Quorum Quenchers from Reynoutria japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
2. Results
2.1. Physiochemical Characterization of Proteins
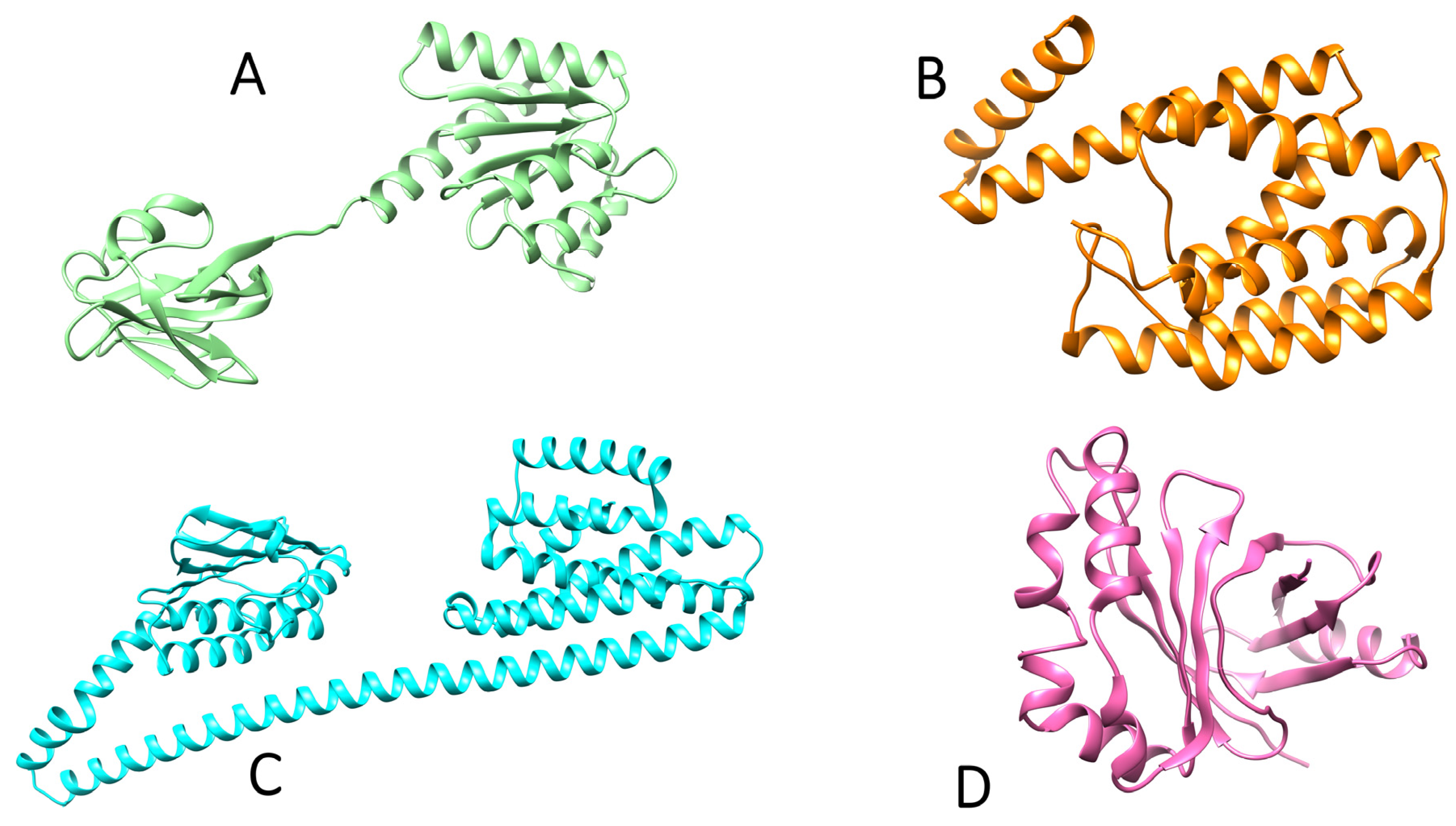
2.2. 3D Structural Prediction of Proteins
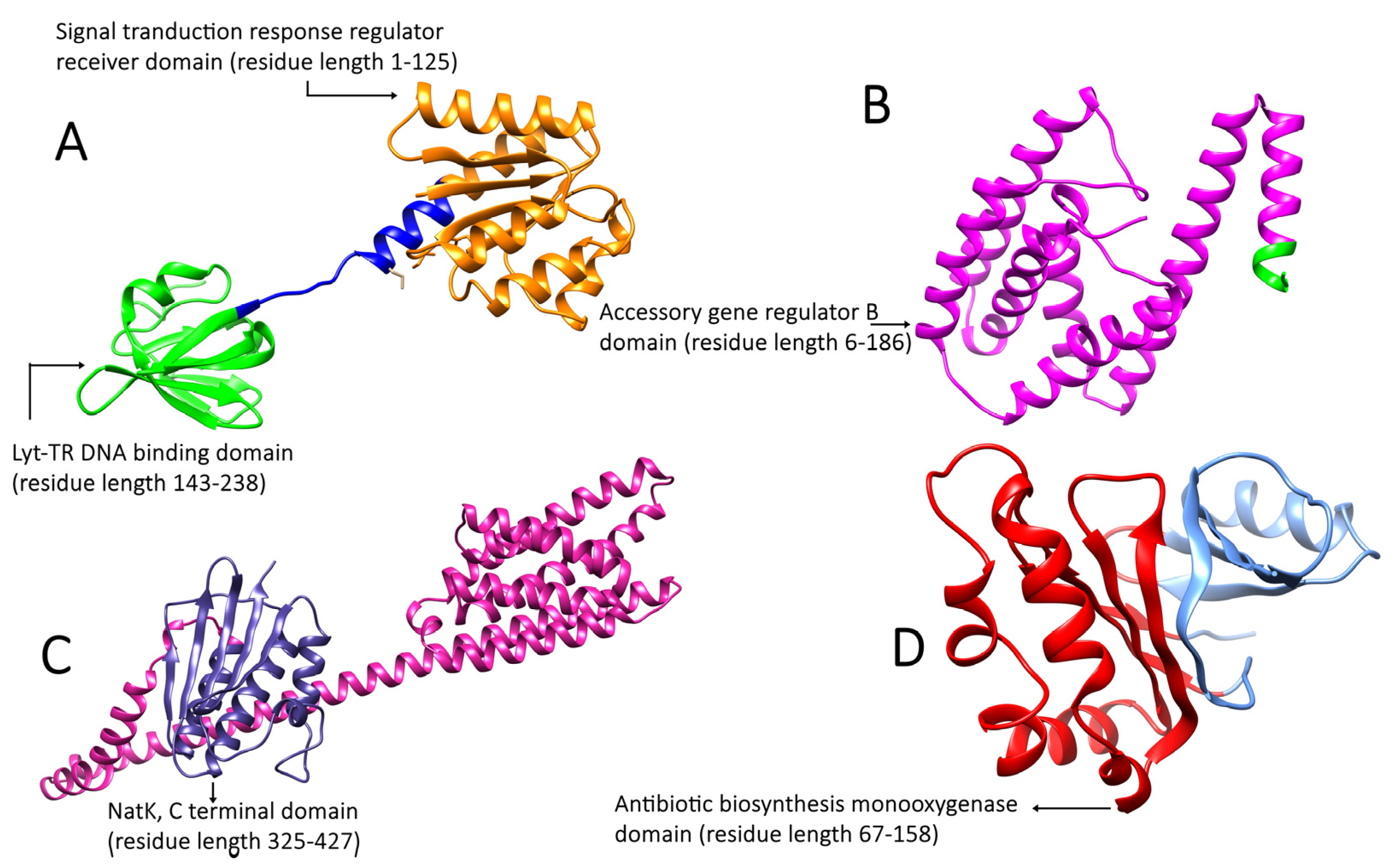
2.3. Functional Domain Identification of Proteins
2.4. Ligand Selection
2.5. Molecular Docking
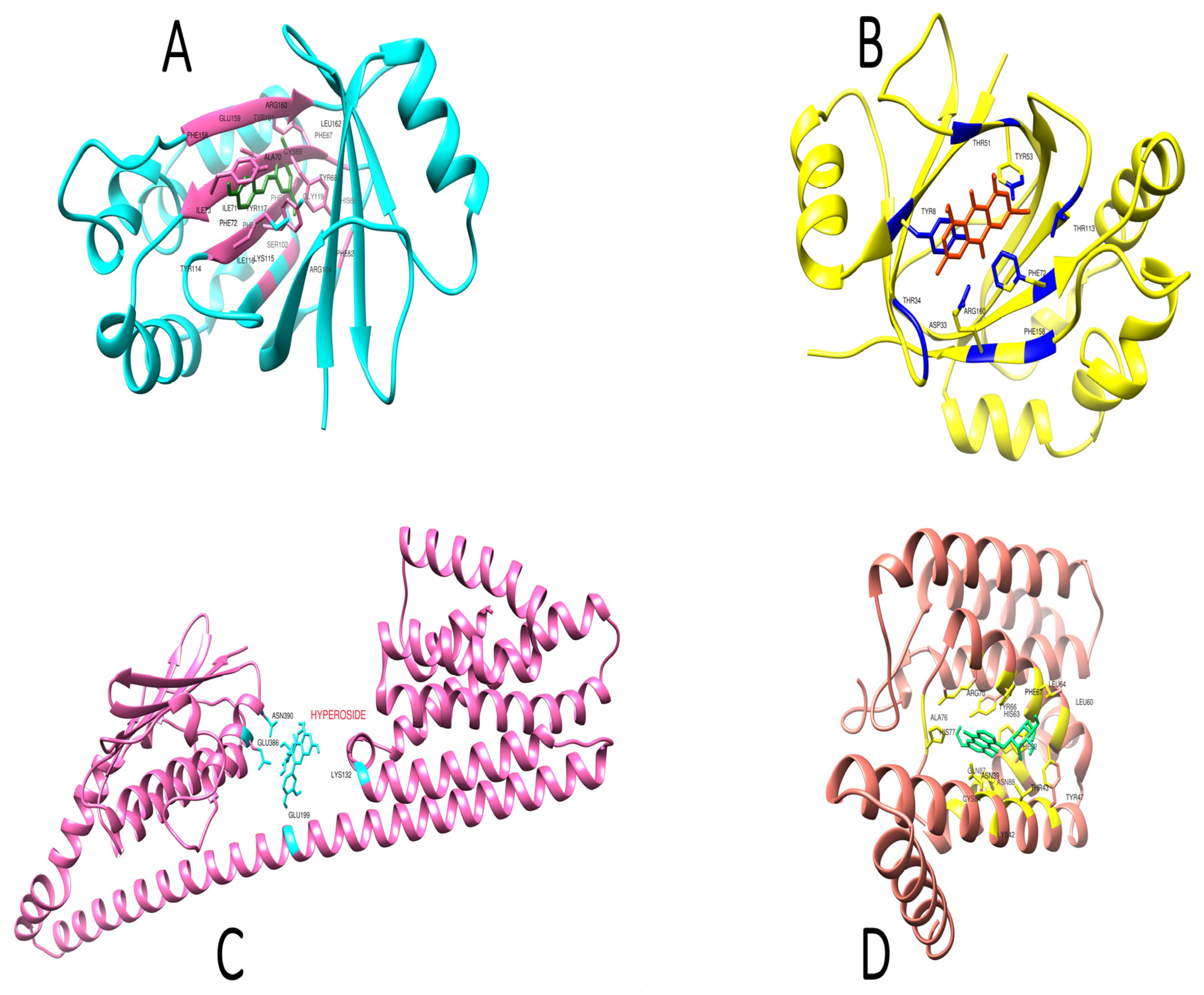
2.6. Active Site Identification
2.7. Interaction of Ligands and Target Proteins
2.8. Ligands’ ADMET Properties
2.9. Distribution, Metabolic, and Excretion Properties of Ligands
2.10. Ligand Toxicity
2.11. Lipinski Rule of Five
2.12. Lead Compound Identification
2.13. Comparative Investigation of Lead Compound vs. Penicillin
2.14. Comparison of Absorption Properties
2.15. Comparison of Distribution Properties
2.16. Comparison of Metabolic Properties
2.17. Comparison of Excretion Properties
2.18. Comparison of Toxicity
2.19. Lipinski Rule of Five
2.20. Docking Score Comparison
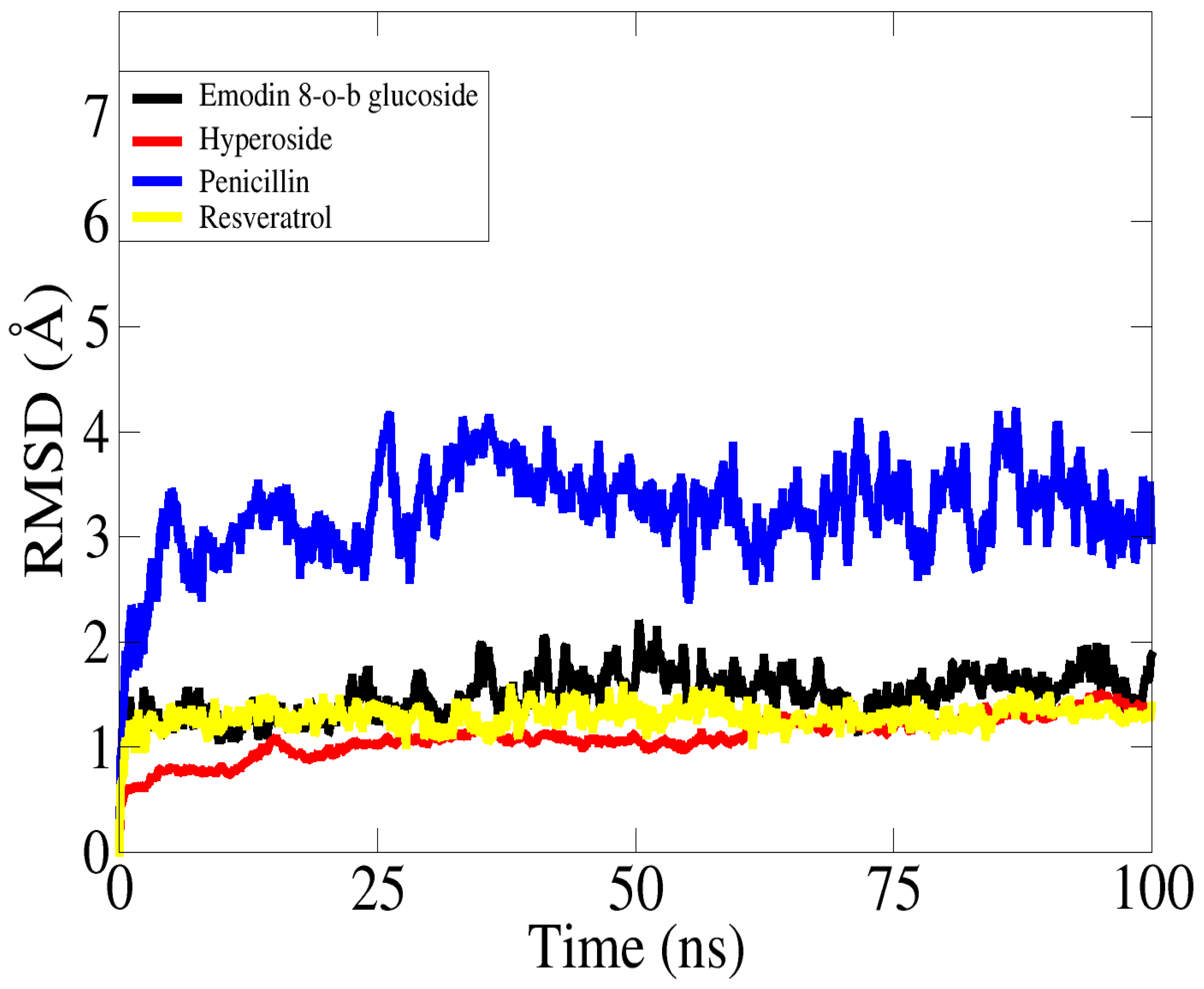
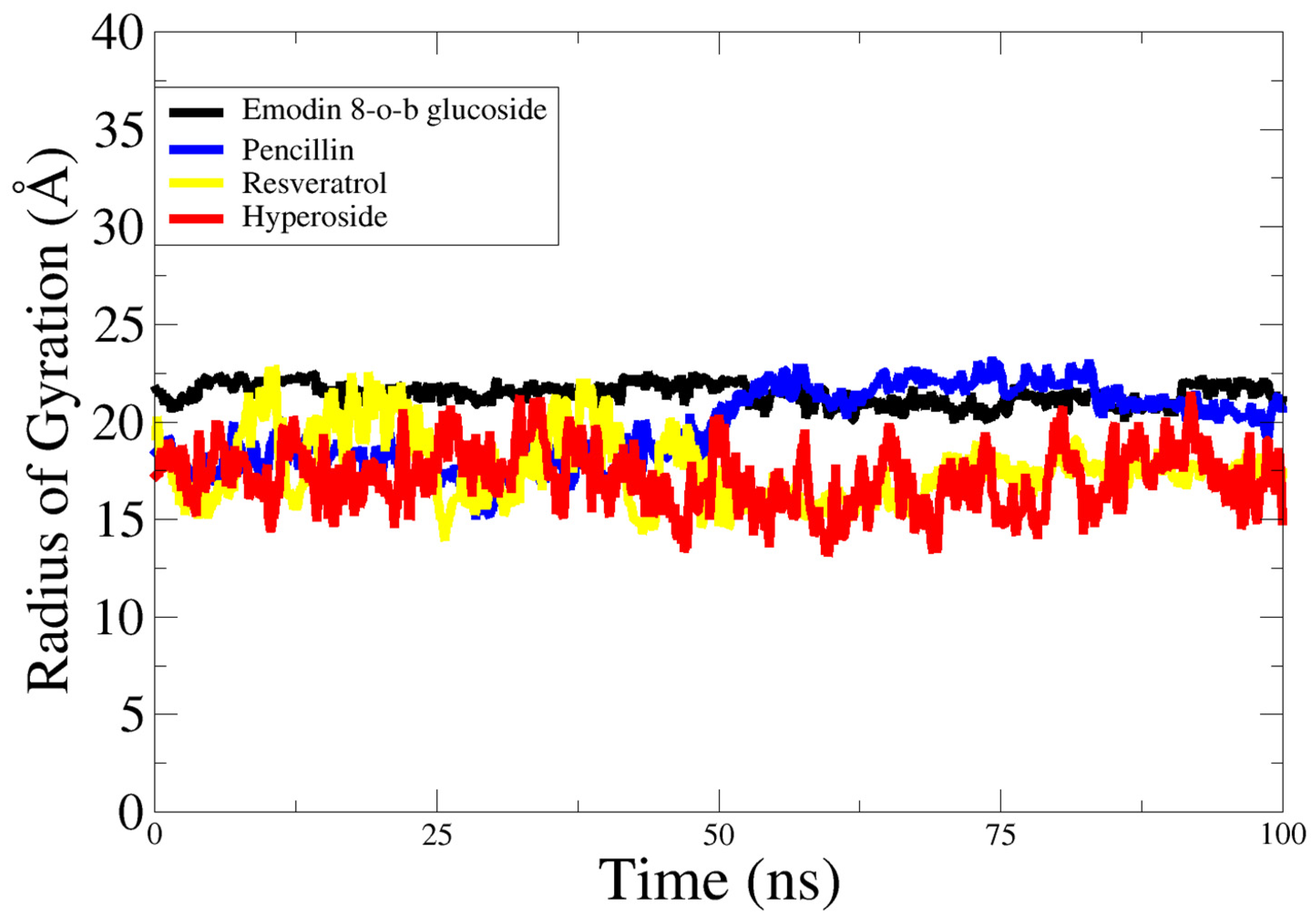
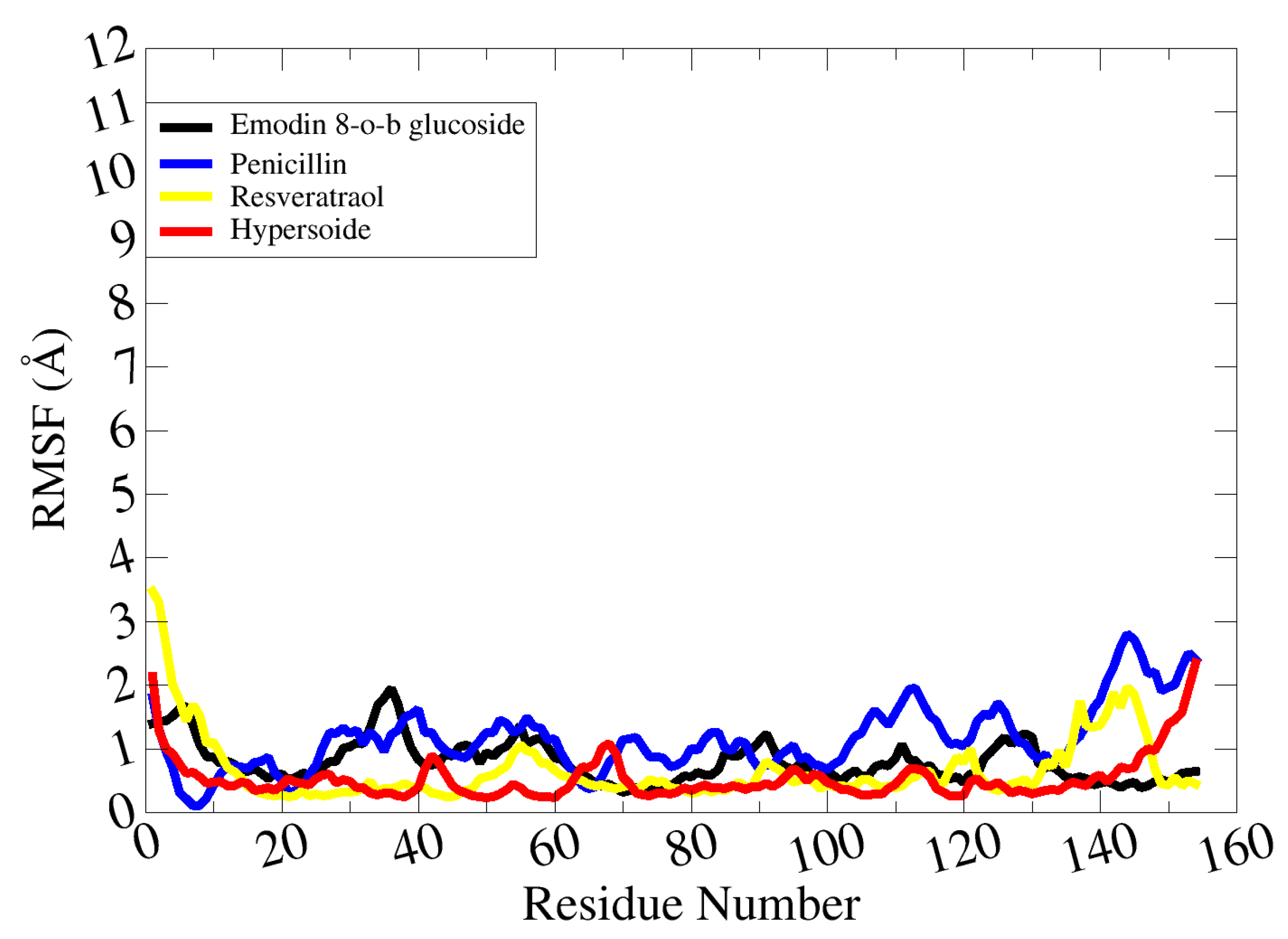
2.21. Molecular Dynamic Simulations
3. Discussion
4. Materials and Methods
4.1. Ligand Preparation and Selection
4.2. Bioactivity Analysis of Ligands and Toxicity Measurement
4.3. Target Protein Selection and Primary Sequence Retrieval
4.4. Physiochemical Properties and 3D Structures of Proteins
4.5. Structure Analysis and Functional Domain Identification
4.6. Active Site Identification
4.7. Molecular Docking of Targeted Proteins
4.8. Lead Compound Identification
4.9. Reference Antibacterial Drug Identification and Selection
4.10. Prediction of Different Parameters of Selected Drugs
4.11. Reference Drug and Lead Compound Comparison
4.12. Molecular Dynamic Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Mali, S.N.; Thorat, B.R.; Gupta, D.R.; Pandey, A. Mini-Review of the Importance of Hydrazides and Their Derivatives—Synthesis and Biological Activity. Eng. Proc. 2021, 11, 21. [Google Scholar] [CrossRef]
- Appelbaum, P.C. Microbiology of Antibiotic Resistance in Staphylococcus aureus. Clin. Infect. Dis. 2007, 45, 323–334. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.H.; Zhang, L.H. Quorum-Quenching Microbial Infections: Mechanisms and Implications. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.; Williams, P. Quorum Sensing and Social Networking in the Microbial World. