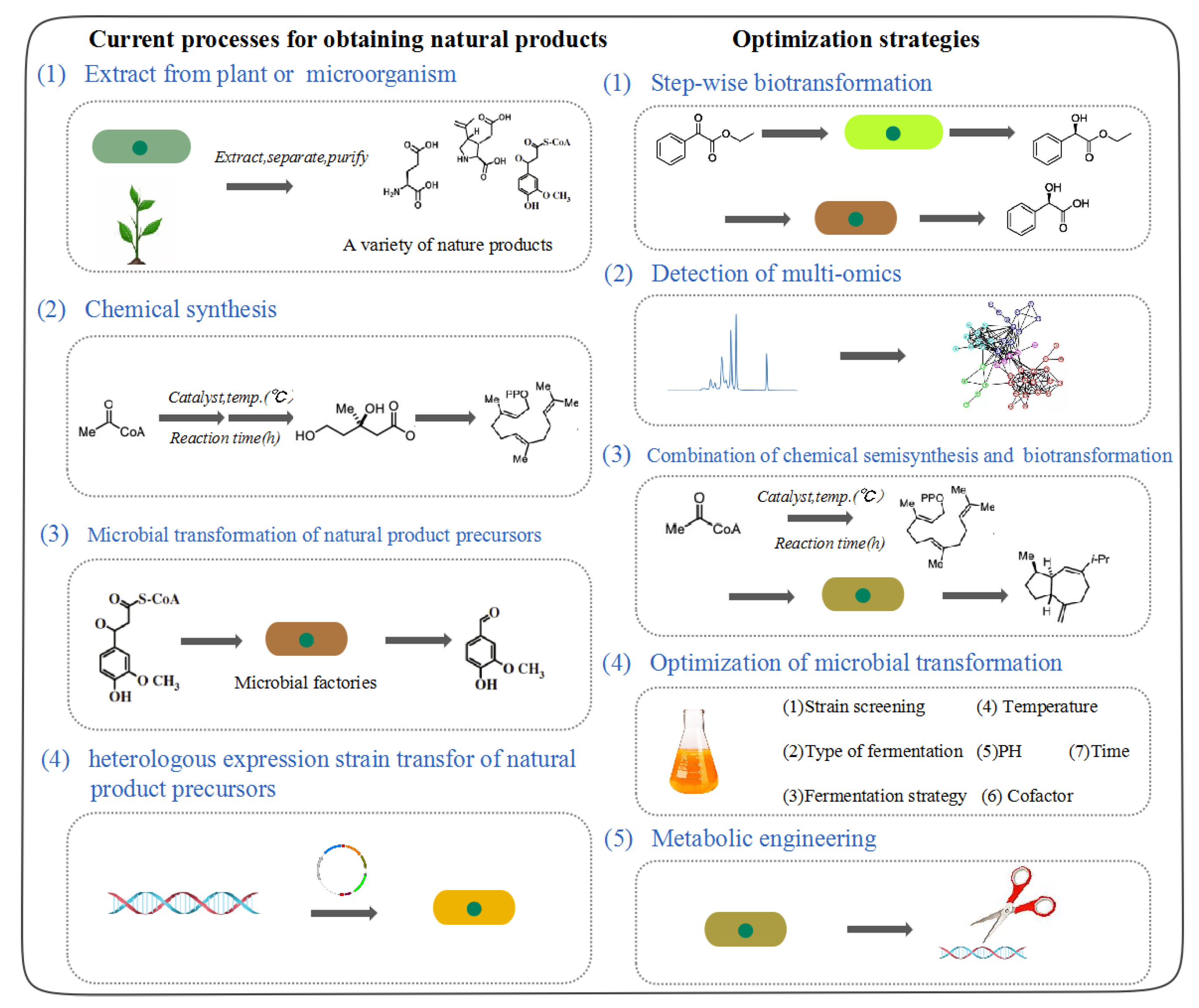
Multi-Level Optimization and Strategies in Microbial Biotransformation of Nature Products
Abstract
1. Introduction
2. Select Microbial Strains for Biotransformation
2.1. Isolation of Transformed Microbial Strains
2.2. Selection of Microbial Strains by Selective Medium
2.3. High-Throughput Screening of Transformed Microbial Strains
2.4. Selection of Microbial Strains by Bioinformation and Genomics
3. Optimization of Microbial Transformation Conditions
3.1. Solid Fermentation and Liquid Fermentation
3.2. Optimization of Fermentation Conditions
3.3. Fermentational Strategy
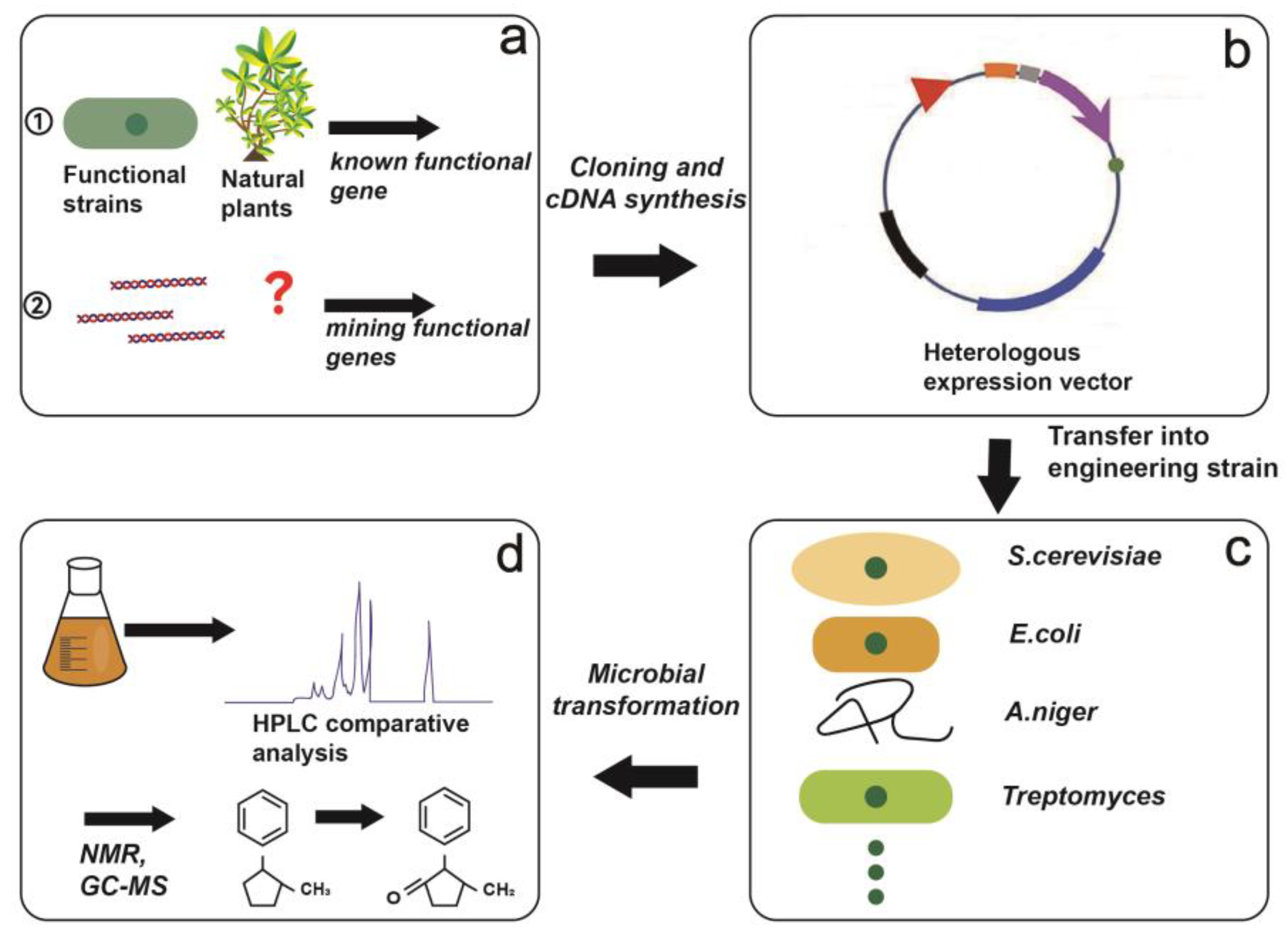
4. Microbial Transformation Based on Synthetic Biology
4.1. Heterologous Biosynthesis Based on Gene Engineering and Enzyme Engineering
4.2. Reduce Cytotoxicity through Metabolic Engineering
4.3. Increase Metabolic Flux of Products through Metabolic Engineering
4.4. Multi-Strain Collaborative Biotransformation Strategy
5. Combination of Chemical Semisynthesis and Microbial Transformation
6. Detection of Transformed Metabolites by Multi-Omics
7. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murthy, H.N.; Georgiev, M.I.; Park, S.Y.; Dandin, V.S.; Paek, K.Y. The safety assessment of food ingredients derived from plant cell, tissue and organ cultures: A review. Food Chem. 2015, 176, 426–432. [Google Scholar] [CrossRef]
- Hegazy, M.-E.F.; Mohamed, T.A.; ElShamy, A.I.; Abou-El-Hamd, H.M.; Mahalel, U.A.; Reda, E.H.; Shaheen, A.M.; Tawfik, W.A.; Shahat, A.A.; Shams, K.A.; et al. Microbial biotransformation as a tool for drug development based on natural products from mevalonic acid pathway: A review. J. Adv. Res. 2015, 6, 17–33. [Google Scholar] [CrossRef]
- Lilies, G. Gambling on marine biotechnology. Bioscience 1996, 46, 250–253. [Google Scholar] [CrossRef]
- Collins, A.M.; Kennedy, M.J. Biotransformations and bioconversions in New Zealand: Past endeavours and future potential. Austral Biotechnol. 1999, 9, 86–94. [Google Scholar]
- James, E.L.; Meyer, H.P. Chemocatalysis and biocatalysis (biotransformation): Some thoughts of a chemist and a biotechnologist. Org. Proc. Res. Dev. 2006, 10, 572–580. [Google Scholar] [CrossRef]
- Kanako, S.; Yusuke, T.; Kazufumi, Y. Prenylation of flavonoids by biotransformation of yeast expressing plant membrane-bound prenyltransferase SfN8DT-1. Biosci. Biotechnol. Biochem. 2009, 73, 759–761. [Google Scholar] [CrossRef]
- Cardillo, A.B.; Perassolo, M.; Sartuqui, M.; Rodríguez Talou, J.; Giulietti, A.M. Production of tropane alkaloids by biotransformation using recombinant Escherichia coli whole cells. Biochem. Eng. J. 2017, 125, 180–189. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, X.; Wu, J.; Mu, S.; Wu, Z.; Jin, J.M.; Tang, S.Y. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab Eng. 2020, 57, 239–246. [Google Scholar] [CrossRef]
- Pandey, A.; Soccol, C.R.; Nigam, P.; Soccol, V.T. Biotechnological potential of agro-industrial residues. I: Sugarcane bagasse. Bioresour. Technol. 2000, 74, 69–80. [Google Scholar] [CrossRef]
- Dhiman, S.; Mukherjee, G.; Singh, A.K. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: A review. Int. Microbiol. 2018, 21, 175–195. [Google Scholar] [CrossRef]
- Esmaeili, A.; Rohany, S.; Safaiyan, S.; Amir, Z.S. Microbial transformation of citral by Aspergillus niger-PTCC 5011 and study of the pathways involved. Czech J. Food Sci. 2011, 6, 610–615. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.; Qi, S.; Wang, H.; Xiu, Z.L. Biotransformation of steriodal saponins in Dioscorea zingiberensis C. H. Wright to diosgenin by Trichoderma harzianum. Appl. Microbiol. Biotechnol. 2010, 85, 933–940. [Google Scholar] [CrossRef]
- Zhou, W.; Yan, Q.; Li, J.Y.; Li, X.C.; Zhang, X.C.; Zhou, P. Biotransformation of Panax notoginseng saponins into ginsenoside compound K production by Paecilomyces bainier sp. 229. J. Appl. Microbiol. 2008, 104, 699–706. [Google Scholar] [CrossRef]
- Wang, F.; Li, B.; Wang, W.; Zhang, C.G.; Wei, D.Z. Biotransformation of diosgenin to nuatigenin-type steroid by a newly isolated strain, Streptomyces virginiae IBL-14. Appl. Microbiol. Biotechnol. 2007, 77, 771–777. [Google Scholar] [CrossRef]
- Qin, D.; Wang, L.; Han, M.; Wang, J.Q.; Song, H.C.; Yan, X.; Duan, X.X.; Dong, J.Y. Effects of an endophytic fungus Umbelopsis dimorpha on the secondary metabolites of host-Plant Kadsura angustifolia. Front. Microbiol. 2018, 9, 2845. [Google Scholar] [CrossRef] [PubMed]
- Khojasteh, S.C.; Argikar, U.A.; Cho, S.; Crouch, R.; Heck, C.J.S.; Johnson, K.M.; Kalgutkar, A.S.; King, L.; Maw, H.; Seneviratne, H.K.; et al. Biotransformation novel advances-2021 year in review. Drug Metab. Rev. 2022, 54, 207–245. [Google Scholar] [CrossRef]
