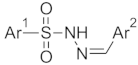
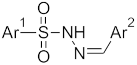
Abstract
Breast cancer (BC) is the second leading cause of cancer death in women, with more than 600,000 deaths annually. Despite the progress that has been made in early diagnosis and treatment of this disease, there is still a significant need for more effective drugs with fewer side effects. In the present study, we derive QSAR models with good predictive ability based on data from the literature and reveal the relationships between the chemical structures of a set of arylsulfonylhydrazones and their anticancer activity on human ER+ breast adenocarcinoma and triple-negative breast (TNBC) adenocarcinoma. Applying the derived knowledge, we design nine novel arylsulfonylhydrazones and screen them in silico for drug likeness. All nine molecules show suitable drug and lead properties. They are synthesized and tested in vitro for anticancer activity on MCF-7 and MDA-MB-231 cell lines. Most of the compounds are more active than predicted and show stronger activity on MCF-7 than on MDA-MB-231. Four of the compounds (1a, 1b, 1c, and 1e) show IC50 values below 1 μM on MCF-7 and one (1e) on MDA-MB-231. The presence of an indole ring bearing 5-Cl, 5-OCH3, or 1-COCH3 has the most pronounced positive effect on the cytotoxic activity of the arylsulfonylhydrazones designed in the present study.
1. Introduction
Breast cancer (BC) is the most common cancer among women worldwide, accounting for 25% of all cancers, and is the second leading cause of cancer death in women after lung cancer [1]. In 2020, an estimated 2.3 million new cases of breast cancer were diagnosed globally, and 627,000 women died from the disease [1]. The most recent data from the American Cancer Society estimate that about one in eight (12%) women in the United States will develop invasive breast cancer at some point in their lives [2].
BC is categorized into three major types based on its molecular characteristics: hormone-based BC (estrogen receptor (ER+) or progesterone receptor (PR+)), human epidermal receptor 2-expressing (HER2+) BC, and triple-negative (ER−, PR−, and HER2−) BC (TNBC) [3]. The type of BC determines the therapeutic approach. The treatment of hormone-based BC involves hormone therapy, which works by inhibiting the production or action of hormones that fuel the growth of cancer cells. Some common types of hormone therapy for BC include tamoxifen (a selective estrogen receptor modulator (SERM) that blocks the effects of estrogen on BC cells) [4] and aromatase inhibitors (a class of drugs that block the production of estrogen by inhibiting the enzyme aromatase) [5]. The CDK4/6 inhibitors (which block the activity of the cyclin-dependent kinases 4 and 6, which play a role in the regulation of the cell cycle) [6], HER2 inhibitors (which block the activity of the HER2 protein, which is overexpressed in some types of BC) [7], and luteinizing hormone-releasing hormone (LHRH) agonists (a class of drugs that lower estrogen levels by inhibiting the production of luteinizing hormone, which is needed for the ovaries to produce estrogen) [8] also have found a place in BC therapy. In addition to hormone therapy, other treatment options for hormone-based BC may include chemotherapy, radiation therapy, surgery, and targeted therapies that block the signaling pathways that promote cancer cell growth [9]. More problematic is the treatment of TNBC, which accounts for 10–15% of BC cases and is characterised by limited possibilities for targeted therapy, as TNBC cells do not overexpress estrogen, progesterone, or HER2/neu receptors. The standard treatment for TNBC typically involves a combination of conventional chemotherapy, surgery, and radiation therapy [10].
Despite the advances in BC therapy, the need for new and more effective drugs with fewer side effects remains. Some promising areas of research in this field include targeted therapies with improved cancer selectivity, that are aimed to specifically target the cancer cells while minimizing harm to normal cells [9], and immunotherapies, which help to boost the body’s own immune system to fight the cancer [11].
Recently, two research groups have independently developed novel arylsulfonylhydrazones as anticancer agents against human BC cells. Senkardes et al. have synthesized and tested a series of sulphonyl hydrazones with anticancer activity on human breast adenocarcinoma cell line MCF-7 and prostate cancer cell line PC-3 [12]. The anticancer activities were in the micromolar range and the selectivity index (SI = IC50 on non-cancer cells/IC50 on cancer cells) has reached 432 for some of the compounds. Additionally, good cyclooxigenase-2 (COX-2) inhibitory activity has been found in vitro for some of the hydrazones. COX-2 is a proinflammatory enzyme and is overexpressed in solid tumours such as BC and prostate cancer. Gaur et al. have synthesized and tested a series of arylsulfonylhydrazones with indole and morpholine moieties [13]. The compounds have shown anticancer activity on MCF in micromolar concentrations, with a SI up to 60. Furthermore, the compounds have been active on the TNBC cell line MDA-MB-468 with IC50 in the lower micromolar range and with a SI up to 37.
In the present study, we analyse the available data for arylsulfonylhydrazones by Quantitative Structure-Activity Relationship (QSAR) modelling. QSAR modelling is a computational technique that has proven to be valuable in the field of anticancer research. QSAR models use mathematical algorithms to analyse and predict the biological activity of chemicals based on their molecular structure. This information can then be used to identify new, promising compounds for further study and development as potential anticancer drugs. Several studies have demonstrated the utility of QSAR in anticancer research by identifying new candidate compounds for specific cancer targets and by facilitating the design of more selective and effective drugs [14,15,16]. In addition, QSAR can provide insights into the molecular mechanisms underlying a compound’s activity, which can help guide the optimization of its structure for improved efficacy and safety [17]. Overall, QSAR modelling represents a powerful tool in the discovery and development of novel anticancer drugs.