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Acuña, L.; Otero, A. Patents on Quorum Quenching: Interfering with Bacterial Communication as a Strategy to Fight Infections. Recent Pat. Biotechnol. 2012, 6, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Painter, K.L.; Krishna, A.; Wigneshweraraj, S.; Edwards, A.M. What Role Does the Quorum-Sensing Accessory Gene Regulator System Play during Staphylococcus aureus Bacteremia? Trends Microbiol. 2014, 22, 676–685. [Google Scholar] [CrossRef]
- Bhakdi, S.; Tranum-Jensen, J. Alpha-Toxin of Staphylococcus aureus. Microbiol. Rev. 1991, 55, 733–751. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Autoinduction and Signal Transduction in the Regulation of Staphylococcal Virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef]
- Junecko, J.M. Transcribing Virulence in Staphylococcus aureus. World J. Clin. Infect. Dis. 2012, 2, 63. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum Sensing Inhibitors: An Overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Ganesh, P.S.; Veena, K.; Senthil, R.; Iswamy, K.; Ponmalar, E.M.; Mariappan, V.; Girija, A.S.S.; Vadivelu, J.; Nagarajan, S.; Challabathula, D.; et al. Biofilm-Associated Agr and Sar Quorum Sensing Systems of Staphylococcus aureus Are Inhibited by 3-Hydroxybenzoic Acid Derived from Illicium Verum. ACS Omega 2022, 7, 14653–14665. [Google Scholar] [CrossRef]
- Peng, W.; Qin, R.; Li, X.; Zhou, H. Botany, Phytochemistry, Pharmacology, and Potential Application of Polygonum Cuspidatum Sieb.et Zucc.: A Review. J. Ethnopharmacol. 2013, 148, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Niesen, D.B.; Hessler, C.; Seeram, N.P. Beyond Resveratrol: A Review of Natural Stilbenoids Identified from 2009–2013. J. Berry Res. 2013, 3, 181–196. [Google Scholar] [CrossRef]
- Abedini, E.; Khodadadi, E.; Zeinalzadeh, E.; Moaddab, S.R.; Asgharzadeh, M.; Mehramouz, B.; Dao, S.; Samadi Kafil, H. A Comprehensive Study on the Antimicrobial Properties of Resveratrol as an Alternative Therapy. Evid. Based Complement. Altern. Med. 2021, 2021, 8866311. [Google Scholar] [CrossRef]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, Agr Groups (Alleles), and Human Disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Muir, T.W. Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles. Cell Chem. Biol. 2016, 23, 214–224. [Google Scholar] [CrossRef]