- Li, Y.; Cai, L.; Dong, J.W.; Xing, Y.; Duan, W.H.; Zhou, H.; Ding, Z.T. Innovative approach to accumulate rubrosterone by fermentation of Asparagus filicinus with Fusarium oxysporum. J. Agric. Food Chem. 2015, 63, 6596–6602. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, S.; Li, Z.; Zhang, J.G.; Fan, K.Q.; Tan, G.Y.; Ai, G.M.; Lam, S.M.; Shui, G.H.; Yang, Z.H.; et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat. Biotechnol. 2020, 38, 76–83. [Google Scholar] [CrossRef]
- Siemon, T.; Wang, Z.; Bian, G.; Bian, G.K.; Seitz, T.; Ye, Z.L.; Lu, Y.; Cheng, S.; Ding, Y.K.; Huang, Y.L.; et al. Semisynthesis of plant-derived englerin a enabled by microbe engineering of guaia-6,10 (14)-diene as building block. J. Am. Chem. Soc. 2020, 142, 2760–2765. [Google Scholar] [CrossRef]
- Becker, J.; Wittmann, C. From systems biology to metabolically engineered cells-an omics perspective on the development of industrial microbes. Curr. Opin. Microbiol. 2018, 45, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Shen, W.; Wang, J.; Han, M.J.; Chai, F.N.; Duan, X.X.; Yan, X.; Guo, J.L.; Gao, T.C.; Zuo, S.H.; et al. Enhanced production of unusual triterpenoids from Kadsura angustifolia fermented by a symbiont endophytic fungus, Penicillium sp. SWUKD4.1850. Phytochemistry 2019, 158, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Shen, W.; Gao, T.C.; Zuo, S.H.; Song, H.C.; Xu, J.R.; Yu, B.H.; Peng, Y.J.; Guo, J.L.; Tang, W.W.; et al. Kadanguslactones A-E, further oxygenated terpenoids from Kadsura angustifolia fermented by a symbiotic endophytic fungus, Penicillium ochrochloron SWUKD4.1850. Phytochemistry 2020, 174, 1122335. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.Y.; Chen, Y.G.; Song, H.C.; He, Y.P.; Li, L.; Zhong, Y.P.; Li, L.; Zhong, Y.P.; Zhu, Y.H.; Cao, J.; et al. Hydroxylation of nigranoic acid to 6β-hydroxynigranoic acid by Caryospora carllicarpa YMF1.01026. Chin. Chem. Lett. 2007, 18, 165–167. [Google Scholar] [CrossRef]
- Dong, J.Y.; Chen, Y.G.; Song, H.C.; Zhu, Y.H.; Zhou, Y.P.; Li, L.; He, H.P.; Cao, J.; Zhang, K.Q. Hydroxylation of the triterpenoid nigranoic acid by the fungus Gliocladium roseum YMF1.00133. Chem. Biodivers. 2007, 4, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Pu, J.X.; Du, X.; Li, X.N.; Sun, H.D. Two new guaianolide-type sesquiterpenoids from Kadsura interior. Chin. Chem. Lett. 2013, 21, 11–113. [Google Scholar] [CrossRef]
- Huang, Q.; An, H.M.; Song, H.C.; Mao, H.Q.; Shen, W.Y.; Dong, J.Y. Diversity and biotransformative potential of endophytic fungi associated with the medicinal plant Kadsura angustifolia. Res. Microbiol. 2015, 166, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Song, H.C.; Yang, Y.H.; Yang, P.; Yang, D.Y.; Shen, K.Z.; Xu, Y.B.; Gao, Y.X.; Chen, Y.G.; Dong, J.Y. Microbiological transformation of the triterpene nigranoic acid by the freshwater fungus Dictyosporium heptasporum. J. Asian. Nat. Prod. Res. 2013, 15, 433–440. [Google Scholar] [CrossRef]
- Ying, Y.M.; Shan, W.G.; Zhan, Z.J. Biotransformation of huperzine a by a fungal endophyte of Huperzia serrata furnished sesquiterpenoid-alkaloid hybrids. J. Nat. Prod. 2014, 77, 2054–2059. [Google Scholar] [CrossRef]
- Ying, Y.M.; Xu, Y.L.; Yu, H.F.; Zhang, G.X.; Mao, W.; Tong, C.P.; Zhang, Z.D.; Tang, O.Y.; Shan, W.G.; Zhan, Z.J. Biotransformation of huperzine A by Irpex lacteus-a fungal endophyte of Huperzia serrata. Fitoterapia 2019, 138, 104341. [Google Scholar] [CrossRef]
- Zhan, Z.J.; Tian, T.; Xu, Y.L.; Yu, H.F.; Zhang, C.X.; Zhang, Z.D.; Tang, Q.Y.; Shan, W.G.; Ying, Y.M. Biotransformation of huperzine B by a fungal endophyte of Huperzia serrata. Chem. Biodivers. 2019, 16, e1900299. [Google Scholar] [CrossRef]
- Kumar, A.; Ahmad, A. Biotransformation of vinblastine to vincristine by the endophytic fungus Fusarium oxysporum isolated from Catharanthus roseus. Biocatal. Biotransfor. 2013, 31, 89–93. [Google Scholar] [CrossRef]
- Shibuya, H.; Kitamura, C.; Maehara, S.; Nagahata, M.; Winarno, H.; Simanjuntak, P.; Kim, H.-S.; Wataya, Y.; Ohashi, K. Transformation of cinchona alkaloids into 1-N-oxide derivatives by endophytic Xylaria sp. isolated from Cinchona pubescens. Chem. Pharm. Bull. 2003, 51, 71–74. [Google Scholar] [CrossRef]
- Luo, S.L.; Dang, L.Z.; Li, J.F.; Zou, C.G.; Zhang, K.Q.; Guo, H.L. Biotransformation of saponins by endophytes isolated from Panax notoginseng. Chem. Biodivers. 2013, 10, 2021–2031. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, X.; Zou, X. Biotransformation of caffeine in oolong tea by Paecilomyces gunnii. Int. Biodeter. Biodegr. 2016, 114, 141–144. [Google Scholar] [CrossRef]
- Shan, L.; Jiao, K.; Yin, M.; Huang, J.J.; Chen, Y.J.; Qin, S.S.; Liu, H.M. Biotransformation of 5-en-3β-ol steroids by Mucor circinelloides Lusitanicus. Biocatal. Biotransfor. 2016, 34, 83–88. [Google Scholar] [CrossRef]
- Wang, Y.C.; Li, X.; Sun, H.; Yi, K.X.; Zheng, J.L.; Zhang, J.; Hao, Z.B. Biotransformation of steroidal saponins in sisal (Agave sisalana Perrine) to tigogenin by a newly isolated strain from a karst area of Guilin, China. Biotechnol. Biotechnol. Equip. 2014, 28, 1024–1033. [Google Scholar] [CrossRef]