We utilize the most effective QSAR models derived in the present study to design a set of potential new anticancer agents. These compounds undergo in silico screening for drug likeness, and the most promising ones are subsequently synthesized and evaluated in vitro on breast cancer cell lines (Figure 1).
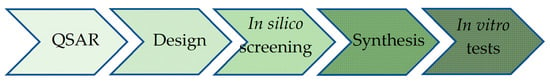
Figure 1.
Flowchart of the present study. QSAR models were derived based on the literature data; the best models were used to design a series of potential new anticancer agents; the compounds were screened in silico for drug likeness and ADME properties; the most prospective ones were synthesized and tested in vitro on BC cell lines.
2. Results
2.1. Quantitative Structure–Activity Relationship (QSAR) Models for Arylsulfonylhydrazones as Breast Anticancer Agents
Two sets of arylsulfonylhydrazones were collected from the literature [12,13] and used as a training set for the derivation of QSAR models. The compounds and their anticancer activities, expressed as ligand efficiency (LE), are given in Table 1. LE measures the ligand activity per non-hydrogen atom and is calculated according to:
where pIC50 is the negative decimal logarithm of IC50 and N is the number of non-hydrogen atoms in the molecule. The LE values ranged from 0.105 to 0.207 and from 0.110 to 0.170 for the activities on MCF-7 and MDA-MB-468, respectively.

Table 1.
Training set used in the study for the derivation of QSAR models. Compounds 3a–o are collected from Senkardes et al. [12], and compounds 5a–k—from Gaur et al. [13]. LE stands for Ligand Efficiency. The anticancer activities of the compounds are measured on human breast adenocarcinoma cell line MCF-7 (n = 26) and on the TNBC cell line MDA-MB-468 (n = 11).
The structures were optimized and described by 70 molecular descriptors, as explained in Materials and Methods. The most relevant descriptors were selected by a genetic algorithm using software tool MDL QSAR v.2.2 (MDL Information Systems Inc., 2004). All possible subset regressions among the selected descriptors were calculated and only models with r2 ≥ 0.6 and q2 ≥ 0.4 were considered.
The best model for anticancer activity on cell line MCF-7 is given below:
where n = 26; r2 = 0.796; SEE = 0.014; q2 = 0.647; CVRSS = 0.007; r2random(mean) = 0.155, morph is a user-defined indicator differentiating the two subsets in the training set; SaaaC_acnt accounts for the number of aromatic aaaC-atoms in the molecule; SaaN_acnt corresponds to the number of aromatic aaN-atoms; ka1 is first order kappa alpha shape index; r2—goodness of fit, SEE—standard error of estimation, q2—leave-one-out cross validation coefficient; CVRSS—cross validation residual sum of squares, and r2random(mean)—the mean value of r2random values calculated for 100 randomizations of the dependent variable among the compounds.
LE (MCF-7) = −0.004 × morph + 0.015 × SaaaC_acnt −0.029 × SaaN_acnt −0.012 × ka1 + 0.367
The values of the descriptors relevant to the cytotoxic activity on MCF-7 are given in Table S1, Supplementary Material. The indicator morph takes 1 for the subset 5a–k and 0 for the subset 3a–o. The negative coefficient for morph means that the substituent 1-(4-morpholinylethyl)-1H- indol-3-yl in 5a–k is not favourable for LE on MCF-7. The descriptor SaaaC_acnt varies from 0 (for 3a–o) to 2 (for most of 5a–k) and 4 (for 5h and 5i, containing fused rings). Its coefficient in the model is positive, i.e., more aaaC-atoms in the molecule correspond to better anticancer activity. The descriptor SaaN_acnt takes value 1 for 3h and 5i, containing pyrazolyl and quinolyl, respectively. For the rest of the compounds, SaaN_acnt takes the value 0. As its coefficient is negative, the presence of aromatic N-atoms of type aaN is not essential for the anticancer activity. The kappa shape indices account for the molecular shape [18]. A higher value for ka1 corresponds to more branched molecules (more paths). In the training set, the values for ka1 vary from 13.666 for 3n to 25.609 for 5h. The average ka1 for the subset 3a–o is 16.701, for the subset 5a–k—22.421. The negative coefficient for ka1 favors the less branched molecules.
The QSAR model for cytotoxic activity on cell line MDA-MB-468 was derived only on the compounds from the subset 5a–k. The best model is given below:
where n = 11; r2 = 0.979; SEE = 0.003; q2 = 0.931; CVRSS = 0.0002; r2random(mean) = 0.155, nelem is the number of chemical elements in the molecule and nvx accounts for the number of graph vertices. The values of the descriptors relevant to the cytotoxic activity on MDA-MB-468 are given in Table S1. The number of elements in the molecules 5a–k is five (C, O, N, S, and H); only 5d has an additional F and 5f has an additional Cl. As the coefficient for nelem is positive, obviously, the presence of F and Cl favours the cytotoxic activity. The range of nvx values is from 29 for 5c to 39 for 5h and 5i. The negative coefficient means that the bulky branched substituents are not favourable for the activity on MDA-MB-468.
