Investigation on Metabolites in Structure and Biosynthesis from the Deep-Sea Sediment-Derived Actinomycete Janibacter sp. SCSIO 52865
Abstract
1. Introduction
2. Results and Discussion
2.1. Analysis of Molecular Networking
2.2. Structural Elucidation of Compounds
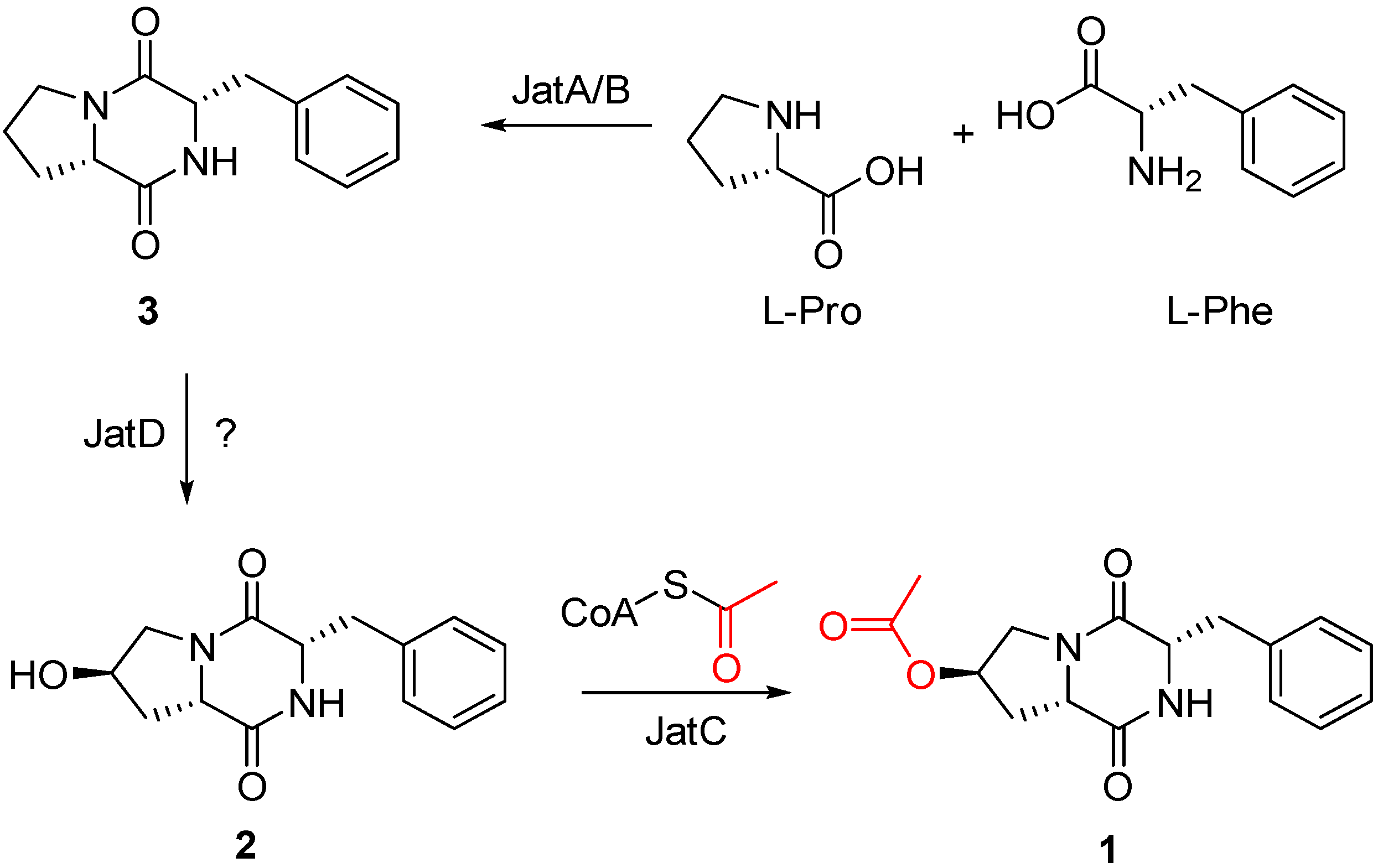
2.3. Putatively Biosynthetic Pathway of Compound 1
2.4. Biological Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Microorganism and Growth Conditions
3.3. Whole Genome Sequencing and Bioinformatic Analysis
3.4. Extraction and Isolation
3.5. Molecular Networking
3.6. Marfey’s Analysis
3.7. GC-MS Analysis
3.8. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022, 39, 1122–1171. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Meng, L.H.; Wang, B.G. Progress in research on bioactive secondary metabolites from deep-sea derived microorganisms. Mar. Drugs 2020, 18, 614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, B.; Cai, L.; Liu, L. New dibenzo-α-pyrone derivatives with α-glucosidase inhibitory activities from the marine-derived fungus Alternaria alternata. Mar. Drugs 2022, 20, 778. [Google Scholar] [CrossRef]
- Xu, D.; Nepal, K.K.; Chen, J.; Harmody, D.; Zhu, H.; McCarthy, P.J.; Wright, A.E.; Wang, G. Nocardiopsistins A-C: New angucyclines with anti-MRSA activity isolated from a marine sponge-derived Nocardiopsis sp. HB-J378. Syn. Syst. Biotechnol. 