OGT Binding Peptide-Tagged Strategy Increases Protein O-GlcNAcylation Level in E. coli
Abstract
1. Introduction
2. Results
2.1. OBPs Increased O-GlcNAc Level of Tau
2.2. O-GlcNAc Sites Identified in Tau and Tagged Tau
2.3. P1Tau Aggregated More Slowly than Tau
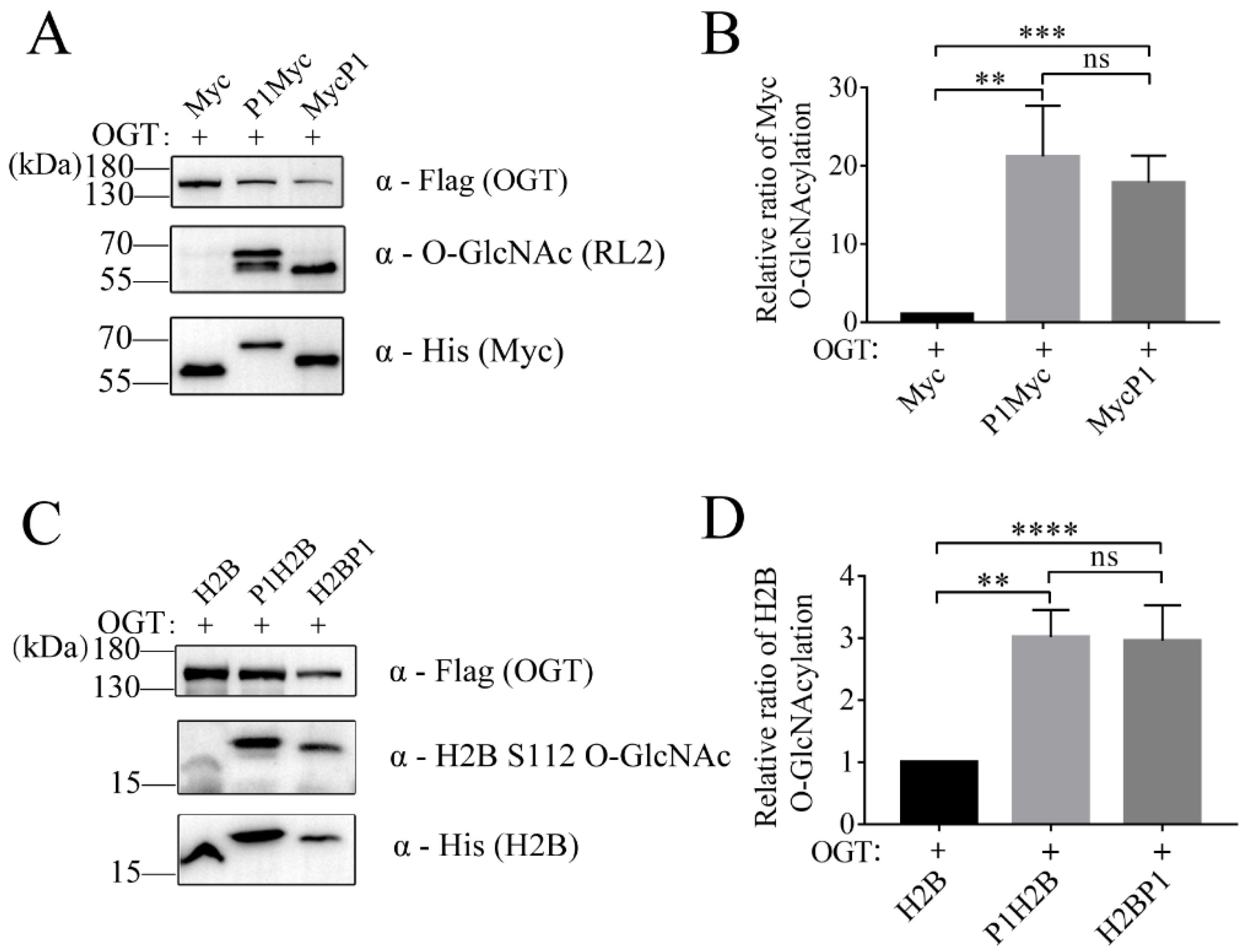
2.4. P1 Tag also Improved the O-GlcNAc Level of c-Myc and H2B
3. Discussion
4. Materials and Methods
4.1. Preparing Gene Constructs in pET-4CDS
4.2. Production of O-GlcNAcylated Protein
4.2.1. Ni-NTA Column
4.2.2. SP Sepharose Column
4.3. Congo Red Binding Assay
4.4. ThT Binding Assays
4.5. SDS-PAGE and Western Blot
4.6. Mass Spectrometry Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torres, C.R.; Hart, G.W. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes—Evidence for O-Linked Glcnac. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [CrossRef]
- Zhao, L.; Shah, J.A.; Cai, Y.; Jin, J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules 2018, 23, 1967. [Google Scholar] [CrossRef] [PubMed]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins—Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wells, L.; Comer, F.I.; Parker, G.J.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins—Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001, 276, 9838–9845. [Google Scholar] [CrossRef]
- Ciraku, L.; Esquea, E.M.; Reginato, M.J. O-GlcNAcylation regulation of cellular signaling in cancer. Cell. Signal. 2022, 90, 110201. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.R.; Dias, T.B.; Natov, P.S.; Zachara, N.E. Stress-induced O-GlcNAcylation: An adaptive process of injured cells. Biochem. Soc. Trans. 2017, 45, 237–249. [Google Scholar] [CrossRef]
- Bacigalupa, Z.A.; Bhadiadra, C.H.; Reginato, M.J. O-GlcNAcylation: Key regulator of glycolytic pathways. J. Bioenerg. Biomembr. 2018, 50, 189–198. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef]
- Chatham, J.C.; Young, M.E.; Zhang, J.H. Role of O-linked N-acetylglucosamine (O-GlcNAc) modification of proteins in diabetic cardiovascular complications. Curr. Opin. Pharm. 2021, 57, 1–12. [Google Scholar] [CrossRef]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 1–14. [Google Scholar] [CrossRef]
- Dassanayaka, S.; Jones, S.P. O-GlcNAc and the cardiovascular system. Pharm. Ther. 2014, 142, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lai, M.K.P.; Arumugam, T.V.; Jo, D.G. O-GlcNAcylation as a Therapeutic Target for Alzheimer’s Disease. Neuromolecular Med. 2020, 22, 171–193. [Google Scholar] [CrossRef] [PubMed]
- Graeber, M.B.; Kupke, K.G.; Muller, U. Delineation of the dystonia-parkinsonism syndrome locus in Xq13. Proc. Natl. Acad. Sci. USA 1992, 89, 8245–8248. [Google Scholar] [CrossRef] [PubMed]
