Supermolecule—Drug Conjugates Based on Acid-Degradable Polyrotaxanes for pH-Dependent Intracellular Release of Doxorubicin
Abstract
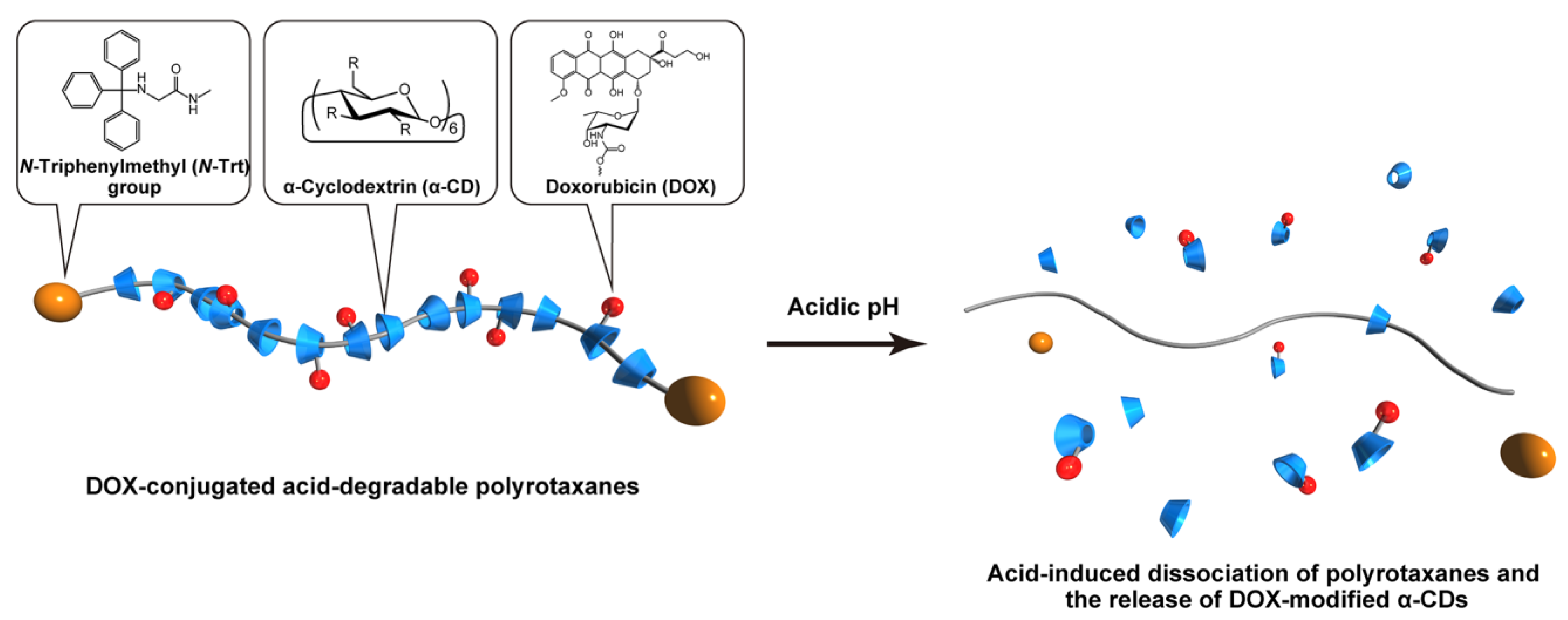
1. Introduction
2. Results
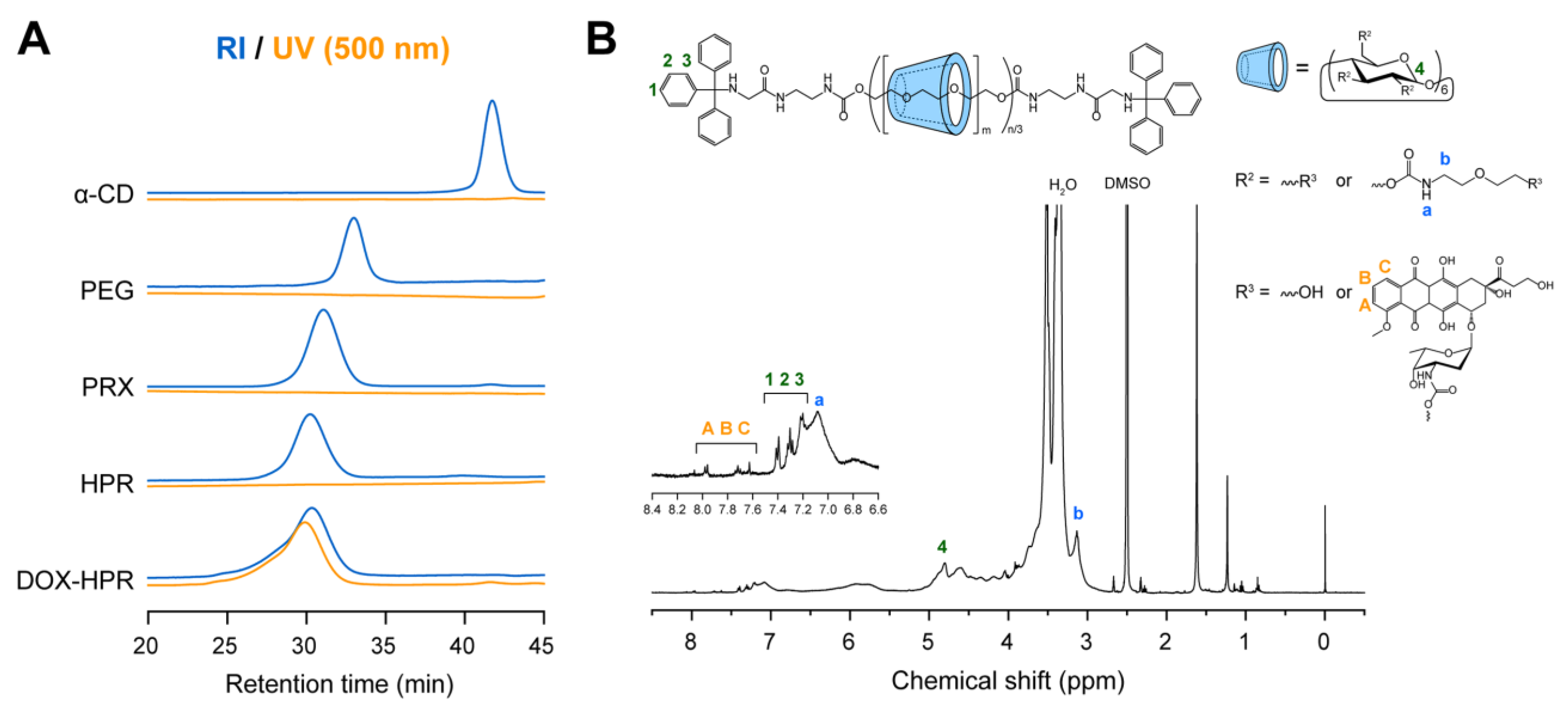
2.1. Synthesis and Characterization of DOX-HPR
2.2. pH-Dependent Dissociation of DOX-HPR
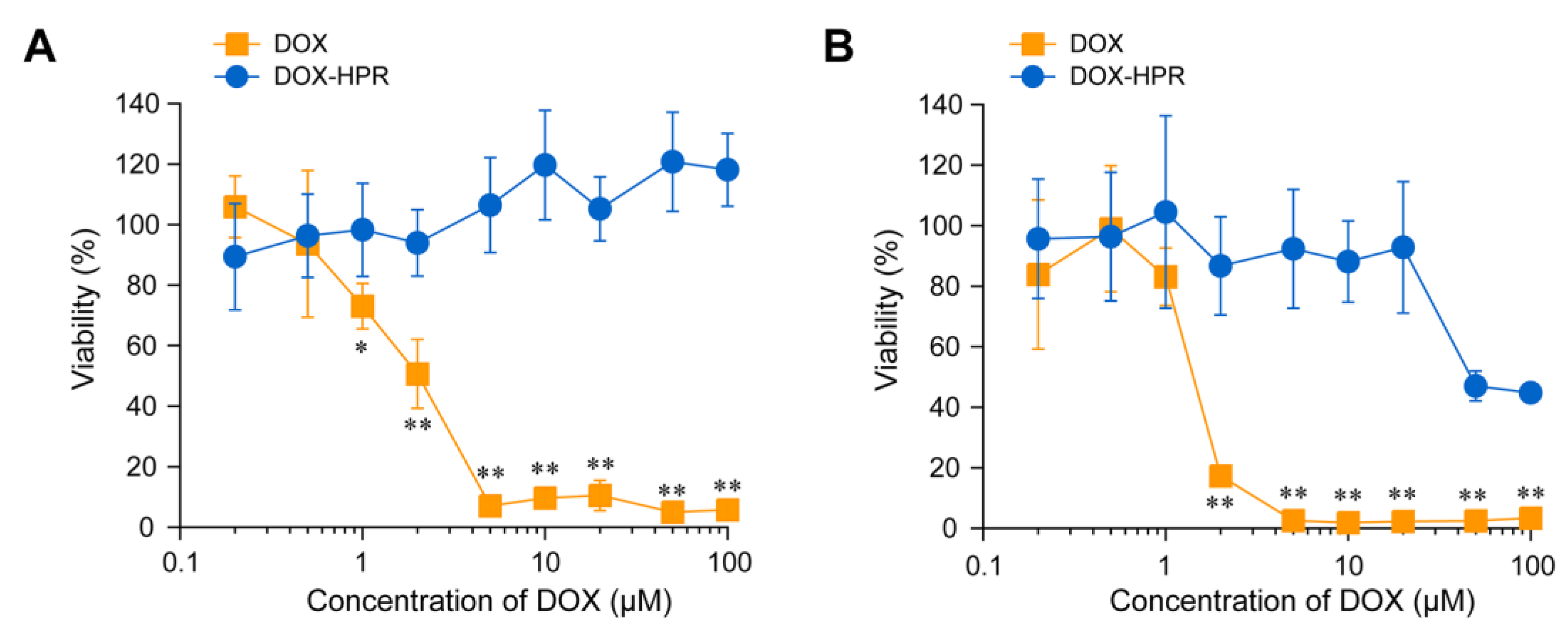
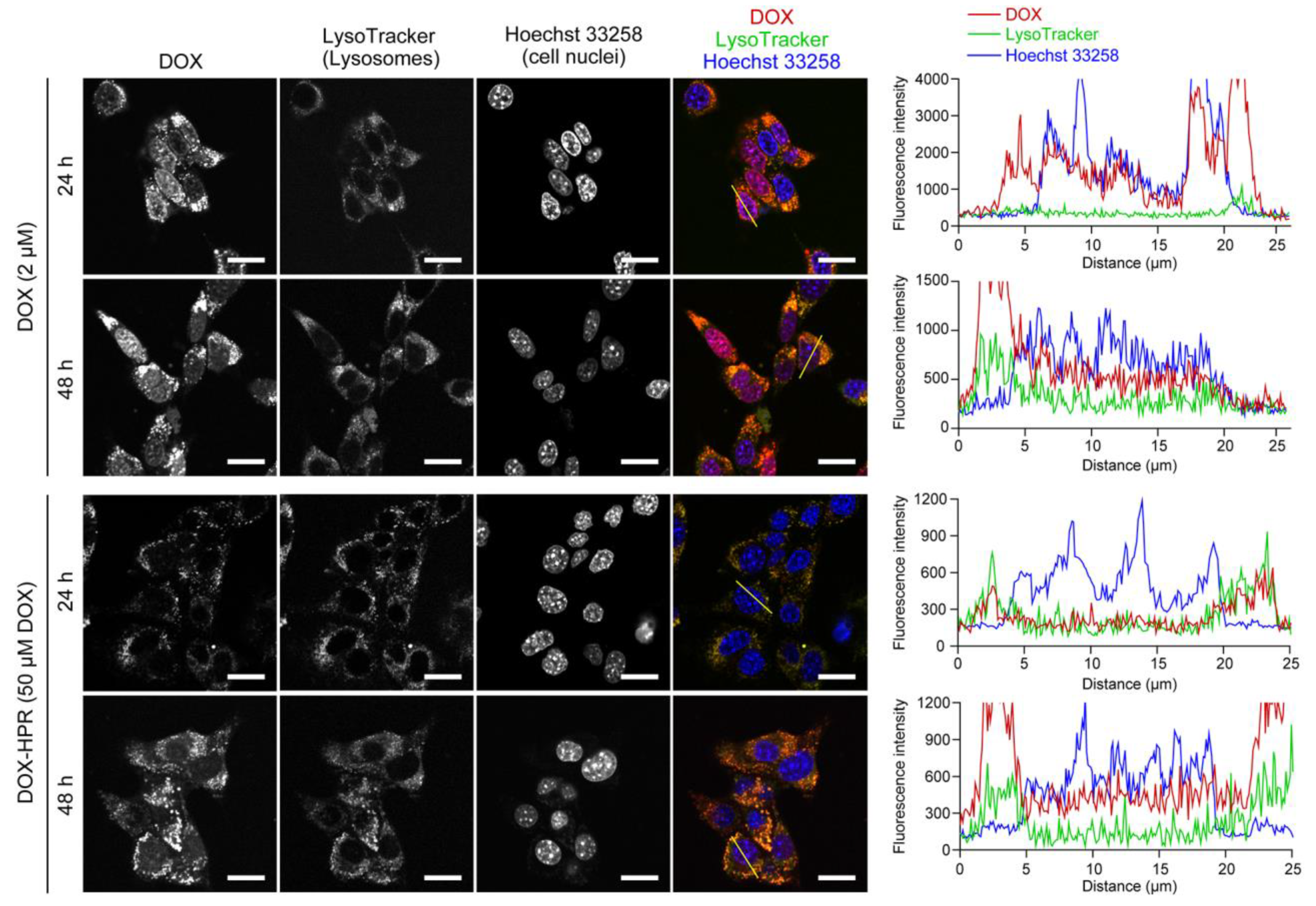
2.3. Cell Viability and Intracellular Localization of DOX-HPR
3. Materials and Methods
3.1. Instrumentation
3.2. Synthesis of α-CD-/PEG-Based Acid-Degradable PRXs
3.3. Synthesis of 2-(2-Hydroxyethoxy)ethoxy Carbamate-Modified PRX (HPR)
3.4. Synthesis of DOX-Conjugated HPR (DOX-HPR)
3.5. Release of DOX-Modified α-CD from DOX-HPR
3.6. Cell Viability
3.7. Intracellular Distribution Analysis
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vargason, A.M.; Anselmo, A.C.; Mitragotri, S. The Evolution of Commercial Drug Delivery Technologies. Nat. Biomed. Eng. 2021, 5, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Otte, A.; Park, K. Evolution of Drug Delivery Systems: From 1950 to 2020 and Beyond. J. Control. Release 2022, 342, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted Drug Delivery Strategies for Precision Medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Tewey, K.M.; Rowe, T.C.; Yang, L.; Halligan, B.D.; Liu, L.F. Adriamycin-Induced DNA Damage Mediated by Mammalian DNA Topoisomerase II. Science 1984, 226, 466–468. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altmana, R.B. Doxorubicin Pathways: Pharmacodynamics and Adverse Effects. Pharmacogenet. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef]
