulation 2017, 135, e146–e603. [Google Scholar] [CrossRef] [PubMed]
- Bjorkegren, J.L.M.; Lusis, A.J. Atherosclerosis: Recent developments. Cell 2022, 185, 1630–1645. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Wu, Q.; Guo, J.; Ares, I.; Rodriguez, J.L. Statins: Adverse reactions, oxidative stress and metabolic interactions. Pharmacol. Ther. 2019, 195, 54–84. [Google Scholar] [CrossRef]
- Yuan, R.; Shi, W.L.; Xin, Q.Q.; Chen, K.J.; Cong, W.H. Holistic regulation of angiogenesis with Chinese herbal medicines as a new option for coronary artery disease. Evid. Based Complement. Altern. Med. 2018, 2018, 3725962. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; Han, J.M.; Kim, H.G.; Choi, M.K.; Son, C.G.; Yoo, H.R.; Jo, H.K.; Seol, I.C. Anti-atherosclerosis and hyperlipidemia effects of herbal mixture, Artemisia iwayomogi Kitamura and Curcuma longa Linne, in apolipoprotein E-deficient mice. J. Ethnopharmacol. 2014, 153, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Memarzia, A.; Khazdair, M.R.; Behrouz, S.; Gholamnezhad, Z.; Jafarnezhad, M.; Saadat, S.; Boskabady, M.H. Experimental and clinical reports on anti-inflammatory, antioxidant, and immunomodulatory effects of Curcuma longa and curcumin, an updated and comprehensive review. Biofactors 2021, 47, 311–350. [Google Scholar] [CrossRef]
- Jyotirmayee, B.; Mahalik, G. A review on selected pharmacological activities of Curcuma longa L. Int. J. Food Prop. 2022, 25, 1377–1398. [Google Scholar] [CrossRef]
- Hosseini, A.; Hosseinzadeh, H. Antidotal or protective effects of Curcuma longa (turmeric) and its active ingredient, curcumin, against natural and chemical toxicities: A review. Biomed. Pharmacother. 2018, 99, 411–421. [Google Scholar] [CrossRef]
- Ayati, Z.; Ramezani, M.; Amiri, M.S.; Moghadam, A.T.; Rahimi, H.; Abdollahzade, A.; Sahebkar, A.; Emami, S.A. Ethnobotany, phytochemistry and traditional uses of Curcuma spp. and pharmacological profile of two important species (C. longa and C. zedoaria): A review. Curr. Pharm. Des. 2019, 25, 871–935. [Google Scholar] [CrossRef]
- Chen, J.F.; Liu, F.; Qiao, M.M.; Shu, H.Z.; Li, X.C.; Peng, C.; Xiong, L. Vasorelaxant effect of curcubisabolanin A isolated from Curcuma longa through the PI3K/Akt/eNOS signaling pathway. J. Ethnopharmacol. 2022, 294, 115332. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, F.; Qiao, M.M.; Guo, L.; Chen, M.H.; Peng, C.; Xiong, L. Curcumanes A and B, two bicyclic sesquiterpenoids with significant vasorelaxant activity from Curcuma longa. Org. Lett. 2019, 21, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.M.; Liu, F.; Liu, Y.; Guo, L.; Zhou, Q.M.; Peng, C.; Xiong, L. Curcumane C and (+/−)-curcumane D, an unusual seco-cadinane sesquiterpenoid and a pair of unusual nor-bisabolane enantiomers with significant vasorelaxant activity from Curcuma longa. Bioorg. Chem. 2019, 92, 103275. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Tang, Y.; Huang, T.; Wang, L.; Wei, J.; Ma, G.; Xiong, J.; Hu, J. Two new bisabolane-type sesquiterpenoids from Curcuma longa. Chin. J. Org. Chem. 2018, 38, 1562–1565. [Google Scholar] [CrossRef]
- Ti, H.; Mai, Z.; Wang, Z.; Zhang, W.; Xiao, M.; Yang, Z.; Shaw, P. Bisabolane-type sesquiterpenoids from Curcuma longa L. exert anti-influenza and anti-inflammatory activities through NF-κB/MAPK and RIG-1/STAT1/2 signaling pathways. Food Funct. 2021, 12, 6697–6711. [Google Scholar] [CrossRef]
- Chen, J.J.; Tsai, C.S.; Hwang, T.L.; Shieh, P.C.; Chen, J.F.; Sung, P.J. Sesquiterpenes from the rhizome of Curcuma longa with inhibitory activity on superoxide generation and elastase release by neutrophils. Food Chem. 2010, 119, 974–980. [Google Scholar] [CrossRef]