- Heaslet, H.; Harris, M.; Fahnoe, K.; Sarver, R.; Putz, H.; Chang, J.; Subramanyam, C.; Barreiro, G.; Miller, J.R. Structural Comparison of Chromosomal and Exogenous Dihydrofolate Reductase from Staphylococcus aureus in Complex with the Potent Inhibitor Trimethoprim. Proteins Struct. Funct. Bioinform. 2009, 76, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Hooft, R.W.W.; Sander, C.; Vriend, G. Objectively Judging the Quality of a Protein Structure from a Ramachandran Plot. Bioinformatics 1997, 13, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Sladek, V.; Yamamoto, Y.; Harada, R.; Shoji, M.; Shigeta, Y.; Sladek, V. PyProGA-A PyMOL Plugin for Protein Residue Network Analysis. PLoS ONE 2021, 16, e0255167. [Google Scholar] [CrossRef] [PubMed]
- Kuyper, L.F.; Baccanari, D.P.; Jones, M.L.; Hunter, R.N.; Tansik, R.L.; Joyner, S.S.; Boytos, C.M.; Rudolph, S.K.; Knick, V.; Wilson, H.R.; et al. High-Affinity Inhibitors of Dihydrofolate Reductase: Antimicrobial and Anticancer Activities of 7,8-Dialkyl-1,3-Diaminopyrrolo[3,2-f]Quinazolines with Small Molecular Size. J. Med. Chem. 1996, 39, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ji, G. Identification of a Staphylococcal AgrB Segment(s) Responsible for Group-Specific Processing of AgrD by Gene Swapping. J. Bacteriol. 2004, 186, 6706–6713. [Google Scholar] [CrossRef] [PubMed]
- Cousins, K.R. Computer Review of ChemDraw Ultra 12.0; ACS Publications: Washington, DC, USA, 2011. [Google Scholar] [CrossRef]
- Kshatriya, R.; Shelke, P.; Mali, S.; Yashwantrao, G.; Pratap, A.; Saha, S. Synthesis and Evaluation of Anticancer Activity of Pyrazolone Appended Triarylmethanes (TRAMs). ChemistrySelect 2021, 6, 6230–6239. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A Program to Generate Schematic Diagrams of Protein-Ligand Interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Desale, V.J.; Mali, S.N.; Thorat, B.R.; Yamgar, R.S. Synthesis, AdmetSAR Predictions, DPPH Radical Scavenging Activity, and Potent Anti-Mycobacterial Studies of Hydrazones of Substituted 4-(Anilino Methyl) Benzohydrazides (Part 2). Curr. Comput. Aided Drug Des. 2021, 17, 493–503. [Google Scholar] [CrossRef]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum Sensing in Bacteria: The LuxR-LuxI Family of Cell Density- Responsive Transcriptional Regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef]