- Carvalho, C.; Santos, R.X.; Cardoso, S.; Correia, S.; Oliveira, P.J.; Santos, M.S.; Moreira, P.I. Doxorubicin: The Good, the Bad and the Ugly Effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12. [Google Scholar] [CrossRef]
- Barenholz, Y. Doxil—The First FDA-Approved Nano-Drug: Lessons Learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Dawidczyk, C.M.; Kim, C.; Park, J.H.; Russell, L.M.; Lee, K.H.; Pomper, M.G.; Searson, P.C. State-of-the-Art in Design Rules for Drug Delivery Platforms: Lessons Learned from FDA-Approved Nanomedicines. J. Control. Release 2014, 187, 133–144. [Google Scholar] [CrossRef]
- Borandeh, S.; van Bochove, B.; Teotia, A.; Seppälä, J. Polymeric Drug Delivery Systems by Additive Manufacturing. Adv. Drug Deliv. Rev. 2021, 173, 349–373. [Google Scholar] [CrossRef]
- Harada, A.; Hashidzume, A.; Yamaguchi, H.; Takashima, Y. Polymeric Rotaxanes. Chem. Rev. 2009, 109, 5974–6023. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Li, J.; Kamachi, M. The Molecular Necklace: A Rotaxane Containing Many Threaded α-Cyclodextrins. Nature 1992, 356, 325–327. [Google Scholar] [CrossRef]
- Tamura, A.; Yui, N. Threaded Macromolecules as a Versatile Framework for Biomaterials. Chem. Commun. 2014, 50, 13433–13446. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ye, L.; Xi, J.; Wang, J.; Feng, Z.-G. Cyclodextrin Polymers: Structure, Synthesis, and Use as Drug Carriers. Prog. Polym. Sci. 2021, 118, 101408. [Google Scholar] [CrossRef]