2018, 3, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhou, Y.X.; Tang, X.X.; Liu, X.X.; Yi, Z.W.; Fang, M.J.; Wu, Z.; Jiang, F.Q.; Qiu, Y.K. Macrolactins from marine-derived Bacillus subtilis B5 bacteria as inhibitors of inducible nitric oxide and cytokines expression. Mar. Drugs 2016, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Abdel-Mageed, W.M.; Ebel, R.; Bull, A.T.; Goodfellow, M.; Fiedler, H.P.; Jaspars, M. Dermacozines H–J isolated from a deep-sea strain of Dermacoccus abyssi from mariana trench sediments. J. Nat. Prod. 2014, 77, 416–420. [Google Scholar] [CrossRef]
- Gustafson, K.; Roman, M.; Fenical, W. The macrolactins, a novel class of antiviral and cytotoxic macrolides from a deep-sea marine bacterium. J. Am. Chem. Soc. 1989, 111, 7519–7524. [Google Scholar] [CrossRef]
- Hughes, C.C.; MacMillan, J.B.; Gaudêncio, S.P.; Jensen, P.R.; Fenical, W. The ammosamides: Structures of cell cycle modulators from a marine-derived Streptomyces species. Angew. Chem. Int. Ed. 2009, 48, 725–727. [Google Scholar] [CrossRef]
- Lim, Y.K.; Kweon, O.J.; Kim, H.R.; Kim, T.H.; Lee, M.K. First case of bacteremia caused by Janibacter hoylei. APMIS 2017, 125, 665–668. [Google Scholar] [CrossRef]
- Yamazoe, A.; Yagi, O.; Oyaizu, H. Degradation of polycyclic aromatic hydrocarbons by a newly isolated dibenzofuran-utilizing Janibacter sp. strain YY-1. Appl. Microbiol. Biotechnol. 2004, 65, 211–218. [Google Scholar] [CrossRef]