- Fleitas Martínez, O.; Rigueiras, P.O.; Pires, Á.D.S.; Porto, W.F.; Silva, O.N.; de la Fuente-Nunez, C.; Franco, O.L. Interference with Quorum-Sensing Signal Biosynthesis as a Promising Therapeutic Strategy Against Multidrug-Resistant Pathogens. Front. Cell. Infect. Microbiol. 2019, 8, 444. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.A.K.; Akter, K.M.; Kim, H.J.; Park, W.S.; Kang, D.M.; Koo, K.A.; Ahn, M.J. Comparative Inner Morphological and Chemical Studies on Reynoutria Species in Korea. Plants 2020, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Singh, R.V.; Yadav, D.B. Synthesis and Evaluation of Novel 1,4-Naphthoquinone Derivatives as Antiviral, Antifungal and Anticancer Agents. Bioorg. Med. Chem. Lett. 2004, 14, 2901–2904. [Google Scholar] [CrossRef] [PubMed]
- Bradford, C. The Use of Commercially Available Alpha-Amylase Compounds to Inhibit and Remove Staphylococcus aureus Biofilms. Open Microbiol. J. 2011, 5, 21–31. [Google Scholar] [CrossRef]
- Neves, R.A.; Lucio, M.; Lima, L.C.J.; Reis, S. Resveratrol in Medicinal Chemistry: A Critical Review of Its Pharmacokinetics, Drug-Delivery, and Membrane Interactions. Curr. Med. Chem. 2012, 19, 1663–1681. [Google Scholar] [CrossRef]
- Smoliga, J.M.; Baur, J.A.; Hausenblas, H.A. Resveratrol and Health—A Comprehensive Review of Human Clinical Trials. Mol. Nutr. Food Res. 2011, 55, 1129–1141. [Google Scholar] [CrossRef]
- Janzon, L.; Arvidson, S. The Role of the δ-Lysin Gene (Hld) in the Regulation of Virulence Genes by the Accessory Gene Regulator (Agr) in Staphylococcus aureus. EMBO J. 1990, 9, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Tegmark, K.; Morfeldt, E.; Arvidson, S. Regulation of Agr-Dependent Virulence Genes in Staphylococcus aureus by RNAIII from Coagulase-Negative Staphylococci. J. Bacteriol. 1998, 180, 3181–3186. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial Interference Caused by Autoinducing Peptide Variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the Agr and/or SarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Butkiewicz, M.; Lowe, E.W.; Mueller, R.; Mendenhall, J.L.; Teixeira, P.L.; Weaver, C.D.; Meiler, J. Benchmarking Ligand-Based Virtual High-Throughput Screening with the Pubchem Database. Molecules 2013, 18, 735–756. [Google Scholar] [CrossRef]
- Mali, S.N.; Pandey, A.; Thorat, B.R.; Lai, C.-H. Greener Synthesis, In-silico and Theoretical Analysis of Hydrazides as Potential Antituberculosis Agents (Part 1). Chem. Proc. 2021, 8, 86. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PK(a) Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of Three-Dimensional Structural Information of Biological Macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Vendeville, A.; Winzer, K.; Heurlier, K.; Tang, C.M.; Hardie, K.R. Making “sense” of Metabolism: Autoinducer-2, LuxS and Pathogenic Bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. [Google Scholar] [CrossRef]