- Jung, S.; Swamy, B.Y.; Moon, J.-B.; Kim, D.-H.; Chung, I. Anti-AIDS Active Polyrotaxane-AZT Conjugates with Bioactive Bulky Stoppers and Their Nanoparticles. J. Polym. Sci. Polym. Chem. 2012, 50, 4895–4901. [Google Scholar] [CrossRef]
- Jiang, L.; Gao, Z.-M.; Ye, L.; Zhang, A.-Y.; Feng, Z.-G. A pH-Sensitive Nano Drug Delivery System of Doxorubicin-Conjugated Amphiphilic Polyrotaxane-Based Block Copolymers. Biomater. Sci. 2013, 1, 1282–1291. [Google Scholar] [CrossRef]
- Zhou, Z.; Mondjinou, Y.; Hyun, S.-H.; Kulkarni, A.; Lu, Z.-R.; Thompson, D.H. Gd3+-1,4,7,10-Tetraazacyclododecane-1,4,7-triacetic-2-hydroxypropyl-β-cyclodextrin/Pluronic Polyrotaxane as a Long Circulating High Relaxivity MRI Contrast Agent. ACS Appl. Mater. Interfaces 2015, 7, 22272–22276. [Google Scholar] [CrossRef]
- Moon, C.; Kwon, Y.M.; Lee, W.K.; Park, Y.J.; Yang, V.C. In Vitro Assessment of a Novel Polyrotaxane-Based Drug Delivery System Integrated with a Cell-Penetrating Peptide. J. Control. Release 2007, 124, 43–50. [Google Scholar] [CrossRef]
- Moon, C.; Kwon, Y.M.; Lee, W.K.; Park, Y.J.; Chang, L.-C.; Yang, V.C. A Novel Polyrotaxane-Based Intracellular Delivery System for Camptothecin: In Vitro Feasibility Evaluation. J. Biomed. Mater. Res. Part A 2008, 84, 238–246. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, Y.; Wang, X.; Zhen, X.; Zhang, Z.; Wu, W.; Jiang, X. Synthesis of Paclitaxel-Conjugated β-Cyclodextrin Polyrotaxane and Its Antitumor Activity. Angew. Chem. Int. Ed. 2013, 52, 7272–7277. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, X.-M.; Zhang, S.; Ding, L.-S.; Li, B.-J. pH-Responsive Dendritic Polyrotaxane Drug-Polymer Conjugates Forming Nanoparticles as Efficient Drug Delivery System for Cancer Therapy. Polym. Chem. 2015, 6, 2098–2107. [Google Scholar] [CrossRef]