- Khessairi, A.; Fhoula, I.; Jaouani, A.; Turki, Y.; Cherif, A.; Boudabous, A.; Hassen, A.; Ouzari, H. Pentachlorophenol degradation by Janibacter sp., a new actinobacterium isolated from saline sediment of Arid Land. Biomed. Res. Int. 2014, 2014, 296472. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhu, T.; Xu, X.; Xu, Y. Biodegradation of dibenzofuran by Janibacter terrae strain XJ-1. Curr. Microbiol. 2006, 53, 30–36. [Google Scholar] [CrossRef]
- Iwai, S.; Yamazoe, A.; Takahashi, R.; Kurisu, F.; Yagi, O. Degradation of mono-chlorinated dibenzo-p-dioxins by Janibacter sp. strain YA isolated from river sediment. Curr. Microbiol. 2005, 51, 353–358. [Google Scholar] [PubMed]
- Ezzat, S.M.; Ahmed, N.A. Short-term biodegradation of crude petroleum oil in water by photostimulated Janibacter terrae strain S1N1. ACS Omega 2022, 7, 13976–13984. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Knaan, T.L.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Ding, W.; Li, Y.; Tian, X.; Chen, M.; Xiao, Z.; Chen, R.; Yin, H.; Zhang, S. Investigation on metabolites in structural diversity from the deep-sea sediment-derived bacterium Agrococcus sp. SCSIO 52902 and their biosynthesis. Mar. Drugs 2022, 20, 431. [Google Scholar] [CrossRef]
- Ding, W.; Li, Y.; Chen, M.; Chen, R.; Tian, X.; Yin, H.; Zhang, S. Structures and antitumor activities of ten new and twenty known surfactins from the deep-sea bacterium Limimaricola sp. SCSIO 53532. Bioorg. Chem. 2022, 120, 105589. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.; Wang, J.