- Kapale, S.S.; Mali, S.N.; Chaudhari, H.K. Molecular Modelling Studies for 4-Oxo-1,4-Dihydroquinoline-3-Carboxamide Derivatives as Anticancer Agents. Med. Drug Discov. 2019, 2, 100008. [Google Scholar] [CrossRef]
- Levitt, M.; Lifson, S. Refinement of Protein Conformations Using a Macromolecular Energy Minimization Procedure. J. Mol. Biol. 1969, 46, 269–279. [Google Scholar] [CrossRef]
- Gill, S.C.; von Hippel, P.H. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data [Published Erratum Appears in Anal Biochem 1990 Sep;189(2):283]. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- King, A.D.; Pržulj, N.; Jurisica, I. Protein Complex Prediction with RNSC. Methods Mol. Biol. 2012, 804, 297–312. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R. Tools and Resources for Identifying Protein Families, Domains and Motifs. Genome Biol. 2002, 3, 1–8. [Google Scholar]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.; Liang, J. CASTp: Computed Atlas of Surface Topography of Proteins with Structural and Topographical Mapping of Functionally Annotated Residues. Nucleic Acids Res. 2006, 34, 116–118. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive Gene and Pathway Analysis of Cervical Cancer Progression. Oncol. Lett. 2020, 19, 3316–3332. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.; Darden, T.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Götz, A.W.; Homeyer, N.; et al. Amber 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham III, T.E.; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. The FF14SB Force Field. Amber 2014, 14, 29–31. [Google Scholar]
- Schafmeister, C.; Ross, W.S.; Romanovski, V. The Leap Module of AMBER; University of California: San Francisco, CA, USA, 1995. [Google Scholar]
- Petersen, H.G. Accuracy and Efficiency of the Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 3668–3679. [Google Scholar] [CrossRef]


| Target Proteins | MW | PI | NR | PR | Ext.Co1 | Ext.Co2 | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| AgrA | 27,905.90 | 5.78 | 37 | 31 | 15,150 | 14,900 | 36.25 | 91.30 | −0.379 |
| AgrB | 21,929.69 | 9.85 | 8 | 19 | 18,910 | 18,910 | 45.16 | 147.04 | 0.828 |
| AgrC | 49,896.91 | 5.19 | 45 | 38 | 38,405 | 38,280 | 39.15 | 127.16 | 0.494 |
| TRAP | 19,547.47 | 6.12 | 22 | 18 | 20,860 | 20,860 | 20.68 | 60.78 | −0.580 |
| S. No | Ligand Name | Molecular Formula | Molecular Weight | Structure |
|---|---|---|---|---|