- Chiang, C.M.; Wang, T.Y.; Ke, A.N.; Chang, T.S.; Wu, J.Y. Biotransformation of ergostane triterpenoid antcin K from Antrodia cinnamomea by soil-isolated Psychrobacillus sp. AK 1817. Catalysts 2017, 7, 299. [Google Scholar] [CrossRef]
- Dong, Q.; Yuan, Y.; Zhou, Y.; Zhang, Y.X.; Zhang, J.P.; Yu, H.B.; Jiao, B.H.; Liu, X.Y.; Lu, X.L. Biotransformation of total coumarins of Radix Glehniae by Lecanicillium attenuatum W-1-9. J. Asian Nat. Prod. Res. 2018, 20, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Hadibarata, T.; Zubir, M.M.F.A.; Rubiyatno, T.Z.C.; Yusoff, A.R.M.; Salim, M.R.; Fulazzaky, M.A.; Seng, B.; Nugroho, A.E. Degradation and transformation of anthracene by white-rot fungus Armillaria sp. F022. Folia Microbiol. 2013, 58, 85–391. [Google Scholar] [CrossRef]
- Kozlowska, E.; Hoc, N.; Sycz, J.; Urbaniak, M.; Dymarska, M.; Grzeszcuzuk, J.; Kostrzewa-Suslow, E.; Stepien, L.; Plaskowska, E.; Janeczko, T. Biotransformation of steroids by entomopathogenic strains of Isaria farinosa. Microb. Cell Fact. 2018, 17, 71. [Google Scholar] [CrossRef]
- Morahem, A. Salinivibrio costicola GL6, a novel isolated strain for biotransformation of caffeine to theobromine under hypersaline conditions. Curr. Microbiol. 2017, 74, 34–41. [Google Scholar] [CrossRef]
- Ye, H.; Yuan, S.; Cong, X. Biotransformation of puerarin into 3’-hydroxypuerarin by Trichoderma harzianum NJ01. Enzyme Microb. Tech. 2007, 40, 594–597. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, O.P.; Dawra, R.K.; Kanwar, S.S.; Mahato, S.B. Biotransformation of lantadene A (22beta-angeloyloxy-3-oxoolean-12-en-28-oic acid), the pentacyclic triterpenoid, by Alcaligenes faecalis. Biodegradation 1999, 10, 373–381. [Google Scholar] [CrossRef]
- Dogra, N.; Qazi, G.N. Steroid biotransformation by different strains of Micrococcus sp. Folia Microbiol. 2001, 46, 17–20. [Google Scholar] [CrossRef]
- Yu, L.; Gao, F.; Yang, L.; Yang, L.P.; Xu, L.; Wang, Z.H.; Ye, H. Biotransformation of puerarin into puerarin-6″-O-phosphate by Bacillus cereus. J. Ind. Microbiol. Biotechnol. 2012, 39, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Dong, J.W.; Zhao, L.X.; Zhou, H.; Xing, Y.; Li, Y.; Li, Z.J.; Duan, W.H.; Li, X.J.; Ding, Z.T. An improved water-soluble/stereospecific biotransformation of aporphine alkaloids in Stephania epigaea to 4R-hydroxyaporphine alkaloids by Clonostachys rogersoniana. Process Biochem. 2016, 51, 933–940. [Google Scholar] [CrossRef]
- Wang, C.; Huo, X.K.; Zhang, B.J.; Sun, C.P.; Tian, X.G.; Deng, S.; Li, B.; Wang, W.; Dong, P.P.; Ma, X.C. Highly regioselective glucosylation of alcoholic hydroxyls of protostane triterpenoids mediated by fungal biotransformation. Catal. Communicat. 2017, 89, 40–43. [Google Scholar] [CrossRef]
- Gonda, S.; Kiss-Szikszai, A.; Szucs, Z.; Balla, B.; Vasas, G. Efficient biotransformation of non-steroid anti-inflammatory drugs by endophytic and epiphytic fungi from dried leaves of a medicinal plant, Plantago lanceolata L. Int. Biodeteriorat. Biodegradat. 2016, 108, 115–121. [Google Scholar] [CrossRef]
- Sripalakit, P.; Wichai, U.; Saraphanchotiwitthaya, A. Biotransformation of various natural sterols to androstenones by Mycobacterium sp. and some steroid-converting microbial strains. J. Mol. Catal. B Enzymatic. 2006, 41, 49–54. [Google Scholar] [CrossRef]
- Marotti, I.; Bonetti, A.; Biavati, B.; Catizone, P.; Dinelli, G. Biotransformation of common bean (Phaseolus vulgaris l.) flavonoid glycosides by Bifidobacterium species from human intestinal origin. J. Agric. Food Chem. 2007, 55, 3913–3919. [Google Scholar] [CrossRef] [PubMed]
- Mayr, L.M.; Bojanic, D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 2009, 9, 580–588. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, G.; Shi, J.; Koffas, M.A.G.; Xu, Z. Microbial production of l-serine from renewable feed-stocks. Trends Biotechnol. 2018, 36, 700–712. [Google Scholar] [CrossRef]
- Lin, J.L.; Wagner, J.M.; Alper, H.S. Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol. Advan. 2017, 35, 950–970. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, S.; Dai, Y.J.; Yang, Y.; Yuan, S.; Dai, Y.J. Microplate for high throughput screening of 6-hydroxynicotinic acid transforming strains. Weishengwu Xuebao. 2008, 48, 112–115. [Google Scholar] [CrossRef]
- Gao, H.; Liu, M.; Zhou, X.L.; Liu, J.T.; Zhou, Y.; Guo, Z.X.; Xu, B.; Zhang, W.Q.; Liu, X.Y.; Luo, A.Q.; et al. Identification of avermectin-high-producing strains by high-throughput screening methods. Appl. Microbiol. Biotechnol. 2010, 85, 1219–1225. [Google Scholar] [CrossRef]
- Yu, H.; Wang, N.; Huo, W.B.; Zhang, Y.H.; Zhang, W.; Yang, Y.; Chen, Z.Y.; Huo, Y.X. Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microbio Cell Factor. 2019, 18, 30. [Google Scholar] [CrossRef]