- Stephen, H.M.; Adams, T.M.; Wells, L. Regulating the Regulators: Mechanisms of Substrate Selection of the O-GlcNAc Cycling Enzymes OGT and OGA. Glycobiology 2021, 31, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Estevez, A.; Zhu, D.; Blankenship, C.; Jiang, J. Molecular Interrogation to Crack the Case of O-GlcNAc. Chemistry 2020, 26, 12086–12100. [Google Scholar] [CrossRef] [PubMed]
- Riu, I.H.; Shin, I.S.; Do, S.I. Sp1 modulates ncOGT activity to alter target recognition and enhanced thermotolerance in E. coli. Biochem. Biophys. Res. Commun. 2008, 372, 203–209. [Google Scholar] [CrossRef]
- Ma, J.; Hart, G.W. O-GlcNAc profiling: From proteins to proteomes. Clin. Proteom. 2014, 11, 8. [Google Scholar] [CrossRef]
- Zhang, N.; Jiang, H.L.; Zhang, K.X.; Zhu, J.X.; Wang, Z.Z.; Long, Y.T.; He, Y.J.; Feng, F.; Liu, W.Y.; Ye, F.; et al. OGT as potential novel target: Structure, function and inhibitors. Chem.-Biol. Interact. 2022, 357, 109886. [Google Scholar] [CrossRef]
- Groenevelt, J.M.; Corey, D.J.; Fehl, C. Chemical Synthesis and Biological Applications of O-GlcNAcylated Peptides and Proteins. ChemBioChem 2021, 22, 1854–1870. [Google Scholar] [CrossRef]
- Gorelik, A.; van Aalten, D.M.F. Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem. Biol. 2020, 1, 98–109. [Google Scholar] [CrossRef]
- Kamemura, K.; Hayes, B.K.; Comer, F.I.; Hart, G.W. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: Alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J. Biol. Chem. 2002, 277, 19229–19235. [Google Scholar] [CrossRef]
- Hirosawa, M.; Hayakawa, K.; Yoneda, C.; Arai, D.; Shiota, H.; Suzuki, T.; Tanaka, S.; Dohmae, N.; Shiota, K. Novel O-GlcNAcylation on Ser(40) of canonical H2A isoforms specific to viviparity. Sci. Rep. 2016, 6, 31785. [Google Scholar] [CrossRef]
- Cameron, A.; Giacomozzi, B.; Joyce, J.; Gray, A.; Graham, D.; Ousson, S.; Neny, M.; Beher, D.; Carlson, G.; O’Moore, J.; et al. Generation and characterization of a rabbit monoclonal antibody site-specific for tau O-GlcNAcylated at serine 400. FEBS Lett. 2013, 587, 3722–3728. [Google Scholar] [CrossRef]
- Yuzwa, S.A.; Yadav, A.K.; Skorobogatko, Y.; Clark, T.; Vosseller, K.; Vocadlo, D.J. Mapping O-GlcNAc modification sites on tau and generation of a site-specific O-GlcNAc tau antibody. Amino Acids 2011, 40, 857–868. [Google Scholar] [CrossRef]
- Fujiki, R.; Hashiba, W.; Sekine, H.; Yokoyama, A.; Chikanishi, T.; Ito, S.; Imai, Y.; Kim, J.; He, H.H.; Igarashi, K.; et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011, 480, 557–560. [Google Scholar] [CrossRef]