| 1 | 2-methoxy-6-acetyl-7-methyljuglone | C14H12O5 | 260.24 g/mol | 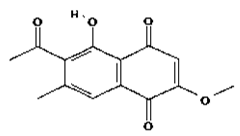 |
| 2 | Emodin | C15H10O5 | 270.24 g/mol | 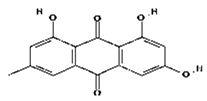 |
| 3 | Emodin 8-o-b glucoside | C21H20O10 | 432.4 g/mol | 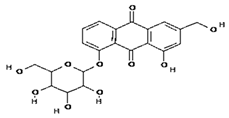 |
| 4 | Polydatin | C20H22O8 | 390.4 g/mol |  |
| 5 | Resveratrol | C14H12O3 | 228.24 g/mol | 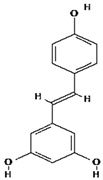 |
| 6 | Physcion | C16H12O5 | 284.26 g/mol |  |
| 7 | Citreorosein | C15H10O6 | 286.24 g/mol | 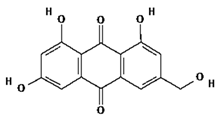 |
| 8 | Quercetin | C15H10O7 | 302.23 g/mol | 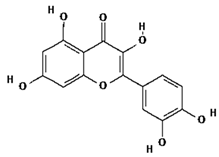 |
| 9 | Hyperoside | C21H20O12 | 464.4 g/mol |  |
| 10 | Coumarin | C9H6O2 | 146.14 g/mol |  |
| S. No | Ligand Name | Binding Score kcal/mol |
|---|---|---|
| 1 | 2-methoxy-6-acetyl-7-methyljuglone | −7.1 |
| 2 | Emodin | −8.4 |
| 3 | Emodin 8-o-b glucoside | −9.9 |
| 4 | Polydatin | −8.8 |
| 5 | Resveratrol | −8.9 |
| 6 | Physcion | −8.6 |
| 7 | Citreorosein | −8.4 |
| 8 | Quercetin | −8.8 |
| 9 | Hyperoside | −9.1 |
| 10 | Coumarin | −6.6 |
| Ligands | Target Proteins with Interactive Residues |
|---|---|
| 2-methoxy-6-acetyl-7-methyljuglone | 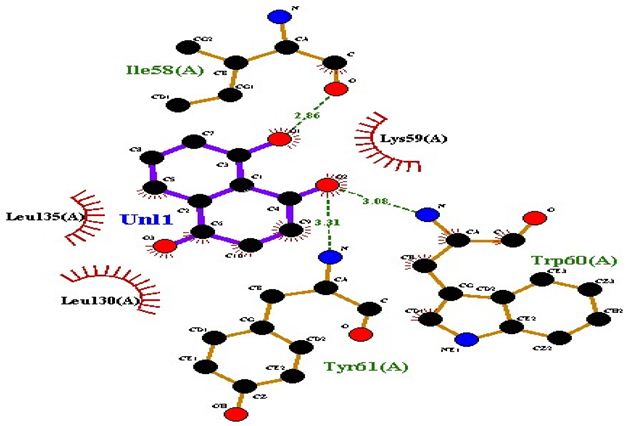 AgrA |
| Emodin | 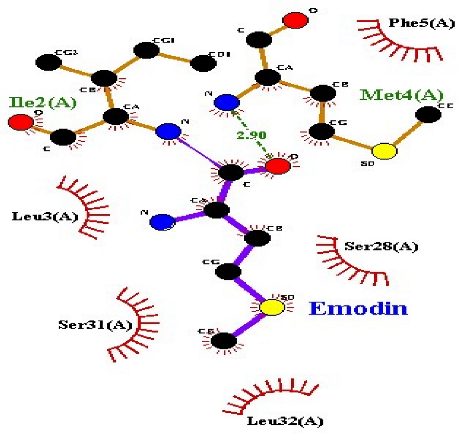 TRAP |
| Emodin 8-o-b glucoside | 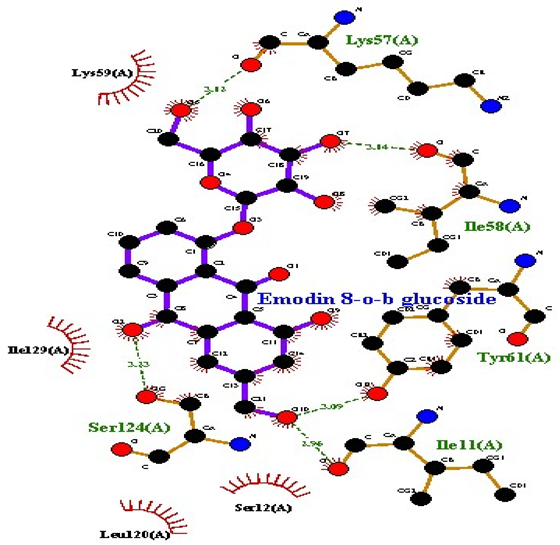 AgrB |
| Polydatin |  TRAP |
| Resveratrol | 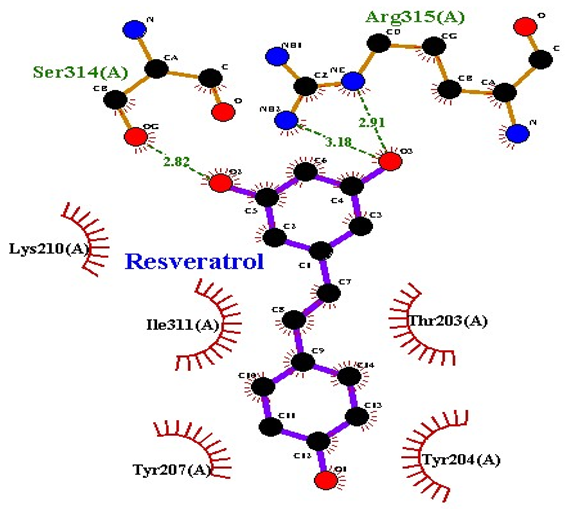 TRAP |
| Physcion |  AgrC |
| Citreorosein |  AgrC |
| Quercetin | 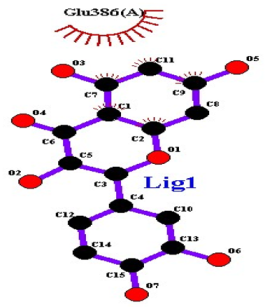 AgrC |
| Hyperoside |  AgrC |
| Coumarin | 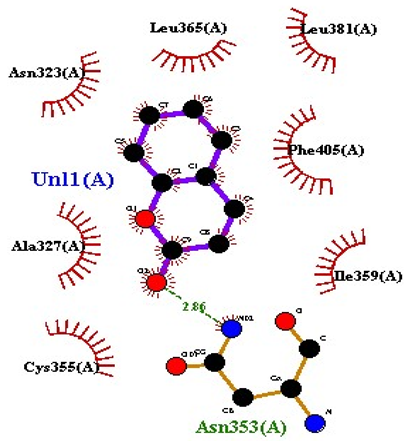 AgrC |
| S. No | Ligand Name | Binding Energy | No of HBs | Amino Acids | Hydrogen Bonding Distance | Hydrophobic Interactions |
|---|---|---|---|---|---|---|
| 1 | 2-methoxy-6-acetyl-7-methyljuglone | −7.1 | 3 | Ile58 Trp60 Tyr61 | 2.86 3.08 3.31 | Lys59 Leu135 Leu130 |
| 2 | Emodin | −8.4 | 2 | Met4 Ile2 | 2.90 2.91 | Phe5 Leu3 Ser28 Ser31 Leu32 |
| 3 | Emodin 8-o-b glucoside | −9.9 | 5 | Lys57 Ile58 Tyr61 Ile11 Ser124 | 3.12 3.14 3.09 2.96 3.23 | Lys59 Ile129 Leu120 Ser12 |
| 4 | Polydatin | −8.8 | 5 | Arg315 Lys210 Ser314 Glu276 Asn215 | 3.10 3.17 3.03 2.76 3.26 | Glu206 Tyr207 Leu280 Ile214 Ile211 |
| 5 | Resveratrol | −8.9 | 2 | Arg315 Ser314 | 2.91 2.82 | Lys210 Ile311 Thr203 Tyr207 Tyr204 |
| 6 | Physcion | −8.6 | 3 | Ser185 Thr68 Ser178 | 2.97 3.01 2.83 | Phe134 Leu120 Ile123 Leu64 Leu142 Thr181 Phe182 |
| 7 | Citreorosein | −8.4 | 0 | - | - | Glu386 |
| 8 | Quercetin | −8.8 | 0 | - | - | Glu386 |