LE (MDA-MB-468) = 0.020 × nelem − 0.004 × nvx + 0.151
The structure–activity relationships found in the derived QSAR models are used next in the design of novel arylsulfonylhydrazones with anticancer activity.
2.2. Design of Novel Arylsulfonylhydrazones Based on QSAR Models
The requirements obtained from the above QSAR models were implemented in the design of novel arylsulfonylhydrazones as anticancer agents, i.e.:
- For Ar1: Single aromatic rings
- For Ar2: Aromatic rings containing aaaC and Cl but no aaN.
The structures of the designed molecules are given in Table 2. For Ar1, we selected phenyl or 4-methylphenyl substituents. The N-tosyl hydrazones (p-Me-Ph-SO2-NH-N=Ar(R)) are a special class of hydrazones with proven anticancer activity against TNBC cell lines [19].

Table 2.
Newly designed arylsulfonylhydrazones. The LE values for MCF-7 and MDA-MB-468 cell lines are predicted by the QSAR models derived in the study. The experimental LE values are obtained in vitro by MTT tests.
For Ar2, we selected indole and phenyl substituents. The indole ring possesses anti-BC activity [20] due several different signaling pathways [21]. The indole system contains aaaC and no aaN. The N-atoms in the selected indole substituents are aaNH or daaN, with slight NH-acidic (pKa 16.2) properties. Six of the nine new hydrazones contain mono-substituted indole moiety (compounds 1a–e, 1i). For comparison, we included three compounds with bi-substituted phenyl rings (compounds 1f–h). Two of the compounds contain the favourable Cl atom (compounds 1e and 1h).
The LE values of the designed compounds were predicted by the derived models. All of them are close to or higher than the maximum LE of the compounds from the training set on both cell lines. At this stage of the study, all designed compounds appeared to be prospective anticancer agents.
2.3. In Silico Screening of the Designed Compounds for Drug Likeness
Prior to synthesis, the designed structures were screened in silico for drug likeness considering their physicochemical and ADME properties and pharmacokinetic (PK) parameters.
2.3.1. Physicochemical Properties
The main physicochemical properties calculated for the designed arylsulfonylhydrazones are given in Table 3. They are molecular weight, Mw; pKa value; fraction of the ionized molecules, fA; logP; distribution coefficient at pH 7.4 logD7.4; polar surface area, PSA; count of free rotatable bonds, FRB; hydrogen bond donors, HBD; hydrogen bond acceptors, HBA; count of the violations of Lipinski’s Rule of 5, R5.

Table 3.
Physicochemical properties of the designed arylsulfonyl hydrazones: Mw—molecular weight; pKa, fA—fraction of the ionized molecules; logP; logD7.4—distribution coefficient at pH 7.4; PSA—polar surface area; FRB—free rotatable bonds; HBD—hydrogen bond donors; HBA—hydrogen bond acceptors; R5—violation of Lipinski’s Rule of 5.
The molecular weights are around 300 g/mol (295–355 g/mol), which is in a good agreement with the recommended Mw for lead compounds [22]. The compounds are weak acids with pKa values between 8.59 and 9.09. At pH 7.4, the neutral molecules dominate as indicated by the negligible fraction of ionized molecules fA and the close values between logP and logD7.4. The logP values are around 3, which is, again, in good agreement with the requirements for lead compounds. PSAs range from 67 to 92 Å, suggesting good oral absorption and inability to cross the blood–brain barrier (BBB) [23]. The number of free rotatable bonds is between 3 and 5; however, the single bonds in the Ar1–S–N–N = fragment are quite rigid due to p-π conjugation. The number of hydrogen bond donors obeys the ‘Rule of 3’; however, the hydrogen bond acceptors exceed it. Regarding Lipinski’s rule of 5, all compounds meet the four criteria and there is no violation.
2.3.2. ADME Properties
The ADME properties calculated in the study are given in Table 4. The water solubility was calculated by three methods [24] and the average value in mol/L is presented as logS. According to the logS scale [24], compounds with logS between −6 and −4 are considered as moderately soluble, while those with logS between −4 and −2—as soluble. According to the BOILED-Egg diagram [23] (Figure 2), all compounds have good oral permeability, one of them (compound 1h) is able to cross the blood–brain barrier (BBB), and none of the compounds are a substrate of the P-glycoprotein (P-gp) transporter. The parameter oral BA summarizes six criteria which definine the suitable physicochemical space for oral bioavailability [23]. These are lipophilicity (logP), size (Mw), polarity (PSA), solubility (logS), insaturation (fraction of Csp3 atoms), and flexibility (number of rotatable bonds). Each criterion has a certain range. The designed compounds violate in insaturation, i.e., the fraction of Csp3 atoms is below the lower limit of 0.25. This violation was expected as most of the C-atoms in the structures are in sp2-hybridization. The BA score indicates the probability of bioavailability being higher than 10% in rats [25]. In our case, the probability is 55%. The CYP inhibition considers the five enzymes that most-commonly take part in drug metabolism: 1A2, 2C19, 2C9, 2D6, and 3A4. The studied compounds are able to inhibit between 2 and 4 of the CYPs. Apart from following Lipinski’s rule, all compounds demonstrate drug likeness filtered by the criteria of Ghose [26], Veber [27], Egan [28], and Muegge [29]. The lead likeness is defined by three criteria: Mw in the range 250–350 g/mol, logP up to 3.5, and up to 7 rotatable bonds in the molecule [30]. Here, again, our compounds fit well in the ranges. Finally, the synthetic feasibility of the designed compounds was assessed by the synthetic accessibility score, which ranges from 1 (very easy synthesis) to 10 (very difficult synthesis). A score between 2.56 and 2.80 points to relatively easy synthesis.