- Shan, H.; Sun, J.; Shi, M.; Liu, X.; Shi, Z.; Yu, W.; Gu, Y. Generation and characterization of a site-specific antibody for SIRT1 O-GlcNAcylated at serine 549. Glycobiology 2018, 28, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.L.; Berkaw, M.N.; Buse, M.G.; Ball, L.E. O-linked N-acetylglucosamine modification of insulin receptor substrate-1 occurs in close proximity to multiple SH2 domain binding motifs. Mol. Cell. Proteom. 2009, 8, 2733–2745. [Google Scholar] [CrossRef] [PubMed]
- Pathak, S.; Borodkin, V.S.; Albarbarawi, O.; Campbell, D.G.; Ibrahim, A.; van Aalten, D.M. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 2012, 31, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; He, Z.; Shao, Z.; Lu, H. TAB3 O-GlcNAcylation promotes metastasis of triple negative breast cancer. Oncotarget 2016, 7, 22807–22818. [Google Scholar] [CrossRef]
- Shen, D.L.; Gloster, T.M.; Yuzwa, S.A.; Vocadlo, D.J. Insights into O-linked N-acetylglucosamine ([0-9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 2012, 287, 15395–15408. [Google Scholar] [CrossRef]
- Han, C.; Shan, H.; Bi, C.; Zhang, X.; Qi, J.; Zhang, B.; Gu, Y.; Yu, W. A highly effective and adjustable dual plasmid system for O-GlcNAcylated recombinant protein production in E. coli. J. Biochem. 2015, 157, 477–484. [Google Scholar] [CrossRef]
- Gao, H.; Shi, M.; Wang, R.; Wang, C.; Shao, C.; Gu, Y.; Yu, W. A widely compatible expression system for the production of highly O-GlcNAcylated recombinant protein in Escherichia coli. Glycobiology 2018, 28, 949–957. [Google Scholar] [CrossRef]
- Pathak, S.; Alonso, J.; Schimpl, M.; Rafie, K.; Blair, D.E.; Borodkin, V.S.; Albarbarawi, O.; van Aalten, D.M.F. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 2015, 22, 744–750. [Google Scholar] [CrossRef]
- Yuzwa, S.A.; Shan, X.; Macauley, M.S.; Clark, T.; Skorobogatko, Y.; Vosseller, K.; Vocadlo, D.J. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 2012, 8, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Yuzwa, S.A.; Cheung, A.H.; Okon, M.; McIntosh, L.P.; Vocadlo, D.J. O-GlcNAc modification of tau directly inhibits its aggregation without perturbing the conformational properties of tau monomers. J. Mol. Biol. 2014, 426, 1736–1752. [Google Scholar] [CrossRef] [PubMed]
- Yuzwa, S.A.; Vocadlo, D.J. Production of O-GlcNAc Modified Recombinant Tau in E. coli and Detection of Ser400 O-GlcNAc Tau In Vivo. Tau Protein Methods Protoc. 2017, 1523, 237–248. [Google Scholar]