- Kim, T.; Park, S.Y.; Lee, M.-H.; Kim, D.-H.; Chung, I. Syntheses of Polyrotaxane Conjugated with 5-Fluorouracil and Vitamins with Improved Antitumor Activities. J. Bioact. Compat. Polym. 2019, 34, 25–38. [Google Scholar] [CrossRef]
- Yu, G.; Yang, Z.; Fu, X.; Yung, B.C.; Yang, J.; Mao, Z.; Shao, L.; Hua, B.; Liu, Y.; Zhang, F.; et al. Polyrotaxane-Based Supramolecular Theranostics. Nat. Commun. 2018, 9, 766. [Google Scholar] [CrossRef]
- Matsunaga, S.; Tamura, A.; Fushimi, M.; Santa, H.; Arisaka, Y.; Nikaido, T.; Tagami, J.; Yui, N. Light-Embrittled Dental Resin Cements Containing Photodegradable Polyrotaxane Cross-Linkers for Attenuating Debonding Strength. ACS Appl. Polym. Mater. 2020, 2, 5756–5766. [Google Scholar] [CrossRef]
- Tamura, A.; Nishida, K.; Yui, N. Lysosomal pH-Inducible Supramolecular Dissociation of Polyrotaxanes Possessing Acid-Labile N-Triphenylmethyl End Groups and Their Therapeutic Potential for Niemann-Pick Type C Disease. Sci. Technol. Adv. Mater. 2016, 17, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Ooya, T.; Choi, H.S.; Yamashita, A.; Yui, N.; Sugaya, Y.; Kano, A.; Maruyama, A.; Akita, H.; Kogure, K.; Harashima, H. Biocleavable Polyrotaxane−Plasmid DNA Polyplex for Enhanced Gene Delivery. J. Am. Chem. Soc. 2006, 128, 3852–3853. [Google Scholar] [CrossRef]
- Ooya, T.; Mori, H.; Terano, M.; Yui, N. Synthesis of a Biodegradable Polymeric Supramolecular Assembly for Drug Delivery. Macromol. Rapid Commun. 1995, 16, 259–263. [Google Scholar] [CrossRef]