| 9 | Hyperoside | −9.1 | 7 | Arg70 Gln131 Asn39 His77 Asn88 Thr43 Tyr66 | 3.06 3.13 2.94 2.80 3.02 2.80 2.22 | Arg78 Phe67 Cys54 Lys43 Glu37 Phe92 |
| 10 | Coumarin | −6.6 | 1 | Asn353 | 2.86 | Leu381 Leu365 Asn323 Phe405 Ile359 Ala327 Cys355 |
| S. No | Ligand Name | Water Solubility (mol/L) | CaCO2 Permeability (cm/S) | Intestinal Absorption (Human) % | Skin Permeability Log/Kp | P-Glycoprotein Substrate | P-Glycoprotein I Inhibtor | P-Glycoprotein II Inhibitor |
|---|---|---|---|---|---|---|---|---|
| 1 | 2methoxy- 6-acetyl- 7-methyljugl one | −0.835 | 1.232 | 94.085 | −2.77 | No | No | No |
| 2 | Emodin | −3.271 | 0.259 | 71.316 | −2.741 | Yes | No | No |
| 3 | Emodin 8-o-b glucoside | −2.972 | 0.367 | 43.072 | −2.735 | Yes | No | No |
| 4 | Polydatin | −3.113 | 0.167 | 42.758 | −2.735 | Yes | No | No |
| 5 | Resveratrol | −3.235 | 1.196 | 87.933 | −2.748 | Yes | No | No |
| 6 | Physcion | −3.156 | 1.26 | 95.924 | −2.8 | Yes | No | No |
| 7 | Citreorosein | −3.186 | −0.368 | 62.631 | −2.74 | Yes | No | No |
| 8 | Quercetin | −3.097 | −0.277 | 76.081 | −2.735 | Yes | No | No |
| 9 | Hyperoside | −2.894 | 0.173 | 44.847 | −2.735 | Yes | No | No |
| 10 | Coumarin | −1.486 | 1.642 | 97.171 | −1.911 | Yes | No | No |
| Ligands | Logp Value | Molecular Weight | H-Bond Acceptor | H-Bond Donor |
|---|---|---|---|---|
| Juglone | 1.3274 | 174.155 | 3 | 1 |
| Emodin | 1.88722 | 270.24 | 5 | 3 |
| Emodin 8-o-b | −1.1614 | 432.381 | 10 | 6 |
| Polydatin | 0.4469 | 390.388 | 8 | 6 |
| Resveratrol | 2.9738 | 228.247 | 3 | 3 |
| Physcion | 2.19022 | 284.267 | 5 | 2 |
| Citreorosein | 1.0711 | 286.239 | 6 | 4 |
| Quercetin | 1.988 | 302.238 | 7 | 5 |
| Hyperoside | −0.5389 | 464.379 | 12 | 8 |
| Coumarin | 1.793 | 146.145 | 2 | 0 |
| S. No | Compound Name | Water Solubility (mol/L) | CaCO2 Permeability (cm/S) | Intestinal Absorption (Human) % | Skin Permeability Log/Kp | P-Glycoprotein Substrate | P-Glycoprotein Inhibitor | P-Glycoprotein II Inhibitor |
|---|---|---|---|---|---|---|---|---|
| 1 | Penicillin | −2.199 | 0.293 | 58.344 | −2.735 | Yes | No | No |
| 2 | Resveratrol | −3.233 | 1.196 | 87.933 | −2.748 | No | No | No |
| S. No | Compound Name | VDss (Human) (L/kg) | Fraction Unbound (Human) (Fu) | BBB Permeability (Human) (Log BB) | CNS Permeability (Log PS) |
|---|---|---|---|---|---|
| 1 | Penicillin | −1.681 | 0.32 | −0.741 | −2.936 |
| 2 | Resveratrol | 0.022 | 0.089 | −0.152 | −2.113 |
| Compound Name | CYP-2D6 Substrate | CYP-3A4 Substrate | CYP-2D6 Inhibitor | CYP-2619 Inhibitor | CYP-269 Inhibitor |