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Cody, D.R.; DeWitt, S.H.H.; Hodges, J.C.; Kiely, J.S.; Moos, W.H.; Pavia, M.R.; Roth, B.D.; Schroeder, M.C.; Stankovic, C.J. Apparatus for Multiple Simultaneous Synthesis. U.S. Patent 5,324,483, 28 June 1994. [Google Scholar]
- Xiang, W.X.; Liu, Q.; Li, X.M.; Lu, C.H.; Shen, Y.M. Four pairs of proline-containing cyclic dipeptides from Nocardiopsis sp. HT88, an endophytic bacterium of Mallotus nudiflorus L. Nat. Prod. Res. 2020, 34, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Park, A.R.; Jeong, S.I.; Jeon, H.W.; Kim, J.; Kim, N.; Ha, M.T.; Mannaa, M.; Kim, J.; Lee, C.W.; Min, B.S.; et al. A diketopiperazine, Cyclo-(L-Pro-L-Ile), derived from Bacillus thuringiensis JCK-1233 controls pine wilt disease by elicitation of moderate hypersensitive reaction. Front. Plant. Sci. 2020, 11, 1023. [Google Scholar] [CrossRef]
- Kumar, N.; Mohandas, C.; Nambisan, B.; Kumar, D.R.S.; Lankalapalli, R.S. Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microbiol. Biotechnol. 2013, 29, 355–364. [Google Scholar] [CrossRef]
- Hanai, K.; Kuwae, A.; Takai, T.; Senda, H.; Kunimoto, K.K. A comparative vibrational and NMR study of cis-cinnamic acid polymorphs and trans-cinnamic acid. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2001, 57, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kim, G.J.; Shin, M.S.; Moon, J.; Kim, S.; Nam, J.W.; Kang, K.S.; Choi, H. Chemical investigation of diketopiperazines and N-phenethylacetamide isolated from Aquimarina sp. MC085 and their effect on TGF-β-induced epithelial–mesenchymal transition. Appl. Sci. 2021, 11, 8866. [Google Scholar] [CrossRef]