Table 4.
ADME properties of the designed arylsulfonyl hydrazones: water solubility; GI abs—gastrointestinal absorption; oral BA—oral bioavailability; BA score—bioavailability score; BBB perm—blood—brain barrier permeability; CYP inh—inhibition of CYP enzymes; P-gp substr—substrate of P-gp; drug likeness; lead likeness; synth access—synthetic accessibility.
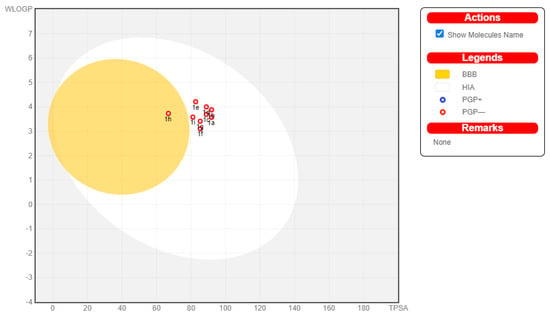
Figure 2.
BOILED-Egg diagram for the designed arylsulfonyl hydrazones. Presence in the white area corresponds to good oral permeability, while presence in the yellow area—to BBB permeability. The red circles indicate that the compounds are not substrates of P-gp.
2.3.3. Pharmacokinetic Parameters
The main pharmacokinetic parameters, fraction of the unbound-to-plasma-proteins molecules, fu; total clearance, CL; steady-state volume of distribution, VDss; half-life, t1/2, of the designed arylsulfonylhydrazones were calcuculated by QSPkR models previously derived in our Lab [31,32,33]. The predicted values are given in Table 5.

Table 5.
PK parameters of the designed arylsulfonyl hydrazones: fu—fraction of the compound unbound to plasma proteins; CL—total clearance in L/h/kg; VDss—steady-state volume of distribution in L/kg; t1/2—half-life in h.
The fu values ranged from 0.010 to 0.074, suggesting high plasma protein binding of all compounds (>90%). It is generally accepted that neutral drugs bind with variable affinity to both human serum albumin and alpha−1-acid glycoprotein [34]. Lipoproteins also contribute to plasma protein binding, especially for highly lipophilic drugs [35].
Total CL values ranged between 0.017 and 0.647 L/h/kg. Most of the compounds can be classified as low CL drugs, while 1c and 1d have medium CL. Analysis of a data set of 754 drugs with different ionization states revealed that 78% of anionic and zwitterionic drugs have low CL (<0.24 L/h/kg) and only 1–2% have high CL (>0.96 L/h/kg). For neutral drugs, these percentages were as follows: 45% low CL, 39% moderate CL, and 16% high CL [36]. Considering the relatively high lipophilicity of the compounds and the negligible ionization at pH 7.4, clearance can be considered to be dominated by metabolism. Neutral drugs have a low renal CLR unless their logD7.4 is negative. For drugs with logD7.4 > 0, the CLR decreases with lipophilicity due to tubular reabsorption [37].
Values for VDss vary between 0.587 and 0.953 L/kg, which is in the order of total body water volume. It is likely that the compounds are evenly distributed throughout the body without significant accumulation in certain tissues and organs.
The half-life (t1/2) is determined by CL and VDss. Therefore, compounds 1a, 1c, 1d, and 1i, with medium CL and/or low VDss, have short t1/2 (0.94–3.07), while compounds 1b, 1e, 1f, 1g, and 1h, with low CL and high VDss, show moderate to long t1/2 (6.20–35.12 h).
2.4. Synthesis of the Novel Arylsulfonyl Hydrazones
The designed compounds showed strong drug and lead likeness in in silico screening procedures and we decided to synthesize and test all of them.
The arylsulfonylhydrazones were prepared by a condensation reaction (Scheme 1) between the corresponding aldehydes and benzenesulfonohydrazide or 4-methylbenzenesulfonohydrazide, at a molar ratio of 1:1, in absolute ethanol for 1–3 h, as described elsewhere [38].
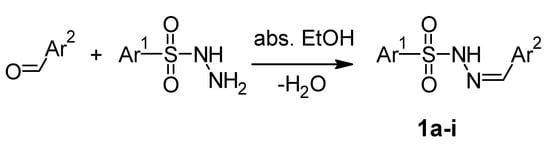
Scheme 1.
Chemical Reaction for preparing arylsulfonylhydrazones.