- Yan, D.; Kim, W.J.; Yoo, S.M.; Choi, J.H.; Ha, S.H.; Lee, M.H.; Lee, S.Y. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 9835–9844. [Google Scholar] [CrossRef]
- Qiu, X.; Xu, P.; Zhao, X.; Du, G.C.; Zhang, J.; Li, J.H. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metabolic. Eng. 2020, 60, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Shen, Y.M.; Liu, Y.; Zhang, K.Q. Production of saponin in fermentation process of Sanchi (Panax notoginseng) and biotransformation of saponin by Bacillus subtilis. Ann. Microbiol. 2006, 56, 151–153. [Google Scholar] [CrossRef]
- Rao, D.G. Introduction to Biochemical Engineering, 2nd ed.; Tata McGraw Hill: Gautam Buddha Nagar, India, 2010. [Google Scholar]
- Furusawa, M.; Hashimoto, T.; Noma, Y.; Asakawa, Y.S. Isolation and structures of new cyclomyltaylane and ent-chamigrane-type sesquiterpenoids from the liverwort reboulia hemishaerica and their biotransformation by the fungus Aspergillus niger. Chem. Pharm. Bull. 2006, 54, 996–1003. [Google Scholar] [CrossRef]
- do Nascimento, J.S.; Nunez, W.E.R.; Dos Santos, V.H.P.; Aleu, J.; Cunha, S.; Oliveria Silva, D. Mapping the biotransformation of coumarins through filamentous fungi. Molecules 2019, 24, 3531. [Google Scholar] [CrossRef]
- Qin, Y.J.; Feng, B.; Song, X.B.; Zhou, W.B.; Yu, H.S.; Zhao, L.L.; Yu, L.Y.; Ma, B.P. Biotransformation of glycyrrhetinic acid by Cunninghamella blakesleeana. Chin. J. Nat. Med. 2010, 8, 373–381. [Google Scholar] [CrossRef]
- Wang, B.Y.; Yang, X.Q.; Hu, M.; Shi, L.J. Biotransformation of natural polyacetylene in red ginseng by Chaetomium globosum. J. Ginseng. Res. 2020, 44, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Li, K.; Hou, Y.; Liu, Y.N.; Ji, S.X.; Qin, H.M.; Lu, F.P. Synergistic effects of components in deep eutectic solvents relieve toxicity and improve the performance of steroid biotransformation catalyzed by Arthrobacter simplex. J. Chem. Technol. Biotechnol. 2018, 93, 2729–2736. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Shen, Y.; Su, L.; Wang, M. Effect of beta-cyclodextrins derivatives on steroids biotransformation by Arthrobacter simplex. Appl. Biochem. Biotechnol. 2018, 185, 1004–1013. [Google Scholar] [CrossRef]
- Awadhiya, P.; Banerjee, T. Tween 80 alters the production ratio of pharmaceutically important steroid intermediates, 4-ad and add during biotransformation of soysterol by Mycobacterium sp. NRRL B-3805. IJPSR 2018, 9, 1935–1941. [Google Scholar] [CrossRef]
- Wang, L.; Shen, Y.; Liu, M.; Tang, R.; Wang, M. Influence of imidazolium-based ionic liquids on steroid biotransformation by Arthrobacter Simplex. J. Chem. Technol. Biotechnol. 2018, 93, 426–431. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, L.; Shen, Y.; Ma, Y.; Zheng, Y.; Luo, J. Effects of hydroxypropyl-beta-cyclodextrin on steroids 1-en -dehydrogenation biotransformation by Arthrobacter simplex TCCC 11037. J. Molecular. Catal. B Enzymatic. 2009, 59, 58–63. [Google Scholar] [CrossRef]
- Mao, S.; Yu, L.; Ji, S.; Liu, X.; Lu, F. Evaluation of deep eutectic solvents as co-solvent for steroids 1-en-dehydrogenation biotransformation by Arthrobacter simplex. J. Chem. Technol. Biotechnol. 2016, 91, 1099–1104. [Google Scholar] [CrossRef]
- Wu, C.; Zhu, H.; van Wezel, G.P.; Chaoi, Y.H. Metabolomics-guided analysis of isocoumarin production by Streptomyces species MBT76 and biotransformation of flavonoids and phenylpropanoids. Metabolomics 2016, 12, 90. [Google Scholar] [CrossRef]
- Haeser, K.; Wenk, H.H.; Schwab, W. Biocatalytic production of dihydrocoumarin from coumarin by Saccharomyces cerevisiae. J. Agric. Food Chem. 2006, 54, 6236–6240. [Google Scholar] [CrossRef]
- Mohammad, Y.M.; Shakya, A.; Al-Bakain, R. New monoterpenoid by biotransformation of thymoquinone using Aspergillus niger. Bioorganic. Chemi. 2018, 80, 212–215. [Google Scholar] [CrossRef]
- Yang, T.W.; Rao, Z.M.; Zhang, X.; Xu, M.J.; Xu, Z.H.; Yang, S.T. Fermentation of biodiesel-derived glycerol by Bacillus amyloliquefaciens: Effects of co-substrates on 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2013, 97, 7651–7658. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Zhang, X.; Lin, Q.; Xia, H.; Xu, Z.; Yang, S. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J. Basic. Microbiol. 2011, 51, 650–658. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Kimani, B.G.; Xu, M.J.; Zhang, X.; Yang, S.T. Two-step production of gamma-aminobutyric acid from cassava powder using Corynebacterium glutamicum and Lactobacillus plantarum. J. Ind. Microbiol. Biotechnol. 2015, 42, 1157–1165. [Google Scholar] [CrossRef]