- Shindo, K.; Asagi, E.; Sano, A.; Hotta, E.; Minemura, N.; Mikami, K.; Tamesada, E.; Misawa, N.; Maoka, T. Diapolycopenedioic acid xylosyl esters A, B, and C, novel antioxidative glyco-C30-carotenoic acids produced by a new marine bacterium Rubritalea squalenifaciens. J. Antibiot. 2008, 61, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Bruns, H.; Thiel, V.; Voget, S.; Patzelt, D.; Daniel, R.; Wagner-Döbler, I.; Schulz, S. N-Acylated alanine methyl esters (NAMEs) from Roseovarius tolerans, structural analogs of quorum-sensing autoinducers, N-acylhomoserine lactones. Chem. Biodivers. 2013, 10, 1559–1573. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.R.; Che, Y.H.; Su, J.S.; Yang, Z.B.; Zhang, Y. Chemical constituents from petroleum ether extract of Yi medicine Blaps rynchopetera. Zhong Yao Cai 2020, 43, 2924–2927. [Google Scholar]
- Rontani, J.F.; Zabeti, N.; Aubert, C. Double bond migration to methylidene positions during electron ionization mass spectrometry of branched monounsaturated fatty acid derivatives. J. Am. Soc. Mass Spectrom. 2009, 20, 1997–2005. [Google Scholar] [CrossRef]
