Abstract
Finding effective antiviral molecular strategies was a main concern in the scientific community when the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged at the end of 2019 as an easily transmissible and potentially deadly β-coronavirus able to cause the coronavirus disease 19 (COVID-19), which famously led to one of the most worrying pandemics in recent times. Other members of this zoonotic pathogenic family were already known before 2019, but apart from the SARS-CoV, which was responsible of severe acute respiratory syndrome (SARS) pandemic in 2002/2003, and Middle East respiratory syndrome coronavirus (MERS-CoV), whose main impact on humans is geographically restricted to Middle Eastern countries, the other human β-coronaviruses known at that time were those typically associated with common cold symptoms which had not led to the development of any specific prophylactic or therapeutic measures. Although SARS-CoV-2 and its mutations are still causing illness in our communities, COVID-19 is less deadly than before and we are returning to normality. Overall, the main lesson learnt after the past few years of pandemic is that keeping our bodies healthy and immunity defenses strong using sport, nature-inspired measures, and using functional foods are powerful weapons for preventing the more severe forms of illness caused by SARS-CoV-2 and, from a more molecular perspective, that finding drugs with mechanisms of action involving biological targets conserved within the different mutations of SARS-CoV-2—and possibly within the entire family of β-coronaviruses—gives more therapeutic opportunities in the scenario of future pandemics based on these pathogens. In this regard, the main protease (Mpro), having no human homologues, offers a lower risk of off-target reactivity and represents a suitable therapeutic target in the search for efficacious, broad-spectrum anti-β-coronavirus drugs. Herein, we discuss on the above points and also report some molecular approaches presented in the past few years to counteract the effects of β-coronaviruses, with a special focus on SARS-CoV-2 but also MERS-CoV.
2. Drug Repositioning
Drug repositioning using previously FDA-approved drugs is a strategy typically employed when a new pathogen emerges in the attempt to treat patients who need urgent cures while waiting for more specific therapies to be developed, which clearly takes a longer time. This was especially true in the context of COVID-19, regarding which countless studies, often based on computational approaches using molecular docking and molecular dynamics simulations, were conducted on different molecular targets of SARS-CoV-2, including the papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp), and SARS-CoV-2 main protease (Mpro), to name only a few. This led to the identification of salinomycin (Figure 1) from Streptomyces albus as a potential inhibitor of SARS-CoV-2 PLpro, while the vegetal toxin ouabain was proposed as a dual inhibitor of the PLpro and Mpro enzymes [10]. By screening 171 candidates obtained from the DrugBank database (http://www.drugbank.ca/ accessed on 17 February 2023), other in silico studies identified possible organic triazole compounds such as bemcentinib, and bisoctrizole, as Mpro inhibitors whose pharmacokinetic characteristics were also evaluated, and their complex stability and conformation were examined using molecular dynamics simulation [11]. Non-steroidal anti-inflammatory drugs, which are frequently used to treat upper airway infections symptomatically, are of crucial importance when administered in the early stages of SARS-CoV-2 infection and, in this context, ketoprofen lysine salt is a non-steroidal anti-inflammatory drug which was suggested to offer notable benefits in early COVID-19 therapy, based on the pharmacodynamic and pharmacokinetic characteristics of this drug [7].
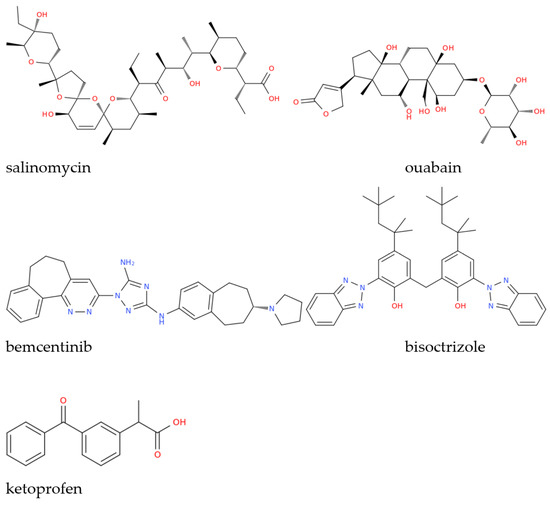
Figure 1.
Structural representation of some of the drugs investigated as anti-COVID-19 therapeutics mentioned in this work.