- Tang, R.; Shen, Y.; Xia, M.; Tu, L.; Luo, J.; Geng, Y.; Gao, T.; Zhou, H.; Zhao, Y.; Wang, M. A highly efficient step-wise biotransformation strategy for direct conversion of phytosterol to boldenone. Bioresour. Technol. 2019, 283, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Ma, T.; Ye, Z.; Li, X.; Huang, Y.; Zhou, Z.; Ding, Y.; Deng, Z.; Liu, T. Systematic metabolic engineering of Saccharomyces cerevisiae for lycopene overproduction. J. Agr. Food Chem. 2019, 67, 11148–11157. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, A.; Redondo, F.J.; Contin, A.; Memelink, J.; Heijden, R.; Verpoore, R. Biotransformation of tryptamine and secologanin into plant terpenoid indole alkaloids by transgenic yeast. Appl. Microbiol. Biotechnol. 2001, 56, 420–424. [Google Scholar] [CrossRef]
- Shen, Z.Q.; Eisenreich, W.; Kutchan, T.M. Bacterial biotransformation of 3 alpha-(S)-Strictosidine to the monoterpenoid indole alkaloid vallesiachotamine. Phytochemistry 1998, 48, 293–296. [Google Scholar] [CrossRef]
- Seeger, M.; Gonzalez, M.; Camara, B.; Munoz, L.; Ponce, E.; Mejisa, L.; Mascayano, C.; Vasquez, Y. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 2003, 69, 5045–5050. [Google Scholar] [CrossRef]
- Zhou, P.; Ye, L.; Xie, W.; Lv, X.; Yu, H. Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Appl. Microbiol. Biotechnol. 2015, 99, 8419–8428. [Google Scholar] [CrossRef] [PubMed]
- Kragl, U.; Gygax, D.; Ghisalba, O.; Wandrey, C. Enzymatic two step synthesis of N-acetyl-neuraminic acid in the enzyme membrane reactor. Angew. Chem. Int. Ed. Engl. 1991, 30, 827–828. [Google Scholar] [CrossRef]
- Kittelmann, M.; Klein, T.; Kragl, U.; Wandrey, C.; Ghisalba, O. CMP-N-acetyl neuraminic-acid synthetase from Escherichia coli: Fermentative production and application for the preparative synthesis of CMP-neuraminic acid. Appl. Microbiol. Biotechnol. 1995, 44, 59–67. [Google Scholar] [CrossRef]
- Watts, K.T.; Lee, P.C.; Schmidt-Dannert, C. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol. 2006, 6, 22. [Google Scholar] [CrossRef]
- Park, S.R.L.; Yoon, J.A.; Paik, J.H.; Paik, J.H.; Jung, W.S.; Ban, Y.H.; Kim, E.J.; Yoo, Y.J.; Yoon, Y.J. Engineering of plant-specific phenylpropanoids biosynthesis in Streptomyces venezuelae. J. Biotechnol. 2009, 144, 181–188. [Google Scholar] [CrossRef]
- Amiri, P.; Shahpiri, A.; Asadollahi, M.A.; Momenbeil, F.; Partow, S. Metabolic engineering of Saccharomyces cerevisiae for linalool production. Biotechnol Lett. 2016, 38, 503–508. [Google Scholar] [CrossRef]
- Nakagawa, A.; Minami, H.; Kim, J.S.; Koyanagi, T.; Katayama, T.; Sato, F.; Kumagai, H. A bacterial platform for fermentative production of plant alkaloids. Nat Commun. 2011, 2, 326. [Google Scholar] [CrossRef] [PubMed]
- Lemuth, K.; Steuer, K.; Albermann, C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microbial. Cell Factor. 2011, 10, 29. [Google Scholar] [CrossRef]
- Lim, C.G.; Fowler, Z.L.; Hueller, T.; Schaffer, S.; Koffsa, M.A.G. High-yield resveratrol production in engineered Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 3451–3460. [Google Scholar] [CrossRef]
- Malla, S.; Koffas, M.A.G.; Kazlauskas, R.J.; Kim, B.G. Production of 7-O-methyl aromadendrin, a medicinally valuable flavonoid, in Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Gao, W.; Rong, Q.; Jin, G.; Chu, H.; Liu, W.; Yang, W.; Zhu, Z.; Li, G.; Zhu, G.; et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Amer. Chem. Soc. 2012, 134, 3234–3241. [Google Scholar] [CrossRef]
- Alonso-Gutierrez, J.; Chan, R.; Batth, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; Mcphee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, Y.J.; Hillwig, M.L.; Shen, Y.; Yang, L.; Wang, Y.; Zhang, X.; Liu, W.; Peters, R.J.; Chen, X.; et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12108–12113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metabolic. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef]
- Zhao, S.; Jones, J.A.; Lachance, D.M.; Bhan, N.; Khalidi, O.; Venkataraman, S.; Wang, Z.; Koffas, M.A.G. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab. Eng. 2015, 28, 43–53. [Google Scholar] [CrossRef]
- Kang, S.Y.; Lee, J.K.; Choi, O.; Kim, C.Y.; Jang, J.H.; Huang, B.Y.; Hong, Y.S. Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol. 2014, 14, 67. [Google Scholar] [CrossRef]
- Thodey, K.; Galanie, S.; Smolke, C.D. A microbial bio-manufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biolo. 2014, 10, 837. [Google Scholar] [CrossRef] [PubMed]
- Galanie, S.; Smolke, C. Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb. Cell Fact. 2015, 14, 144. [Google Scholar] [CrossRef]
- Trenchard, I.J.; Smolke, C.D. Engineering strategies for the fermentative production of plant alkaloids in yeast. Metabolic. Eng. 2015, 30, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wei, Y.; Fan, Y.; Liu, Q.; Wei, W.; Yang, C.; Zhang, L.; Zhao, G.; Yue, J.; Yan, X.; et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metabolic. Eng. 2015, 29, 97–105. [Google Scholar] [CrossRef]