- Cantrelle, F.X.; Loyens, A.; Trivelli, X.; Reimann, O.; Despres, C.; Gandhi, N.S.; Hackenberger, C.P.R.; Landrieu, I.; Smet-Nocca, C. Phosphorylation and O-GlcNAcylation of the PHF-1 Epitope of Tau Protein Induce Local Conformational Changes of the C-Terminus and Modulate Tau Self-Assembly Into Fibrillar Aggregates. Front. Mol. Neurosci. 2021, 14, 661368. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, Z.; Zhang, L.; Sui, R.; Khan, S. Exploring the inhibitory effects of liquiritigenin against tau fibrillation and related neurotoxicity as a model of preventive care in Alzheimer’s disease. Int. J. Biol. Macromol. 2021, 183, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.H.; Aonbangkhen, C.; Wu, H.Y.; Naftaly, J.A.; Tang, S.; O’Meara, T.R.; Woo, C.M. Engineering a Proximity-Directed O-GlcNAc Transferase for Selective Protein O-GlcNAcylation in Cells. ACS Chem. Biol. 2020, 15, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Levine, Z.G.; Fan, C.; Melicher, M.S.; Orman, M.; Benjamin, T.; Walker, S. O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix. J. Am. Chem. Soc. 2018, 140, 3510–3513. [Google Scholar] [CrossRef]
- Joiner, C.M.; Levine, Z.G.; Aonbangkhen, C.; Woo, C.M.; Walker, S. Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. J. Am. Chem. Soc. 2019, 141, 12974–12978. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.H.; Yang, B.; D’Souza, A.K.; Shen, D.; Woo, C.M. Truncation of the TPR domain of OGT alters substrate and glycosite selection. Anal. Bioanal. Chem. 2021, 413, 7385–7399. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.A.; Nosella, M.; Vanama, M.; Ruiz-Arduengo, R.; Forman-Kay, J.D. Exploration of O-GlcNAc-transferase (OGT) glycosylation sites reveals a target sequence compositional bias. bioRxiv 2022. [Google Scholar] [CrossRef]
- Aulak, K.S.; Barnes, J.W.; Tian, L.; Mellor, N.E.; Haque, M.M.; Willard, B.; Li, L.; Comhair, S.C.; Stuehr, D.J.; Dweik, R.A. Specific O-GlcNAc modification at Ser-615 modulates eNOS function. Redox Biol. 2020, 36, 101625. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Liu, H.Y.; Yu, T.J.; Lu, Q.; Zhang, F.L.; Liu, G.Y.; Shao, Z.M.; Li, D.Q. O-GlcNAcylation of MORC2 at threonine 556 by OGT couples TGF-beta signaling to breast cancer progression. Cell Death Differ. 2022, 29, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Qian, M.; Lu, L.; Chen, Y.; Zhang, X.; Wu, Q.; Liu, Y.; Bian, Z.; Yang, Y.; Guo, S.; et al. O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis 2019, 40, 1121–1131. [Google Scholar] [CrossRef]
- Hu, J.Y.; Zhang, D.L.; Liu, X.L.; Li, X.S.; Cheng, X.Q.; Chen, J.; Du, H.N.; Liang, Y. Pathological concentration of zinc dramatically accelerates abnormal aggregation of full-length human Tau and thereby significantly increases Tau toxicity in neuronal cells. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 414–427. [Google Scholar] [CrossRef]