- Bohlmann, F.; Zdero, C.; Robinson, H.; King, R.M. Germacranolides from Lychnophora species. Phytochemistry 1982, 21, 1087–1091. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef]
- Numata, A.; Kawai, K.; Takahashi, C.; Miyamoto, T. Occurrence of epijuvabione-type sesquiterpenoids in Abies sachalinensis. Phytochemistry 1992, 31, 3773–3778. [Google Scholar] [CrossRef]
- Khrimian, A.; Guggilapu, S.D.; Guzman, F.; Blassioli-Moraes, M.C.; Borges, M. Absolute configurations of stink bug- and plant-produced sesquipiperitols via synthesis of all stereoisomers. J. Nat. Prod. 2020, 83, 2281–2286. [Google Scholar] [CrossRef]
- Fotsop, D.F.; Roussi, F.; Leverrier, A.; Breteche, A.; Gueritte, F. Biomimetic total synthesis of meiogynin A, an inhibitor of Bcl-xL and Bak interaction. J. Org. Chem. 2010, 75, 7412–7415. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Wu, T.R.; Sarlah, D.; Shaw, D.M.; Rowcliffe, E.; Burton, D.R. Total synthesis, revised structure, and biological evaluation of biyouyanagin A and analogues thereof. J. Am. Chem. Soc. 2008, 130, 11114–111121. [Google Scholar] [CrossRef] [PubMed]
- Kreiser, W.; Körner, F. Stereospecific synthesis of (+)-β-sesquiphellandrene. Helv. Chim. Acta 1999, 82, 1427–1433. [Google Scholar] [CrossRef]
- Xu, J.; Ji, F.; Kang, J.; Wang, H.; Li, S.; Jin, D.Q.; Zhang, Q.; Sun, H.; Guo, Y. Absolute configurations and NO inhibitory activities of terpenoids from Curcuma longa. J. Agric. Food Chem. 2015, 63, 5805–5812. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Li, Y.L.; Li, S.M.; Shen, Y.H.; Tian, J.M.; Zhu, Z.J.; Feng, L.; Wu, L.; Lin, S.; Wang, N.; et al. Mono- and sesquiterpenoids, flavonoids, lignans, and other miscellaneous compounds of Abies georgei. Planta Med. 2010, 77, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Zhang, C.; Qiu, C.; Xia, G.; Wang, F.; Lin, B.; Li, H.; Chen, L. Chemical constituents from Curcuma longa L. and their inhibitory effects of nitric oxide production. Nat. Prod. Res. 2017, 32, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Helen, H.W.; Gabriella, G.; Min, L.; Piero, P.; David, Q.H.W. Cholesterol and lipoprotein metabolism and atherosclerosis: Recent advances in reverse cholesterol transport. Ann. Hepatol. 2017, 16, s27–s42. [Google Scholar] [CrossRef]
- Mehu, M.; Narasimhulu, C.A.; Singla, D.K. Inflammatory cells in atherosclerosis. Antioxidants 2022, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, T.; Prange, K.H.M.; Glass, C.K.; de Winther, M.P.J. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat. Rev. Cardiol. 2020, 17, 216–228. [Google Scholar] [CrossRef]
- Bäck, M.; Yurdagul, A., Jr.; Tabas, I.; Öörni, K.; Kovanen, P.T. Inflammation and its resolution in atherosclerosis: Mediators and therapeutic opportunities. Nat. Rev. Cardiol. 2019, 16, 389–406. [Google Scholar] [CrossRef]
- Shu, H.Z.; Peng, C.; Bu, L.; Guo, L.; Liu, F.; Xiong, L. Bisabolane-type sesquiterpenoids: Structural diversity and biological activity. Phytochemistry 2021, 192, 112927. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Osawa, E. Corner flapping: A simple and fast algorithm for exhaustive generation of ring conformations. J. Am. Chem. Soc. 1989, 111, 8950–8951. [Google Scholar] [CrossRef]
- Goto, H.; Osawa, E. An efficient algorithm for searching low-energy conformers of cyclic and acyclic molecules. J. Chem. Soc. Perkin Trans. 1993, 2, 187–198. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16. Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR Shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Tenderholt, A.L.; Langner, K.M.J. Cclib: A library for package-independent computational chemistry algorithms. Comput Chem. 2008, 29, 839–845. [Google Scholar] [CrossRef] [PubMed]

| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 1 | 198.6 | 199.9 | ||
| 2 | 5.74 s | 127.3 | 5.73 brs | 127.2 |
| 3 | 161.7 | 162.4 | ||
| 4 | 2.37 m | 31.5 | 2.35 m | 31.1 |
| 5 | (a) 2.00 m | 25.2 | (a) 1.96 m | 23.8 |
| (b) 1.74 m | (b) 1.74 m | |||
| 6 | 2.17 dt (12.6, 4.2) | 51.0 | 2.15 dt (12.6, 4.2) | 50.2 |
| 7 | 2.64 m | 28.3 | 2.79 m | 27.3 |
| 8 | (a) 2.51 dd (16.2, 3.6) | 49.0 | (a) 2.58 dd (17.4, 6.0) | 49.8 |
| (b) 2.37 m | (b) 2.46 dd (17.4, 7.8) | |||
| 9 | 211.9 | 211.5 | ||
| 10 | 2.60 s | 55.5 | (a) 2.60 d (15.0) | 55.0 |
| (b) 2.56 d (15.0) | ||||
| 11 | 69.9 | 69.9 | ||
| 12 | 1.20 s | 29.8 | 1.19 s | 29.8 |
| 13 | 1.20 s | 29.8 | 1.20 s | 29.8 |
| 14 | 0.91 d (6.6) | 17.4 | 0.79 d (7.2) | 16.5 |
| 15 | 1.93 s | 24.0 | 1.92 brs | 24.0 |
| OH-11 | 3.84 s | 3.87 s | ||
| No. | 3 | 4 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 1 | 200.8 | 200.7 | ||
| 2 | 5.83 s | 127.2 | 5.82 s | 126.9 |
| 3 | 161.6 | 161.6 | ||
| 4 | (a) 2.32 dd (7.8, 4.2) | 31.1 | (a) 2.30 m | 30.5 |
| (b) 2.32 dd (7.8, 4.2) | (b) 2.30 m | |||
| 5 | (a) 2.00 m | 24.7 | (a) 1.97 m | 23.2 |
| (b) 1.77 m | (b) 1.81 m | |||
| 6 | 2.19 m | 50.5 | 2.12 m | 49.6 |
| 7 | 2.65 m | 28.8 | 2.79 m | 27.9 |
| 8 | (a) 2.55 m | 48.6 | (a) 2.41 m | 49.3 |
| (b) 2.19 dd (15.0, 4.2) | (b) 2.35 m | |||
| 9 | 201.2 | 200.8 | ||
| 10 | 6.16 s | 124.1 | 6.10 brs | 123.9 |
| 11 | 155.3 | 155.6 | ||
| 12 | 1.88 s | 27.8 | 1.87 brs | 27.9 |
| 13 | 2.14 s | 20.9 | 2.12 brs | 20.9 |
| 14 | 0.93 d (6.6) | 17.5 | 0.87 d (6.6) | 16.8 |
| 15 | 1.93 s | 24.2 | 1.92 s | 24.3 |
| Compounds | Absolute Configuration | Optical Rotations |
|---|---|---|
| a1 [19] | (6S,7S) | [α: +72.7 (c 0.33, EtOH) |
| a2 [19] | (6R,7S) | [α: −34.3 (c 0.33, EtOH) |
| b1 [16] | (6S,7S) | [α: +58.7 (c 0.11, CHCl3) |
| b2 [16] | (6R,7S) | [α: −38.4 (c 0.13, CHCl3) |
| c1 [20] | (6R,7R) | [α: −20.0 (c 1.20, CHCl3) |
| c2 [20] | (6R,7S) | [α: −40 (c 1.1, CHCl3) |
| d1 [21] | (6S,7S) | [α: +88 (c 0.1, CH2Cl2) |
| d2 [21] | (6R,7S) | [α: −50(c 0.1, CH2Cl2) |
| e1 [22] | (6S,7S) | [α: +175.4 (c 0.41, CHCl3) |
| e2 [22] | (6S,7R) | [α: +128.6 (c 0.98, CHCl3) |
| f1 [23] | (6S,7S) | [α: +5.95 (c 1.17, CHCl3) |
| f2 [23] | (6S,7R) | [α: +39.58 (c 0.43, CHCl3) |
| g1 [22] | (6S,7S) | [α: +80.6 (c 1.78, CH2Cl2) |
| g2 [22] | (6S,7R) | [α: +60.2 (c 0.41, CH2Cl2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.-Q.; Wu, G.-X.; Peng, C.; Fan, Y.-Q.; Li, L.; Liu, F.; Xiong, L. New Bisabolane-Type Sesquiterpenoids from Curcuma longa and Their Anti-Atherosclerotic Activity. Molecules 2023, 28, 2704. https://doi.org/10.3390/molecules28062704
Guo Y-Q, Wu G-X, Peng C, Fan Y-Q, Li L, Liu F, Xiong L. New Bisabolane-Type Sesquiterpenoids from Curcuma longa and Their Anti-Atherosclerotic Activity. Molecules. 2023; 28(6):2704. https://doi.org/10.3390/molecules28062704
Chicago/Turabian StyleGuo, Yu-Qin, Guang-Xu Wu, Cheng Peng, Yun-Qiu Fan, Lei Li, Fei Liu, and Liang Xiong. 2023. "New Bisabolane-Type Sesquiterpenoids from Curcuma longa and Their Anti-Atherosclerotic Activity" Molecules 28, no. 6: 2704. https://doi.org/10.3390/molecules28062704
APA StyleGuo, Y.-Q., Wu, G.-X., Peng, C., Fan, Y.-Q., Li, L., Liu, F., & Xiong, L. (2023). New Bisabolane-Type Sesquiterpenoids from Curcuma longa and Their Anti-Atherosclerotic Activity. Molecules, 28(6), 2704. https://doi.org/10.3390/molecules28062704