- Nakagawa, A.; Matsumura, E.; Koyanagi, T.; Katayama, T.; Kawano, N.; Yoshimatsu, K.; Yamamoto, K.; Kumagai, H.; Sato, F.; Mainami, H. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 2016, 7, 10390. [Google Scholar] [CrossRef]
- Li, Y.; Smolke, C.D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 2016, 7, 12137. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, Q.; Zhou, Z.; Zhao, F.; Lu, W. Heterologous biosynthesis of triterpenoid dammarenediol-II in engineered Escherichia coli. Biotechnol. Lett. 2016, 38, 603–609. [Google Scholar] [CrossRef]
- Matsumura, E.; Nakagawa, A.; Tomabechi, Y.; Ikushiro, S.; Sakaki, T.; Katayama, T.; Yamamoto, K.; Kumaga, H.; Sato, F.; Minami, H. Microbial production of novel sulphated alkaloids for drug discovery. Sci. Rep. 2018, 8, 7980. [Google Scholar] [CrossRef]
- Farhi, M.; Marhevka, E.; Masci, T.; Marocos, E.; Eyal, Y.; Ovadia, M.; Abeliovichi, H.; Vainstein, A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011, 13, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Sun, Q.; Zhao, L.; Shi, H.; Tang, F.; Cao, F. Efficient biotransformation of luteolin to isoorientin through adjusting induction strategy, controlling acetic acid, and increasing UDP-Glucose supply in Escherichia coli. J. Agric. Food Chem. 2019, 67, 331–340. [Google Scholar] [CrossRef]
- Liu, M.; Fu, Y.; Gao, W.; Xian, M.; Zhao, G. Highly efficient biosynthesis of hypoxanthine in Escherichia coli and transcriptome-based analysis of the purine metabolism. ACS Synth. Biol. 2020, 9, 525–535. [Google Scholar] [CrossRef]
- Rico, J.; Pardo, E.; Orejas, M. Enhanced production of a plant monoterpene by over-expression of the 3-hydroxy-3-methylglutaryl coenzyme A reductase catalytic domain in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 6449–6454. [Google Scholar] [CrossRef] [PubMed]
- Scheler, U.; Brandt, W.; Porzel, A.; Rothe, K.; Manzano, D.; Bozic, D.; Papaefthimiou, D.; Balcke, G.U.; Henning, A.; Lohse, S.; et al. Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat. Commun. 2016, 7, 12942. [Google Scholar] [CrossRef] [PubMed]
- Biggs, B.W.; Lim, C.G.; Sagliani, K.; Shankar, S.; Stephanopoulos, G.; Mey, M.D.; Ajikmar, P.K. Overcoming heterologous protein interdependency to optimize P450-mediated taxol precursor synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, 3209–3214. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, T.; Lu, J.; Durre, P.; Zhang, W.; Dong, W.; Zhou, J.; Jiang, M.; Xin, F. Microbial co-culturing systems: Butanol production from organic wastes through consolidated bioprocessing. Appl. Microbiol. Biotechnol. 2018, 102, 5419–5425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wu, B. Synthetic microbiome: When “synthetic biology” meets “microbiomics”. Chin. Sci. Bull. 2019, 64, 1781–1798. [Google Scholar] [CrossRef]
- Ou, Z.; Lan, M.; Niu, Y.; Cui, J. Preparation of (R)-(-)-mandelic acid by two-step biotransformation of ethyl benzoylformate. Biocatal. Biotransfor. 2018, 36, 409–416. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Zhang, H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metabolic. Eng. 2019, 54, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ambreen; Haque, S.; Singh, V.; Katiyar, D.; Khan, M.T.A.; Tripathi, V.; Enshasy, H.E.; Pasupuleti, M.; Mishra, B.N. Biotransformation of newly synthesized coumarin derivatives by Candida albicans as potential antibacterial, antioxidant and cytotoxic agents. Process Biochem. 2019, 87, 138–144. [Google Scholar] [CrossRef]
- Garcia-Granados, A.; Gutierrez, M.C.; Rivas, F. Improved microbiological hydroxylation of sesquiterpenoids: Semisynthesis, structural determination and biotransformation studies of cyclic sulfite eudesmane derivatives. Org. Biomol. Chem. 2003, 1, 2314–2320. [Google Scholar] [CrossRef] [PubMed]
- Chekan, J.R.; McKinnie, S.M.K.; Moore, M.L.; Poplawski, S.G.; Michael, T.P.; Moore, B.S. Scalable biosynthesis of the seaweed neurochemical, kainic acid. Angew. Chem. Int. Ed. 2019, 58, 8454–8457. [Google Scholar] [CrossRef]
- Martins, I.; Varela, A.; Frija, L.M.T.; Estevao, M.A.S.; Planchon, S.; Renaut, J.; Afonoso, C.A.M.; Pereira, C.S. Proteomic insights on the metabolism of Penicillium janczewskii during the biotransformation of the plant terpenoid labdanolic acid. Front. Bioeng. Biotechnol. 2017, 5, 45. [Google Scholar] [CrossRef]
- Zhao, M.; Su, X.Q.; Nian, B.; Chen, L.J.; Zhan, D.L.; Duan, S.M.; Wang, L.Y.; Shi, X.Y.; Jiang, B.; Jiang, W.W.; et al. Integrated meta-omits approaches to understand the microbiome of spontaneous fermentation of traditional Chinese puerh tea. mSystems 2019, 4, e00680-19. [Google Scholar] [CrossRef]
- Cao, L.; Shcherbin, E.; Mohimani, H. A metabolome- and metagenome-wide association network reveals microbial natural products and microbial biotransformation products from the human microbiota. mSystems. 2019, 4, e00387-19. [Google Scholar] [CrossRef] [PubMed]