- Ooya, T.; Yui, N. Synthesis of Theophylline–Polyrotaxane Conjugates and Their Drug Release via Supramolecular Dissociation. J. Control. Release 1999, 58, 251–269. [Google Scholar] [CrossRef]
- Tamura, A.; Ohashi, M.; Nishida, K.; Yui, N. Acid-Induced Intracellular Dissociation of β-Cyclodextrin-Threaded Polyrotaxanes Directed toward Attenuating Phototoxicity of Bisretinoids through Promoting Excretion. Mol. Pharm. 2017, 14, 4714–4724. [Google Scholar] [CrossRef]
- Tamura, A.; Yui, N. Lysosomal-Specific Cholesterol Reduction by Biocleavable Polyrotaxanes for Ameliorating Niemann-Pick Type C Disease. Sci. Rep. 2014, 4, 4356. [Google Scholar] [CrossRef]
- Tamura, A.; Yui, N. Polyrotaxane-Based Systemic Delivery of β-Cyclodextrins for Potentiating Therapeutic Efficacy in a Mouse Model of Niemann-Pick Type C Disease. J. Control. Release 2018, 269, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Tamura, A.; Nishida, K.; Zhang, S.; Kang, T.W.; Tonegawa, A.; Yui, N. Cografting of Zwitterionic Sulfobetaines and Cationic Amines on β-Cyclodextrin-Threaded Polyrotaxanes Facilitates Cellular Association and Tissue Accumulation with High Biocompatibility. ACS Biomater. Sci. Eng. 2022, 8, 2463–2476. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, S.; Tamura, A.; Nikiforov, N.; Koike, H.; Kudo, F.; Cheng, Y.; Miyazaki, T.; Kubekina, M.; Kirichenko, T.V.; Orekhov, A.N.; et al. Activated Cholesterol Metabolism is Integral for Innate Macrophage Responses by Amplifying Myd88 Signaling. JCI Insight 2022, 7, e138539. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Li, J.; Kamachi, M. Preparation and Properties of Inclusion Complexes of Polyethylene Glycol with α-Cyclodextrin. Macromolecules 1993, 26, 5698–5703. [Google Scholar] [CrossRef]