- Yang, M.N.; Zhang, H.; Liu, J.; Li, Y.; Li, W.T.; Xia, H.L. Study on chemical constituents from Phyllanthus urinaria. Zhong Cao Yao 2016, 47, 3573–3577. [Google Scholar]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Mishra, A.K.; Choi, J.; Choi, S.J.; Baek, K.H. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 2017, 22, 1796. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Gao, X.; Song, Z.; Li, C.; Lu, F.; Tanokura, M.; Qin, H.M. Development of engineered ferredoxin reductase systems for the efficient hydroxylation of steroidal substrates. ACS Sustain. Chem. Eng. 2020, 8, 16720–16730. [Google Scholar] [CrossRef]
- Okubo, S.; Ena, E.; Okuda, A.; Kozone, I.; Hashimoto, J.; Nishitsuji, Y.; Fujie, M.; Satoh, N.; Ikeda, H.; Shin-ya, K. Identification of functional cytochrome P450 and ferredoxin from Streptomyces sp. EAS-AB2608 by transcriptional analysis and their heterologous expression. Appl. Microbiol. Biotechnol. 2021, 105, 4177–4187. [Google Scholar] [CrossRef]
- Naggert, J.; Narasimhan, M.L.; DeVeaux, L.; Cho, H.; Randhawa, Z.I.; Cronan, J.E.; Green, B.N.; Smith, S. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J. Biol. Chem. 1991, 266, 11044–11050. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Wavdhane, M.; Haque, S.; Govender, T.; Kruger, H.G.; Mishra, M.K.; Chandra, R.; Tiwari, D. A sensitive WST-8-based bioassay for PEGylated granulocyte colony stimulating factor using the NFS-60 cell line. Pharm. Biol. 2015, 53, 849–854. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yang, X.W.; Wang, R.F. Bio-assay guided isolation and identification of α-glucosidase inhibitors from the leaves of Aquilaria sinensis. Phytochemistry 2011, 72, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.B.; Tian, J.Y.; Dai, Z.; Ye, F.; Ma, S.C.; Wang, A.G. α-Glucosidase inhibitors extracted from the roots of Polygonum multiflorum Thunb. Fitoterapia 2017, 117, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Sales-Campos, H.; Souza, P.R.; Peghini, B.C.; da Silva, J.S.; Cardoso, C.R. An overview of the modulatory effects of oleic acid in health and disease. Mini-Rev. Med. Chem. 2013, 13, 201–210. [Google Scholar]
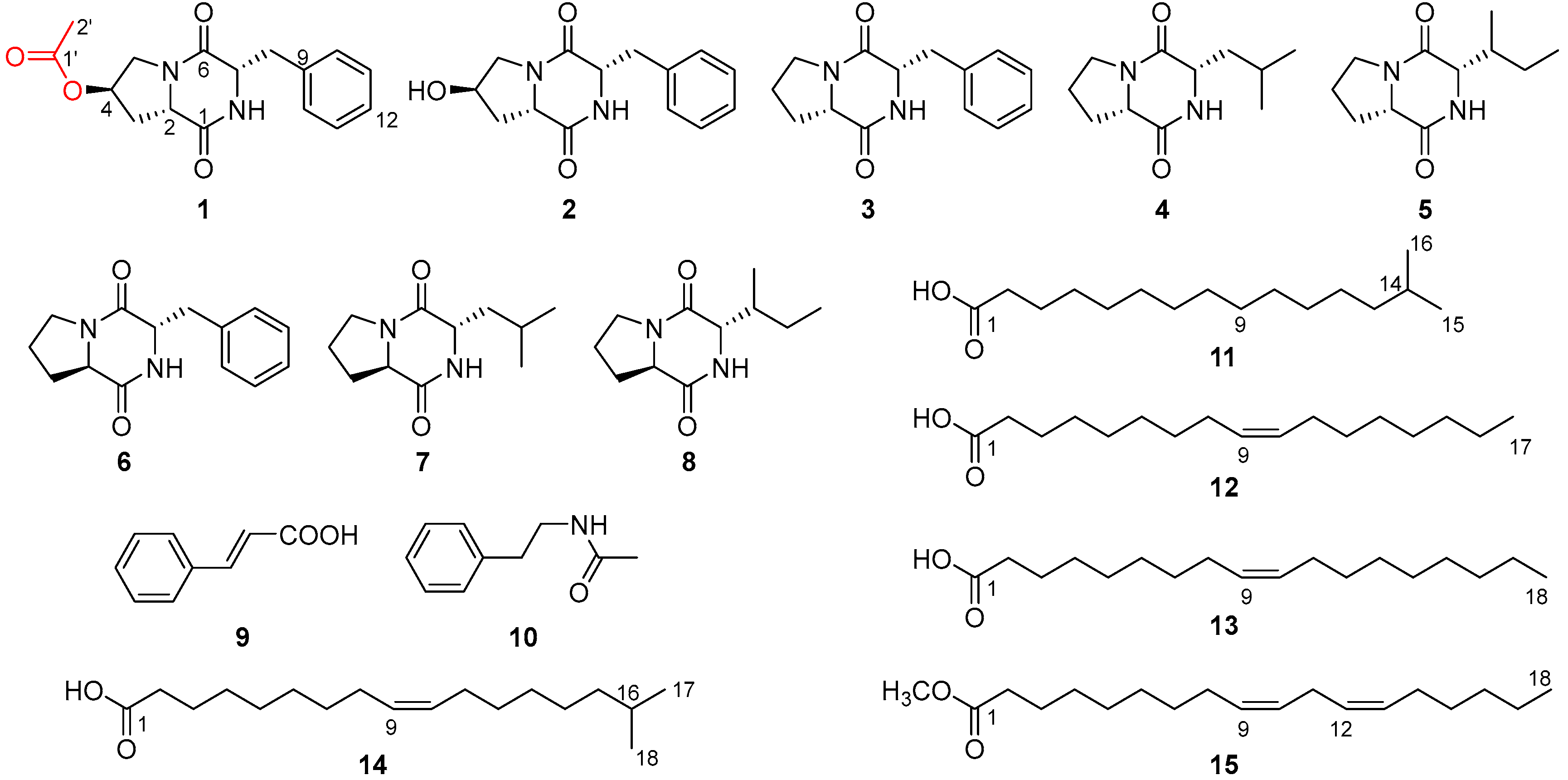
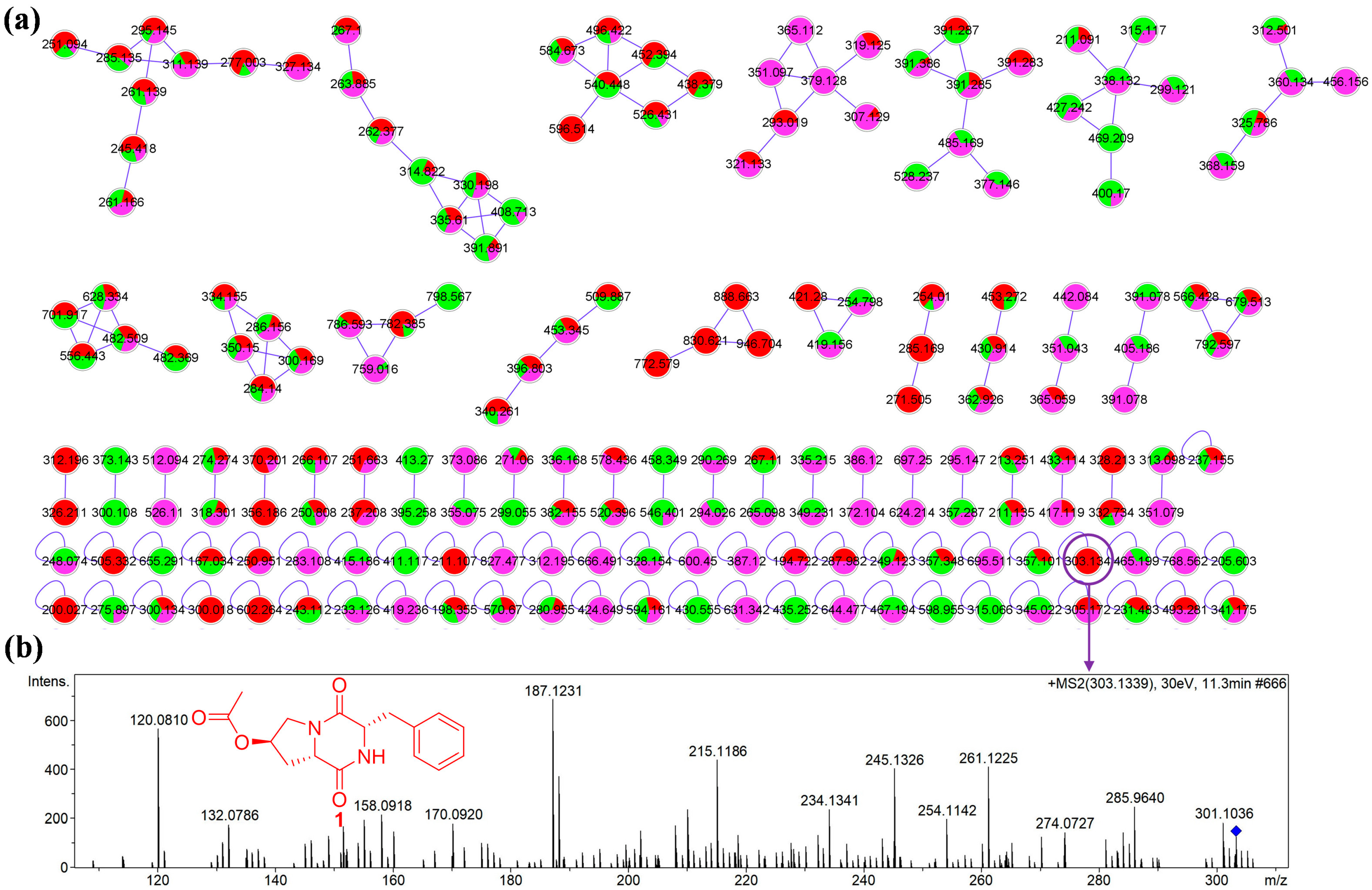
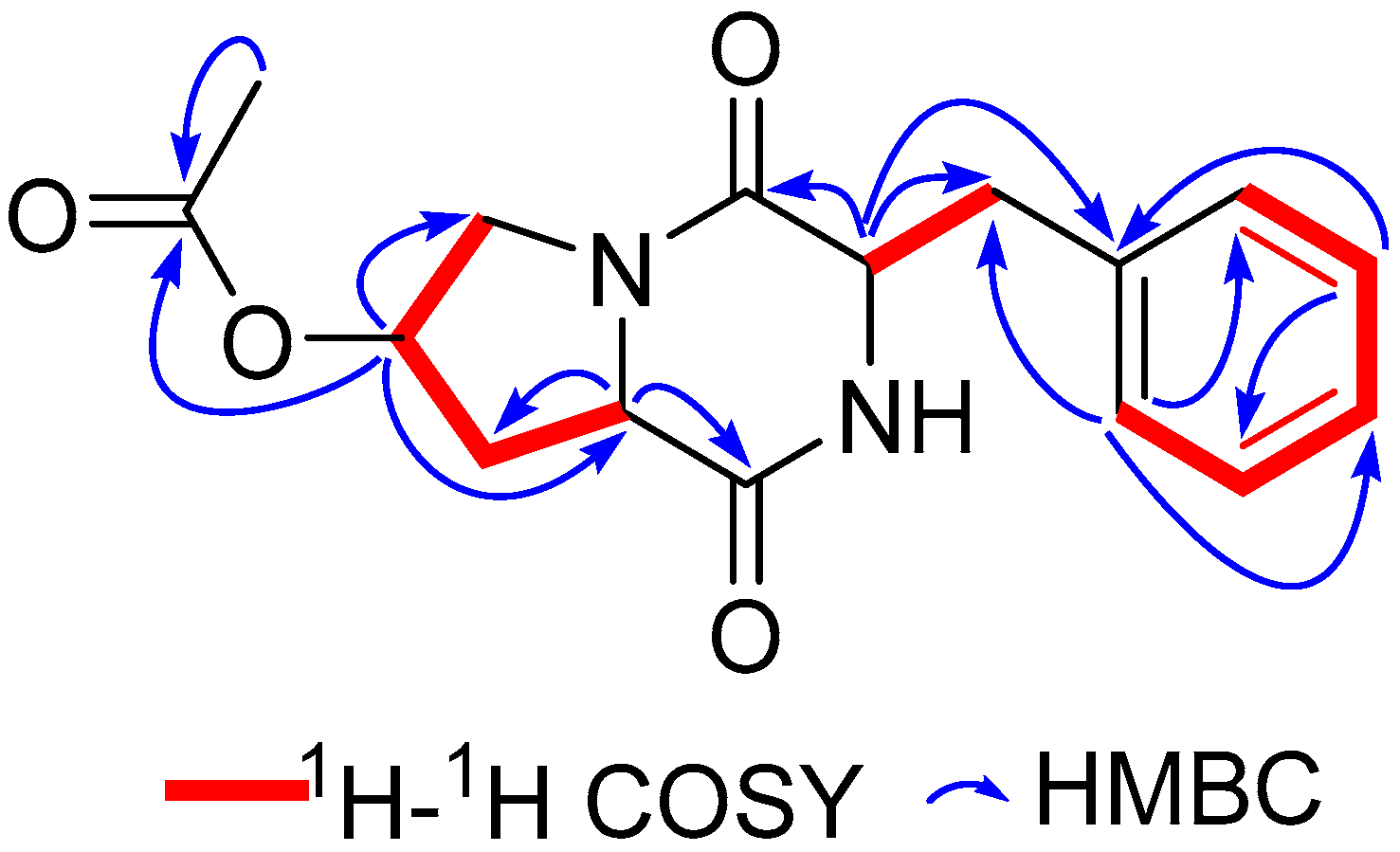
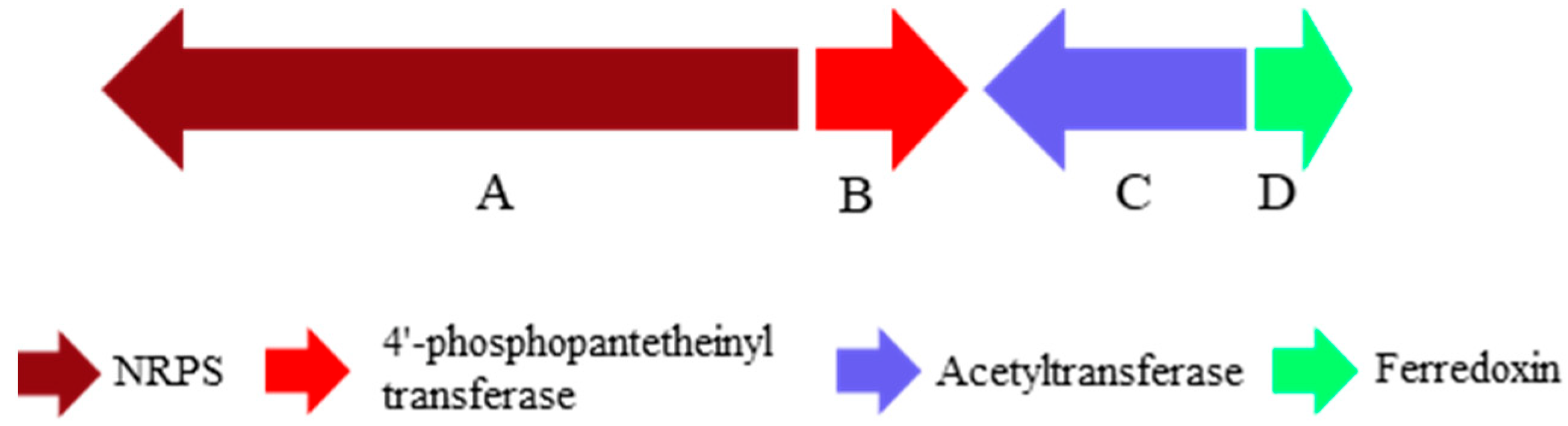
| Position | δH | δC |
|---|---|---|
| 1 | 170.5, s | |
| 2 | 4.34 (ddd, 11.8, 5.9, 1.9) | 58.4, d |
| 3 | 2.18 (dd, 13.7, 6.0), 1.45 (ddd, 13.7, 11.8, 5.3) | 36.1, t |
| 4 | 5.13 (t, 5.4) | 71.0, d |
| 5 | 3.89 (dd, 13.8, 5.6), 3.38 (d, 13.8) | 52.9, t |
| 6 | 167.0, s | |
| 7 | 4.49 (td, 5.1, 1.8) | 57.7, d |
| 8 | 3.17 (m) | 38.2, t |
| 9 | 137.0, s | |
| 10, 14 | 7.24 (overlapped) | 131.1, d |
| 11, 13 | 7.29 (overlapped) | 129.6, d |
| 12 | 7.24 (overlapped) | 128.2, d |
| 1′ | 172.0, s | |
| 2′ | 2.02 (s) | 20.9, q |
| Amino Acid | Standard | Cyclo(L-trans-Hyp-L-Leu) | 1 | 2 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| tRL 1 | Pro | 21.570 | 10.925 4 | 10.496 4 | 10.575 4 | 21.544 | ||
| Phe | 38.816 | 38.353 | 38.486 | 38.037 | ||||