| Substrate | Solvent | Microbial Catalyst | Cell Age | Conditions | Obtained Compounds | Conversion Yields | Ref. |
|---|---|---|---|---|---|---|---|
| Citral | Methanol | Aspergillus niger-PTCC 5011 | 2 day | 27 °C; pH 5.5; 6 days, 15 days; 150 r·min−1 | 6 day:hydroxy citronellal 15 day:citronellol | 37.0%; 36.0% | [11] |
| Genistein | DMSO | Streptomyces sp. MBT76 | 2 day | 30 °C; pH 6.8; 3 days; 220 r·min−1 | Methoxylated isoflavones | - | [71] |
| Coumarin (tonka bean meal) | EtOH | Saccharomyces cerevisiae | 1 day | 30 °C; pH 4.5–6.5; 150 h; 180 r·min−1 | Melilotic acid | 3.6% | [72] |
| Hyoscyamine | - | Escherichiacol (Hyoscyamine-6β-hydroxylase) | - | 30 °C; pH 7.8; 25 h; 250 r·min−1; FeSO4, Ascorbate,2-oxoglutarate | 6-hydroxyhyoscyamine; Scopolamine | 5.22%; 0.67% | [7] |
| Steriodal saponin | Water | Trichoderma harzianum | 1 day | 30 °C; pH 6; 4 days; 150 r·min−1; Na2HPO4, KH2PO4, 0.93 mmol/L Fe2+; 0.07% (w/v) Tween-85 | Diosgenin | 30.05% | [12] |
| GinsenosidesRb1, Rb2, Rb3 | Water | Paecilomyces bainier sp. 229 | 3 day | 28 °C; pH 4.5–5.5; 3 days;150 r·min−1; Mg,Ca; 0.2% Tween80 | Ginsenoside compound K | 82.6% | [13] |
| Sitosterol; cholesterol; stigmasterol; ergosterol | Tween 80 | Mycobacterium sp. | 2 day | 25 °C; 20 days; 200 r·min−1 | 4-androstene-3,17-dion; 1,4-androstadiene-3,17-dione; testosterone | 81.83% (total) | [49] |
| Dihydroalisol A; alisol G, F, B | Acetone | Syncephalastrum racemosum AS 3.264 | 1 day | 4 days | 11-OH-Glucosylation(1a; 2a; 3a; 4a) | 25%; 58%; 55%; 53% | [28] |
| Labdanolic acid | Ethanol | Penicillium janczewskii | - | 60 days; 0 r·min−1 | 3β-hydroxy-labdanolic acid | >90% | [73] |
| Thymoquinone | Acetone | Aspergillus niger | 3 day | 28 °C; 7 days; 128 r·min−1 | 5-isopropyl-2-methyloxepin-1-on; 3-hydroxy-5- isopropyl-2-Methylcyclohexa-2,5-diene-1,4-dione; 5-isopropyl-2-methylbenzene-1,4-diol | 4.9%;7.3%; 28.0% | [73] |
| Diosgenin | Heated ethanol | S.virginiae IBL-14 | 15 h | 30 °C; pH 5.2; 2 days; 200 r·min−1 | Diosgenone; Isonuatigenone | 28.4% | [14] |
| Ginsenoside Rh1,Re | Water | Bacillus subtilis | 3 day | 30 °C; pH 7; 10 days; 120 r·min−1 | Gingsenoside Rh4 | - | [53] |
| Naringenin | - | Yeast (SfN8DT-1) | - | 30 °C; 20 h | 8-dimethylallylnaringenin (8DN) | 1.9% | [6] |
| (R)-dicentrine | MeOH | Clonostachys rogersonia | 3 day | 28 °C; 7 days; 2% (v/v); n-tetradecane | Corresponding (4R,6aR)-4-hydroxydicentrine | 99.5%; 97.4%; 99.9% | [43] |
| Nigranoic acid | - | Umbelopsis dimorpha, Penicillium sp. | 3–5 day | 28 °C; 25 days, 45 days; Solid-state | A series of high oxidation triterpenes | - | [15,21,22] |
| Caffeine | - | Paecilomyces gunnii | - | 25 °C; 35 days; Solid-state | Theophylline; 1,7-dimethylxanthine | - | [34] |
| Products | Precursor | Heterologously Expressed Genes and Sources | Engineering Strains | Ref. |
|---|---|---|---|---|
| Neu5Ac | N-acetylglucosamine (GlcNAc), pyruvic acid | GlcNAc epimerase and Neu5Ac al-dolase | E. coli | [83] |
| CMP-Neu5Ac | Neu5Ac | CMP-Neu5Ac synthetase (N.meningitidis) | E. coli | [84] |
| Vallesiachotamine, Isovallesiachotamine | Tryptamine, Secologanin | Strictosidine synthase (R.serpentina) | Aeromonas sp., Bacillus licheniformis, | [80] |
| Cathenamine; Strictosidine | Tryptamine, Secologanin | STR, SGD (C. roseus) | Saccharomyces cerevisiae | [79] |
| 7,2′3′-trihydroxy-8-methylisoflavone, 7,3′4′-trihydroxyysoflavone | 7-hydroxy-8-methylisoflavone, 7-hydroxyysoflavone | BphA, BphB | Burkholderia sp.LB400 | [81] |
| Resveratrol, Piceatannol, Isorhapontigenin | 4-coumaric acid, Caffeic acid, Ferulic acid | 4CL1 (A. thaliana),STS (A. hypogaea) | E. coli | [85] |
| Naringenin, Pinocembrin, Resveratrol | 4-coumaric acid, cinnamic acid | CCL (S. coelicolor), CHS (A. thaliana) | S. venezuelae | [86] |
| Amorphadiene | Valencene | SfN8DT-1 | Yeast | [6] |
| Linalool | Mevalonic acid | LIS (C. brewer), LIS (L. angustifolia) | Saccharomycesce. cerevisiae | [87] |
| (S)-reticuline | (S)-norcoclaurine | TYR (S.castaneoglobisporus), DODC (P.putida), 6OMT, CNMT, 4′OMT (C.japonica), MAO (M. luteus) | E. coli | [88] |
| Astaxanthin | Isopentenyl diphosphate | CrtW148 (N. punctiforme), crtE, crtB, crtI, crtY, crtZ (P.ananatis) | E. coli BW-ASTA | [89] |
| Resveratrol | p-coumaric acid, Malonyl-CoA | 4CL1 (A. thaliana), STS (V. vinifera) | E. coli | [90] |
| 7-O-Methylaromadendrin | p-coumaric acid | 4CL (P. crispum), CHS (P.hybrida), CHI (M. sativa) | E. coli | [91] |
| Miltiradiene | Glucose | CPS, KSL, BTS1, ERG20 (S. miltiorrhiza) | S. cerevisiae | [92] |
| Perillyl alcohol | Limonene | GPPS (Abies grandis), LS (Mentha spicata), P450 (co) (Mycobacterium HXN 1500) | E. coli | [93] |
| Artemisinic acid | Amorphadiene | CYP71AV1, CPR1, CYB5, ADH1, ALDH1 (A. annua) | S. cerevisiae | [94] |
| Ferruginol | Miltiradiene | CYP76AH1 (A.thaliana) | S. cerevisiae | [95] |
| β-carotene | Pyruvate, glyceraldehyde-3-phosphate | CrtEXYIB (P. agglomerans) | E. coli | [96] |