- Tamura, A.; Ohashi, M.; Yui, N. Oligo(ethylene glycol)-Modified β-Cyclodextrin-Based Polyrotaxanes for Simultaneously Modulating Solubility and Cellular Internalization Efficiency. J. Biomater. Sci. Polym. Ed. 2017, 28, 1124–1139. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, J.; Zheng, Y.; Guo, R.; Wang, S.; Mignani, S.; Caminade, A.-M.; Majoral, J.-P.; Shi, X. Doxorubicin-Conjugated PAMAM Dendrimers for pH-Responsive Drug Release and Folic Acid-Targeted Cancer Therapy. Pharmaceutics 2018, 10, 162. [Google Scholar] [CrossRef]
- Coluccini, C.; Ng, Y.M.; Reyes, Y.I.A.; Chen, H.-Y.T.; Khung, Y.L. Functionalization of Polyethyleneimine with Hollow Cyclotriveratrylene and Its Subsequent Supramolecular Interaction with Doxorubicin. Molecules 2020, 25, 5455. [Google Scholar] [CrossRef]
- Karukstis, K.K.; Thompson, E.H.Z.; Whiles, J.A.; Rosenfeld, R.J. Deciphering the Fluorescence Signature of Daunomycin and Doxorubicin. Biophys. Chem. 1998, 73, 249–263. [Google Scholar] [CrossRef]
- Agrawal, P.; Barthwal, S.K.; Barthwal, R. Studies on Self-Aggregation of Anthracycline Drugs by Restrained Molecular Dynamics Approach Using Nuclear Magnetic Resonance Spectroscopy Supported by Absorption, Fluorescence, Diffusion Ordered Spectroscopy and Mass Spectrometry. Eur. J. Med. Chem. 2008, 44, 1437–1451. [Google Scholar] [CrossRef]