Author Contributions
C.V. and G.N.R. have contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
Caterina Vicidomini and Giovanni N. Roviello, who served as Guest Editors for the Special Issue ‘Potential Anti-SARS-CoV-2 Molecular Strategies’ of Molecules, are grateful to all the authors for their contributions and to all the expert reviewers involved in the Special Issue.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Z.; Wan, X.; Li, X.; Wan, C. Effects of a Shift of the Signal Peptide Cleavage Site in Signal Peptide Variant on the Synthesis and Secretion of SARS-CoV-2 Spike Protein. Molecules 2022, 27, 6688. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Jamsransuren, D.; Makita, Y.; Kaneko, A.; Matsuda, S.; Ogawa, H.; Oh, H. Inactivation Activities of Ozonated Water, Slightly Acidic Electrolyzed Water and Ethanol against SARS-CoV-2. Molecules 2021, 26, 5465. [Google Scholar] [CrossRef] [PubMed]
- Henri, J.; Minder, L.; Mohanasundaram, K.; Dilly, S.; Goupil-Lamy, A.; Di Primo, C.; Slama Schwok, A. Neuropeptides, New Ligands of SARS-CoV-2 Nucleoprotein, a Potential Link between Replication, Inflammation and Neurotransmission. Molecules 2022, 27, 8094. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, V.; Roviello, G.N. The Potential Role of Vaccines in Preventing Antimicrobial Resistance (AMR): An Update and Future Perspectives. Vaccines 2023, 11, 333. [Google Scholar] [CrossRef]
- Islam, A.; Bashir, M.S.; Joyce, K.; Rashid, H.; Laher, I.; Elshazly, S. An Update on COVID-19 Vaccine Induced Thrombotic Thrombocytopenia Syndrome and Some Management Recommendations. Molecules 2021, 26, 5004. [Google Scholar] [CrossRef] [PubMed]
- Villar, M.; Urra, J.M.; Artigas-Jerónimo, S.; Mazuecos, L.; Contreras, M.; Vaz-Rodrigues, R.; Rodríguez-del-Río, F.J.; Gortázar, C.; de la Fuente, J. Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals. Molecules 2022, 27, 5933. [Google Scholar] [CrossRef] [PubMed]
- Mariniello, D.F.; Allocca, V.; D’Agnano, V.; Villaro, R.; Lanata, L.; Bagnasco, M.; Aronne, L.; Bianco, A.; Perrotta, F. Strategies Tackling Viral Replication and Inflammatory Pathways as Early Pharmacological Treatment for SARS-CoV-2 Infection: Any Potential Role for Ketoprofen Lysine Salt? Molecules 2022, 27, 8919. [Google Scholar] [CrossRef] [PubMed]
- Diniz, L.R.L.; Elshabrawy, H.A.; Souza, M.T.d.S.; Duarte, A.B.S.; Datta, S.; de Sousa, D.P. Catechins: Therapeutic Perspectives in COVID-19-Associated Acute Kidney Injury. Molecules 2021, 26, 5951. [Google Scholar] [CrossRef] [PubMed]
- Bouback, T.A.; Pokhrel, S.; Albeshri, A.; Aljohani, A.M.; Samad, A.; Alam, R.; Hossen, M.S.; Al-Ghamdi, K.; Talukder, M.E.K.; Ahammad, F.; et al. Pharmacophore-Based Virtual Screening, Quantum Mechanics Calculations, and Molecular Dynamics Simulation Approaches Identified Potential Natural Antiviral Drug Candidates against MERS-CoV S1-NTD. Molecules 2021, 26, 4961. [Google Scholar] [CrossRef] [PubMed]
- Qayed, W.S.; Ferreira, R.S.; Silva, J.R.A. In Silico Study towards Repositioning of FDA-Approved Drug Candidates for Anticoronaviral Therapy: Molecular Docking, Molecular Dynamics and Binding Free Energy Calculations. Molecules 2022, 27, 5988. [Google Scholar] [CrossRef] [PubMed]
- Sur, V.P.; Sen, M.K.; Komrskova, K. In Silico Identification and Validation of Organic Triazole Based Ligands as Potential Inhibitory Drug Compounds of SARS-CoV-2 Main Protease. Molecules 2021, 26, 6199. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.P.; Terracciano, M.; Oliviero, G.; Roviello, G.N.; Borbone, N. Exploring the Relationship between G-Quadruplex Nucleic Acids and Plants: From Plant G-Quadruplex Function to Phytochemical G4 Ligands with Pharmaceutic Potential. Pharmaceutics 2022, 14, 2377. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.; Musumeci, D.; Borbone, N.; Falanga, A.P.; D’Errico, S.; Terracciano, M.; Piccialli, I.; Roviello, G.N.; Oliviero, G. Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin. Molecules 2022, 27, 2997. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Roviello, G.N. Exploring the Protective Effect of Food Drugs against Viral Diseases: Interaction of Functional Food Ingredients and SARS-CoV-2, Influenza Virus, and HSV. Life 2023, 13, 402. [Google Scholar] [CrossRef]
- Adarshan, S.; Akassh, S.; Avinash, K.; Bharathkumar, M.; Muthuramalingam, P.; Shin, H.; Baskar, V.; Chen, J.-T.; Bhuvaneshwari, V.; Ramesh, M. Transcriptomics, Cheminformatics, and Systems Pharmacology Strategies Unveil the Potential Bioactives to Combat COVID-19. Molecules 2022, 27, 5955. [Google Scholar] [CrossRef] [PubMed]
- Eissa, I.H.; Khalifa, M.M.; Elkaeed, E.B.; Hafez, E.E.; Alsfouk, A.A.; Metwaly, A.M. In Silico Exploration of Potential Natural Inhibitors against SARS-Cov-2 nsp10. Molecules 2021, 26, 6151. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Tamura, K.; Jamsransuren, D.; Matsuda, S.; Ogawa, H. Severe Acute Respiratory Syndrome Coronavirus-2 Inactivation Activity of the Polyphenol-Rich Tea Leaf Extract with Concentrated Theaflavins and Other Virucidal Catechins. Molecules 2021, 26, 4803. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, C.; Roviello, V.; Roviello, G.N. Molecular Basis of the Therapeutical Potential of Clove (Syzygium aromaticum L.) and Clues to Its Anti-COVID-19 Utility. Molecules 2021, 26, 1880. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).