| Catechin, Afzelechin | Eriodictyol, Naringenin | F3H (C. sinensis), DFR (A.andraeanum), LAR (D. uncinatum) | E. coli | [97] |
| Pinostilbene, Resveratrol | Tyrosine | TAL (S. espanaensis), 4CL(S. coelicolor), STS (A. hypogaea), SbOTM1, OTM3 (S. bicolor) | E. coli | [98] |
| Morphine, 14-hydrocodine, | Thebaine | T6ODM, CODM, COR (P.somniferum) | S. cerevisiae | [99] |
| Astaxanthin | Carotene | CrtZ and BKT (H. pluvialis) | S. cerevisiae | [82] |
| Berberine | Norlaudanosoline | 6OMT, 4′OMT, BBE (P.somniferum), S9OMT (T. flavum), CAS (T.flavum), CPR (A. thaliana) | S. cerevisiae | [100] |
| Sanguinarine, Stylopine | Norlaudanosoline | ATR1 (A. thaliana), CFS, STS, P6H (E. californica), 6OMT, OMT, TNMT (P. somniferum) | S. cerevisiae | [101] |
| Ginsenosides Rh2,Rg3 | Protopanaxadiol | PPD, UGT (Panax ginseng) | S. cerevisiae | [102] |
| Thebaine | (R)-reticuline | ATR2 (Arabidopsis thaliana), CPR (Papaver somniferum), CPR (Rattus norvegicus) | E. coli | [103] |
| Nocapine, Noscapine | Norlaudanosoline | CYP82Y1, TNMT, MT1, MT2, MT3, SDR1 (P. somniferum), CAS (C. japonica), ATR1 (A. thaliana) | S. cerevisiae | [104] |
| Dammarenediol-II | Farnesyl diphosphate | SS, SE, CPR (S. cerevisiae), SE (M.capsulatus) CPR (A.thaliana) | E. coli | [105] |
| Scopolamine | Hyoscyamine | H-6-H | E. coli | [7] |
| (S)-reticuline 3-O-sulphate (S)-reticuline 7-O-sulphate | (S)-reticuline | HSULT1A1, hSULT1A3, hSULT1E1 | E. coli | [106] |
| Products | Precursor | Heterologously Expressed Genes and Sources | Engineering Strains | Ref. |
|---|---|---|---|---|
| Valencene and amorphadiene | FDP | Deregulate HMG1, express mitochondrion-targeted FDPS, block other metabolic pathways of FDP, FDP flows into mitochondria in a large amount | S. cerevisiae | [107] |
| Vanillin | Ferulic acid | Dynamic regulatory element (Express Fcs, Ech, hucr mutant), reduces the metabolic burden and toxic side effects during early growth | E. coli | [8] |
| Isoorientin | Luteolin | Expression of synthetic gene Gt6CGT. Express Cep, ugpA to inhibit accumulation of by-product acetic acid, more flow of cellobiose to UDP glucose and less flow to TCA | E. coli | [108] |
| Acti norhodin | TAGs | Genome data mining and physiological and biochemical analysis, ‘dynamic degradation of TAG’: by controlling Sco6196, mobilize the TAG pool and increase polyketide biosynthesis | Streptomyces | [18] |
| Guaia-6,10 (14)-diene | FPP | Expression of synthetic gene FgJ02895, construction of a series of mutants to increase MVA and the downstream synthesis pathway | E. coli, S. cerevisiae | [19] |
| Hypoxanthine | 5-phosphoribosylpyrophosphate, glutamine | Deregulation of PurR (Regulatory proteins), site directed mutation of key enzyme. Increase precursor accumulation and disrupt branch pathways | E. coli, S. cerevisiae | [109] |
| Artemisinic acid | Amorphadiene | Expresion of CYP71AV1, CPR1, CYB5, ADH1, ALDH1, regulate the MVA pathway, optimize the upstream and downstream and reduce the branch pathway | S. cerevisiae | [94] |
| Linalool | Mevalonic acid | Expression of synthetic gene LIS, LIS, overexpression of upstream gene tHMG1 to increase the MVA pathway | S. cerevisiae | [110] |
| Carnosic acid | GGPP | Overexpress tHMGR, knock out LPP1, MVA pathway optimization, increase the vitality of transcription factors, reduce the branch pathway | S. cerevisiae | [111] |
| Racemic naringenin, pinocembrin | 4-coumaric acid,cinnamic acid | Selects a deletion of native pikromycin polyketide synthase gene strain, expression of synthetic gene CCL, CHS under the control of a single ermE promoter | S.venezuelae | [86] |
| Stilbene resveratrol, isorhapontigenin | 4-coumaric acid, Caffeic acid | Expression of 4CL1,STS, optimization of precursor conversion and cyclization of the bulky ferulic acid precursor by metabolic engineering and protein engineering | E. coli | [85] |
| Taxol-5α-l | Glycerol | Expression of CYP725A4, tcCPR, optimize P450 expression, reductase partner interactions, N-terminal modifications | E. coli | [112] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, D.; Dong, J. Multi-Level Optimization and Strategies in Microbial Biotransformation of Nature Products. Molecules 2023, 28, 2619. https://doi.org/10.3390/molecules28062619
Qin D, Dong J. Multi-Level Optimization and Strategies in Microbial Biotransformation of Nature Products. Molecules. 2023; 28(6):2619. https://doi.org/10.3390/molecules28062619
Chicago/Turabian StyleQin, Dan, and Jinyan Dong. 2023. "Multi-Level Optimization and Strategies in Microbial Biotransformation of Nature Products" Molecules 28, no. 6: 2619. https://doi.org/10.3390/molecules28062619
APA StyleQin, D., & Dong, J. (2023). Multi-Level Optimization and Strategies in Microbial Biotransformation of Nature Products. Molecules, 28(6), 2619. https://doi.org/10.3390/molecules28062619