The structures were confirmed by 1H NMR, 13C NMR, and HRMS spectroscopic data and melting points. The 1H-NMR spectra of 1a–i have single signals corresponding to resonances of azomethine protons (CH=N) at 7.82–8.26 ppm. The hydrazide/hydrazone N/H protons are observed at 11.37–11.93 ppm. The 13C-NMR spectra exhibit resonances arising from azomethine (C=N) from 130.94 to 147.45, respectively (Figures S1–S28 in Supplementary Materials).
2.5. Anticancer Activity of the Novel Arylsulfonyl Hydrazones
The anticancer activity of the novel arylsulfonylhydrazones was tested on two BC cell lines: MCF-7 and MDA-MB-231. The cell line MCF-7 originates from human breast adenocarcinoma and expresses estrogen receptor alpha (ER-α) [39], while the cell line MDA-MB-231 represents TNBC adenocarcinoma and lacks any receptor [40]. To test the cytotoxicity of the compounds on healthy cells, they were incubated within Neuro-2a cells, which are mouse neuroblasts isolated from brain tissue [41,42]. The results from the in vitro tests are summarized in Table 6.

Table 6.
Cytotoxicity of the designed arylsulfonylhydrazones on MCF-7, MDA-MB-231, and Neuro-2a cell lines. LE—ligand efficiency; SI—selectivity index (IC50 on Neuro-2a/IC50 on BC cell line).
The differences (errors) between the experimental and the predicted LE values are given in Table 1. The positive values correspond to underpredicted activity, the negative—to overpredicted activity. The errors range between −0.047 and 0.063 for MCF-7 and from −0.027 to 0.081 for MDA-MB-231. Most of the compounds are more active than expected. Only compounds 1i and 1g are less active on MCF-7 and MDA-MB-231, respectively.
The experimental IC50 values of the novel compounds on MCF-7 range from 0.6 μM to 164.9 μM. The LEs are between 0.158 and 0.286, with an average value of 0.230. For comparison, the average LE of the training set on the same cell line is 0.156 (0.171 for the subset 3a–o and 0.135 for the subset 5a–k) with the highest value being 0.207. The selectivity index SI is defined as the ratio of IC50 on healthy cells and IC50 on cancer cells. A SI higher than 10 is considered to belong to a selective compound [43]. The SIs of the novel compounds span from 0.747 to 46 on MCF-7. Four of the nine compounds show cytotoxic activities on MCF-7 below 1 μM. These are compounds 1e, 1a, 1b, and 1c. The most efficient compounds on MCF-7 are 1e and 1a, while the most selective are compounds 1d and 1c.
The most active, efficient, and selective compound on MDA-MB-231 is 1e, with an IC50 of 0.9 μM, LE of 0.275, and SI of 7.222. Compounds 1a, 1b, and 1c have IC50s in the lower micromolar range with LEs around and above 0.2; however, they have low SIs.
3. Discussion
Based on data from the literature, QSAR models were obtained in the present study to reveal the relationship between the structures of arylsulfonylhydrazones and their anticancer activity against BC. It was found that, for the activity against ER+ BC, measured on a MCF-7 cell line, a less-branched aromatic substituent with more aaaC-atoms, Cl, and no aaN-atoms performed better as anticancer agents. Less-branched aromatic moieties bearing F and Cl are required for activity against TNBC, as measured in the MDA-MB-231 cell line. These findings were implemented in the design of nine arylsulfonyl hydrazones. The structures contain mono- and/or bi-substituted phenyl and indolyl moieties. Cl atoms were included in two of them. The anticancer activities on both cell lines, expressed as LE, were predicted by the derived QSAR models. All compounds demonstrated higher than or close to the maximal LEs of the compounds from the training set. Prior to synthesis, the structures were screened in silico for drug likeness by calculating their physicochemical and ADME properties and main PK parameters, such as fraction of the unbound to plasma protein molecules, fu; total clearance, CL; steady-state volume of distribution, VDss; and half-life, t1/2. In terms of drug likeness, all nine of the designed compounds were suitable as leads. They were synthesized and tested. The in vitro tests confirmed the predicted activities. What is more, seven and eight of the compounds are more active on MCF-7 and MDA-MB-231, respectively, than predicted. Most of the designed compounds are more active on MCF-7 than on MDA-MB-231. The IC50 values for 1e, 1a, 1b, and 1c on MCF-7 are below 1 μM. On MDA-MB-231, only compound 1e shows activity below 1 μM.
The most active and most efficient compound on both cell lines is 1e, with a SI of 13 for MCF-7 and 7 for MDA-MB-231. It contains a phenyl ring as an Ar1 substituent and 5-chloroindole as an Ar2 subsitituent. Further, 1e obeys drug and lead likeness rules, has high GI absorption, and has no BBB permeability. In terms of PK behavior, 1e is predicted to be extensively bound to plasma proteins (only 1% free fraction), with a total clearance of 5 L/h and a VDss of 54 L for a 70-kg patient, as well as a half-life of 7 h.
The next-most active and efficient arylsulfonylhydrazone on both cell lines is 1a, with a SI of about 9 for MCF-7 and only 1.7 for MDA-MB-231. Further, 1a bears phenyl as Ar1 and 5-methoxyindole as Ar2. This compound is predicted to be a good drug candidate and lead compound in terms of physicochemical and ADME properties, with extensive plasma–protein binding, a total clearance of 13.5 L/h, a VDss of 41 L, and a half-life of 2 h.