|---|---|---|---|---|---|
| Penicillin | No | Yes | No | No | No |
| Resveratrol | No | Yes | Yes | No | No |
| S. No | Compound Name | Total Clearance (mL/Kg) | Renal OCT2 Substrate |
|---|---|---|---|
| 1 | Penicillin | 0.02 | No |
| 2 | Resveratrol | 0.094 | No |
| S. No | Toxicity Parameters | Penicillin | Resveratrol |
|---|---|---|---|
| 1 | Max tolerated dose (human) (mg/kg) | 1.284 | 0.561 |
| 2 | hERGI inhibitor | No | No |
| 3 | hERGII inhibitor | No | No |
| 4 | Oral rat acute toxicity (mol/kg) | 2.04 | 2.216 |
| 5 | Oral rat chronic toxicity (mg/kg) | 2.63 | 1.761 |
| 6 | Hepatoxicity (log μg/L) | Yes | No |
| 7 | Skin sensitization | No | No |
| 8 | T. pyriformis activity (log μg/L) | 0.285 | 0.982 |
| 9 | Minnow toxicity (log mM) | 2.255 | 1.367 |
| S. No | Compound Name | Logp Value | Molecular Weight | H-Bond Acceptor | H-Bond Donor |
|---|---|---|---|---|---|
| 1 | Penicillin | 0.8608 | 334.397 g/mol | 4 | 2 |
| 2 | Resveratrol | 2.9738 | 228.247 g/mol | 3 | 3 |
| S. No | Compound Name | Binding Score | Cavity Size | Grid Map | Minimum Energy (Kcal/mol) | Maximum Energy (Kcal/mol) |
|---|---|---|---|---|---|---|
| 1 | Penicillin | −6.7 | 86 | 23 | 0.00 | 1.6 × 100 |
| 2 | Resveratrol | −8.9 | 1857 | 34 | 0.00 | 1.6 × 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, M.; Amin, A.; Alharbi, M.; Ishtiaq, S.; Sajjad, W.; Ahmad, F.; Ahmad, S.; Hanif, F.; Faheem, M.; Khalil, A.A.K. Quorum Quenchers from Reynoutria japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2023, 28, 2635. https://doi.org/10.3390/molecules28062635
Fatima M, Amin A, Alharbi M, Ishtiaq S, Sajjad W, Ahmad F, Ahmad S, Hanif F, Faheem M, Khalil AAK. Quorum Quenchers from Reynoutria japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules. 2023; 28(6):2635. https://doi.org/10.3390/molecules28062635
Chicago/Turabian StyleFatima, Maliha, Arshia Amin, Metab Alharbi, Sundas Ishtiaq, Wasim Sajjad, Faisal Ahmad, Sajjad Ahmad, Faisal Hanif, Muhammad Faheem, and Atif Ali Khan Khalil. 2023. "Quorum Quenchers from Reynoutria japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA)" Molecules 28, no. 6: 2635. https://doi.org/10.3390/molecules28062635
APA StyleFatima, M., Amin, A., Alharbi, M., Ishtiaq, S., Sajjad, W., Ahmad, F., Ahmad, S., Hanif, F., Faheem, M., & Khalil, A. A. K. (2023). Quorum Quenchers from Reynoutria japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules, 28(6), 2635. https://doi.org/10.3390/molecules28062635