- Beijnen, J.H.; van der Houwen, O.A.G.J.; Underberg, W.J.M. Aspects of the Degradation Kinetics of Doxorubicin in Aqueous Solution. Int. J. Pharm. 1986, 32, 123–131. [Google Scholar] [CrossRef]
- Tamura, A.; Yui, N. Cellular Internalization and Gene Silencing of siRNA Polyplexes by Cytocleavable Cationic Polyrotaxanes with Tailored Rigid Backbones. Biomaterials 2013, 34, 2480–2491. [Google Scholar] [CrossRef] [PubMed]



| Code | Number of Threading α-CDs | Number of Modified HEE Groups | Number of Conjugated DOX | Mn |
|---|---|---|---|---|
| PRX | 58.8 | - | - | 77,600 |
| HPR | 58.8 | 129 | - | 94,600 |
| DOX-HPR | 58.8 | 129 | 8.1 | 99,200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamura, A.; Osawa, M.; Yui, N. Supermolecule—Drug Conjugates Based on Acid-Degradable Polyrotaxanes for pH-Dependent Intracellular Release of Doxorubicin. Molecules 2023, 28, 2517. https://doi.org/10.3390/molecules28062517
Tamura A, Osawa M, Yui N. Supermolecule—Drug Conjugates Based on Acid-Degradable Polyrotaxanes for pH-Dependent Intracellular Release of Doxorubicin. Molecules. 2023; 28(6):2517. https://doi.org/10.3390/molecules28062517
Chicago/Turabian StyleTamura, Atsushi, Mamoru Osawa, and Nobuhiko Yui. 2023. "Supermolecule—Drug Conjugates Based on Acid-Degradable Polyrotaxanes for pH-Dependent Intracellular Release of Doxorubicin" Molecules 28, no. 6: 2517. https://doi.org/10.3390/molecules28062517
APA StyleTamura, A., Osawa, M., & Yui, N. (2023). Supermolecule—Drug Conjugates Based on Acid-Degradable Polyrotaxanes for pH-Dependent Intracellular Release of Doxorubicin. Molecules, 28(6), 2517. https://doi.org/10.3390/molecules28062517