Next in activity and efficiency on MCF-7 line are compounds 1b, 1c, 1d, and 1f. Compounds 1c and 1d demonstrate the highest selectivity of 40 and 46, respectively, followed by 1f with a SI of 18. Compounds 1g, 1h, and 1i are less active, efficient, and selective.
For MDA-MB-231, compounds 1b and 1c show activities in the low micromolar range and efficiencies around 0.2; however, they show poor selectivities (up to 2). The remaining compounds are less active and non-selective.
The analysis of substituents shows that the indole ring has the most pronounced positive effect on the cytotoxic activity of the arylsulfonylhydrazones designed in the present study. The substitution of indole by phenyl dramatically reduces the activity on both cell lines (from 10-fold to more than 300-fold on MCF-7 and from 70-fold to complete loss of activity on MDA-MB-231). Among the substituents on the indole ring, 5-Cl, 5-OCH3, and 1-COCH3 increase the activity between 92- and 330-fold on MCF-7 compared with the 1-CH3 substituent. The effects of these substituents on the activity on MDA-MB-231 are moderate. The Cl atom deserves special attention. Attached to an indole moiety, it increases activity 330-fold on MCF-7 and 70-fold on MDA-MB-231 compared to when it is attached to the phenyl ring.
In conclusion, the QSAR-guided strategy for the design of novel arylsulfonylhydrazones with anticancer activity, applied in the present study, generated several prospective leads with IC50 values below 1 μM and SI values up to 46. The newly designed compounds were more active than the compounds from the training set and represent a starting point for further lead optimization.
4. Materials and Methods
4.1. Materials and Reagents
The reagents for the synthesis were analytical or chemically pure and obtained from Sigma–Aldrich (Steinheim, Germany). The solvents used were of analytical grade. The structures of the new molecules were proven by 1H-NMR, 13CNMR, and HRMS spectral data. Their purity was determined by TCL characteristics and melting points.
The in vitro antineoplastic activity of the newly synthesized compounds was evaluated against human BC cell lines of different molecular types: the triple negative MDA-MB-231 cell line and the ER/PR/Her2 positive variant MCF-7, as well as against mouse neuroblast cells, Neuro-2a. All cell lines were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ GmbH, Braunschweig, Germany) and cultivated according to supplier’s instructions. Cells were cultured in an RPMI 1640 growth medium supplemented with 10% fetal bovine serum (FBS) and 5% L-glutamine, and incubated under standard conditions of 37 °C and 5% humidified CO2 atmosphere.
4.2. QSAR Protocol
The training set for the development of QSAR models consisted of 26 compounds. Fifteen compounds were derivatives of 4-methylphenyl hydrazone [12]. The remaining 11 compounds were morpholinylethylindolyl derivatives [13]. The anticancer activities of both subsets were measured in vitro by MTT tests on MCF-7 cell line. The second set was tested on MDA-MB-468 cell line as well. The chemical structures were modeled and optimized by MM+ force field, steepest descent algorithm, and RMS gradient of 0.1 kcal/A.mol using HyperChem 7.52 (Hypercube Inc., Gainesville, FL, USA, 2005).
The chemical structures were described by 70 descriptors divided into eight groups: atom-type E-state indices, atom-type E-state accounts, hydrogen E-state categories, internal H-bonds E-state indices, kappa shape indices, molecular properties (logP, molecular weight, number of elements, number of rings, number of hydrogen-bond donors and acceptors, etc.), 3D descriptors (dipole, polarizability, surface, volume, etc.), and user-defined (morph). The descriptor morph accounts for the presence of an indole-morpholine fragment in the molecule. If an indole-morpholine is presented in the molecule, morph takes 1, otherwise it takes 0. The relevant descriptors were selected by genetic algorithm (GA) at the following settings: size of initial population 32, tournament selection, uniform crossover, one-point mutation, and Friedman’s lack-of-fit scoring function with parameter 2. All possible subset regressions among the selected descriptors were calculated and only models with r2 (goodness of fit) ≥ 0.6 and q2 (leave-one-out cross validation coefficient) ≥ 0.4 were considered. To check the validity of the selected descriptor set, 100 randomizations of the dependent variable among the compounds were carried out and r2random values were calculated for each regression. If the mean value of r2random was lower than r2, the selected descriptor set was considered as valid. QSAR models were derived by MDL QSAR v.2.2 (MDL Information Systems Inc., 2004).
4.3. In Silico Screening for Drug Likeness
The physicochemical properties of the designed compounds were calculated by ACD/LogD tool v. 9.08 (ACD/Labs, Toronto, Canada). The ADME properties were calculated by SwissADME tool [20]. The PK parameters were calculated by previously derived QSPkR models [31,32,33]. As the fraction of the ionized molecules of most of the designed arylsulfonylhydrazones was below 3%, the predictions were based on the QSPkR models derived for neutral molecules. Separate QSPkR models have been derived for the fraction of neutral molecules unbound to plasma proteins, fu; unbound clearance of neutral drugs, Clu; and steady state volume of distribution of basic and neutral drugs, VDss. The datasets consisted of 117 neutral molecules or 407 basic and neutral drugs, respectively, extracted from Obach’s database—the largest and best curated source of data for the key pharmacokinetic parameters after iv administration [44]. The chemical structures of the compounds have been encoded by more than 113 to 138 molecular descriptors calculated by ACD/LogD tool v. 9.08 and MDL QSAR version 2.2. Genetic algorithm and step-wise multiple linear regression have been applied for variable selection and model derivation. The QSPkRs have been evaluated by internal and external validation procedures.