- Liu, X.L.; Hu, J.Y.; Hu, M.Y.; Zhang, Y.; Hong, Z.Y.; Cheng, X.Q.; Chen, J.; Pang, D.W.; Liang, Y. Sequence-dependent abnormal aggregation of human Tau fragment in an inducible cell model. Biochim. Biophys. Acta 2015, 1852, 1561–1573. [Google Scholar] [CrossRef]
- Chatani, E.; Yamamoto, N. Recent progress on understanding the mechanisms of amyloid nucleation. Biophys. Rev. 2018, 10, 527–534. [Google Scholar] [CrossRef]
- Sidhu, A.; Vaneyck, J.; Blum, C.; Segers-Nolten, I.; Subramaniam, V. Polymorph-specific distribution of binding sites determines thioflavin-T fluorescence intensity in alpha-synuclein fibrils. Amyloid 2018, 25, 189–196. [Google Scholar] [CrossRef]
- Pan, K.; Yi, C.W.; Chen, J.; Liang, Y. Zinc significantly changes the aggregation pathway and the conformation of aggregates of human prion protein. Biochim. Biophys. Acta 2015, 1854, 907–918. [Google Scholar] [CrossRef] [PubMed]



| O-GlcNAc Sites | P1Tau | TauP1 | Tau |
|---|---|---|---|
| T76 * | + | ||
| T181 * | +++ | ||
| S184 * | +++ | +++ | +++ |
| S185 | +++ | +++ | +++ |
| S191 | +++ | +++ | + |
| S195 * | ++ | ||
| S198 * | ++ | ||
| T205 | ++ | ||
| S208 | +++ | ||
| S241/T245 * | + | ||
| S305 * | +++ | ||
| S316 * | + | ||
| T386 * | + | + | |
| S396 | +++ | +++ | +++ |
| S400 | +++ | +++ | +++ |
| T403 * | + | ||
| S400/T403 * | + | ||
| S412 | +++ | ||
| S413 | +++ | ||
| T414 * | + | ||
| S416 * | +++ | ||
| S422 | ++ | ||
| T427 * | ++ |
| Protein | A + ct | Lag Time (min) | k (h−1) |
|---|---|---|---|
| Tau (no OGT) | 31935.7 ± 1678.8 | 120.3 ± 5.3 | 1.99 ± 0.35 |
| Tau | 37526.3 ± 1634.5 | 138.9 ± 2.2 | 2.36 ± 0.36 |
| P1Tau | 37937.9 ± 1709.7 | 233.1 ± 4.8 | 3.30 ± 0.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yang, Z.; Chen, J.; Chen, Y.; Jiang, C.; Zhong, T.; Su, Y.; Liang, Y.; Sun, H. OGT Binding Peptide-Tagged Strategy Increases Protein O-GlcNAcylation Level in E. coli. Molecules 2023, 28, 2129. https://doi.org/10.3390/molecules28052129
Li Y, Yang Z, Chen J, Chen Y, Jiang C, Zhong T, Su Y, Liang Y, Sun H. OGT Binding Peptide-Tagged Strategy Increases Protein O-GlcNAcylation Level in E. coli. Molecules. 2023; 28(5):2129. https://doi.org/10.3390/molecules28052129
Chicago/Turabian StyleLi, Yang, Zelan Yang, Jia Chen, Yihao Chen, Chengji Jiang, Tao Zhong, Yanting Su, Yi Liang, and Hui Sun. 2023. "OGT Binding Peptide-Tagged Strategy Increases Protein O-GlcNAcylation Level in E. coli" Molecules 28, no. 5: 2129. https://doi.org/10.3390/molecules28052129
APA StyleLi, Y., Yang, Z., Chen, J., Chen, Y., Jiang, C., Zhong, T., Su, Y., Liang, Y., & Sun, H. (2023). OGT Binding Peptide-Tagged Strategy Increases Protein O-GlcNAcylation Level in E. coli. Molecules, 28(5), 2129. https://doi.org/10.3390/molecules28052129