| Ile | 35.863 | 35.533 | ||||||
| Leu | 36.490 | 36.621 | 36.157 | |||||
| tRD 2 | Pro | 23.835 | 23.250 | 23.708 | ||||
| Phe | 44.059 | |||||||
| Ile | 42.390 | |||||||
| Leu | 42.851 | |||||||
| tR 3 | L-FDAA | 22.463 | 23.277 | 22.560 | 22.699 | 22.821 | 22.218 | 22.747 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, W.; Li, Y.; Tian, X.; Xiao, Z.; Li, R.; Zhang, S.; Yin, H. Investigation on Metabolites in Structure and Biosynthesis from the Deep-Sea Sediment-Derived Actinomycete Janibacter sp. SCSIO 52865. Molecules 2023, 28, 2133. https://doi.org/10.3390/molecules28052133
Ding W, Li Y, Tian X, Xiao Z, Li R, Zhang S, Yin H. Investigation on Metabolites in Structure and Biosynthesis from the Deep-Sea Sediment-Derived Actinomycete Janibacter sp. SCSIO 52865. Molecules. 2023; 28(5):2133. https://doi.org/10.3390/molecules28052133
Chicago/Turabian StyleDing, Wenping, Yanqun Li, Xinpeng Tian, Zhihui Xiao, Ru Li, Si Zhang, and Hao Yin. 2023. "Investigation on Metabolites in Structure and Biosynthesis from the Deep-Sea Sediment-Derived Actinomycete Janibacter sp. SCSIO 52865" Molecules 28, no. 5: 2133. https://doi.org/10.3390/molecules28052133
APA StyleDing, W., Li, Y., Tian, X., Xiao, Z., Li, R., Zhang, S., & Yin, H. (2023). Investigation on Metabolites in Structure and Biosynthesis from the Deep-Sea Sediment-Derived Actinomycete Janibacter sp. SCSIO 52865. Molecules, 28(5), 2133. https://doi.org/10.3390/molecules28052133