4.4. Synthesis
4.4.1. General Information
The nuclear magnetic resonance (NMR) experiments were carried out on a Bruker Avance spectrometer at 600 MHz at 20 °C in deuterated dimethyl sulfoxide (DMSO-d6) as a solvent, and tetramethylsilane (TMS) as an internal standard. The precise assignment of the 1H and 13CNMR spectra was accomplished by measurement of two-dimensional (2D) homonuclear correlation (correlation spectroscopy (COSY)), DEPT-135, and 2D inverse detected heteronuclear (C–H) correlations (heteronuclear single-quantum correlation spectroscopy (HMQC) and heteronuclear multiple bond correlation spectroscopy (HMBC)). Mass spectra were measured on a Q Exactive Plus mass spectrometer (ThermoFisher Scientific) equipped with a heated electrospray ionization (HESI-II) probe (Thermo Scientific, Bremen, Germany). The melting points were determined using a Buchi 535 apparatus and melting point meter M5000 apparatus. We used IUPAC nomenclature for naming of the newly synthesized compounds.
4.4.2. General Procedure for the Synthesis of the Compounds 1a–i
The solution of 20 mmol of the corresponding carbonyl compounds in 10 mL of absolute ethanol was mixed with a hot solution of 20 mmol (60 °C) benzenesulfonohydrazide or 4-methylbenzenesulfonohydrazide in 10 mL of absolute ethanol and stirred for 1–3 h. Upon cooling, the obtained crystalline precipitates were filtered, washed with ethanol-ether, recrystallized from ethanol, and dried. The new compounds were colorless, white, and light-yellow crystalline solids, stable at normal conditions and soluble in methanol, acetonitrile, and DMSO; poorly soluble in water and ethanol.
N’-[(Z)-(5-methoxy-1H-indol-3-yl)methylidene]benzenesulfonohydrazide, 1a
Yellow solid. Yield: 90%; m.p. 174–175 °C. 1H NMR (600 MHz, DMSO-d6) δ 3.74 (s, 3H, CH3), 6.79 (dd, J = 2.6, 8.8 Hz, 1H, H-6), 7.28 (d, J = 8.8 Hz, 1H, H-7), 7.43 (d, J = 2.5 Hz, 1H, H-4), 7.60 (tt, J = 1.7, 7.1 Hz, 2H, H-m), 7.64 (tt, J = 1.8, 7.3 Hz, 1H, H-p), 7.67 (d, J = 2.8 Hz, 1H, H-2), 7.93 (td, J = 1.6, 6.5 Hz, 2H, H-o), 8.08 (s, 1H, CH=N), 10.94 (s, 1H, NH), 11.40 (d, J = 2.0 Hz, 1H, NH-indol). 13C NMR (151 MHz, DMSO-d6) δ 55.15 (CH3), 103.03 (C-4) 110.77 (C-3), 112.55 (C-7), 112.65 (C-6), 124.46 (C-3a), 127.35 (C-o), 129.08 (C-m), 130.95 (C-2), 131.79 (C-7a), 132.85 (C-p), 139.17 (C-i), 145.82 (CH=N), 154.38 (C-5). HREIMS m/z [M + H]+ 330.090688 (calcd for C16H15N3O3S, [M + H]+ 330.09057).
N’-[(E)-(5-methoxy-1H-indol-3-yl)methylidene]-4-methylbenzenesulfonohydrazide,1b
Light-yellow solid. Yield: 86%; m.p. 201–202 °C. 1H NMR (DMSO-d6) δ(ppm): 2.34 (s, 3H, CH3), 3.75 (s, 3H, OCH3), 6.79 (dd, J = 2.6, 8.8 Hz, 1H, H-6), 7.28 (d, J = 8.8 Hz, 1H, H-7), 7.39 (d, J = 8.0 Hz, 2H, H-3′ and H-5′), 7.45 (d, J = 2.5 Hz, 1H, H-4), 7.65 (d, J = 2.8 Hz, 1H, H-2), 7.81 (d, J = 8.3 Hz, 2H, H-2′ and H-6′), 8.06 (s, 1H, CH=N), 10.82 (s, 1H, NH), 11.37 (s, 1H, NH-indol). NOESY: between H-2′(H-6′) and NH-N; CH and NH-N; CH and H-2(H-4), which proves the E orientation. 13C NMR (DMSO-d6) δ(ppm): 20.94 (CH3), 55.10 (OCH3), 103.07 (C-4), 110.81 (C-3), 112.49 (C-7), 112.62 (C-6), 124.45 (C-3a, s), 127.36 (C-2′ and C-6′), 129.44 (C-3′ and C-5′), 130.79 (C-2), 131.77 (C-7a), 136.29 (C-1′), 143.15 (C-4′), 145.52 (CH=N), 154.35 (C-5). HREIMS m/z [M + H]+ 344.10547 (calcd for C17H17N3O3S, [M + H]+ 344.106338).
N’-[(E)-(1-acetyl-1H-indol-3-yl)methylidene]benzenesulfonohydrazide, 1c
Light-yellow solid. Yield: 89%; m.p. 209–210 °C. 1H NMR (DMSO-d6) δ(ppm): 2.63 (s, 3H, CH3), 7.33–7.40 (m, 2H, H-5 and H-6), 7.59–7.66 (m, 3H, H-3′, H4′ and H-5′), 7.93 (dd, J = 1.7, 7.9 Hz, 2H, H-2′ and H-6′), 8.05 (dd, J = 1.6, 7.7 Hz, 1H, H-4), 8.10 (s, 1H, H-2), 8.26 (s, 1H, CH=N), 8.32 (dd, J = 1.4, 6.7 Hz, 1H, H-7), 11.47 (s, 1H, NH-indol). 13C NMR (DMSO-d6) δ(ppm): 23.78 (CH3), 115.87 (C-4), 116.17 (C-3), 122.00 (C-7), 124.21 (C-5), 125.69 (C-6), 126.36 (C-3a), 127.21 (C-2′ and C-6′), 129.21 (C-3′ and C-5′), 131.08 (CH=N), 133.05 (C-4′), 135.69 (C-7a), 138.90 (C-1′), 142.61 (C-2), 169.56 (C=O). HREIMS m/z [M + H]+ 342.08989 (calcd for C17H15N3O3S, [M + H]+ 342.090688).
N’-[(E)-(1-acetyl-1H-indol-3-yl)methylidene]-4-methylbenzenesulfonohydrazide, 1d
Light-yellow solid. Yield: 87%; m.p. 211–212 °C. 1H NMR (DMSO-d6) δ(ppm): 2.33 (s, 3H, CH3), 2.63 (s, 3H, COCH3), 7.33–7.40 (m, 2H, H-5 and H-6), 7.40 (d, J = 8.0 Hz, 2H, H-3′ and H-5′), 7.81 (d, J = 8.3 Hz, 2H, H-2′ and H-6′), 8.07 (dd, J = 1.7, 5.8 Hz, 1H, H-4), 8.08 (s, 1H, CH=N), 8.25 (s, 1H, H-2), 8.32 (dd, J = 1.6, 6.4 Hz, 1H, H-7), 11.39 (bs, 1H, NH-indol). 13C NMR (DMSO-d6) δ(ppm): 20.96 (CH3), 23.79 (COCH3), 115.87 (C-4), 116.26 (C-3), 122.06 (C-7), 124.20 (C-5), 125.68 (C-6), 126.40 (C-3a), 127.26 (C-2′ and C-6′), 129.61 (C-3′ and C-5′), 130.94 (CH=N), 135.70 (C-7a), 136.08 (C-1′), 142.34 (C-2), 143.42 (C-4′), 169.56 (C=O). HREIMS m/z [M + H]+ 356.10542 (calcd for C18H17N3O3S, [M + H]+ 356.106338).
N’-[(E)-(5-chloro-1H-indol-3-yl)methylidene]benzenesulfonohydrazide, 1e
Yellow solid. Yield: 81%; m.p. 183–184 °C. 1H NMR (600 MHz, DMSO-d6) δ 7.17 (dd, J = 2.1, 8.6 Hz, 1H, H-6), 7.41 (d, J = 8.6 Hz, 1H, H-7), 7.62 (t, J = 7.3 Hz, 2H, H-m), 7.66 (t, J = 7.2 Hz, 1H, H-p), 7.80 (d, J = 2.7 Hz, 1H, H-2), 7.89 (d, J = 2.0 Hz, 1H, H-4), 7.92 (d, J = 7.0 Hz, 2H, H-o), 8.08 (s, 1H, CH=N), 11.05 (s, 1H, NH), 11.70 (bs, 1H, NH-indol). 13C NMR (151 MHz, DMSO-d6) δ 110.68 (C-3), 113.47 (C-7), 120.73 (C-4), 122.56 (C-6), 125.02 (C-5), 125.09 (C-3a), 127.37 (C-o), 129.14 (C-m), 131.94 (C-2), 133.02 (C-p), 135.36 (C-7a), 138.98 (C-i), 144.88 (CH=N). HREIMS m/z [M + H]+ 334.041151 (calcd for C15H12ClN3O2S, [M + H]+ 334.04123).
N’-[(E)-(3,4-dimethoxyphenyl)methylidene]benzenesulfonohydrazide,1f
White solid. Yield: 87%; m.p. 150–152 °C. 1H NMR (600 MHz, DMSO-d6) δ 3.76 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 6.95 (d, J = 8.4 Hz, 1H, H-5), 7.08 (dd, J = 1.9, 8.3 Hz, 1H, H-6), 7.12 (d, J = 1.9 Hz, 1H, H-2), 7.61 (tt, J = 1.6, 7.5 Hz, 2H, H-m), 7.66 (tt, J = 1.7, 11.1 Hz, 1H, H-p), 7.83 (s, 1H, CH=N), 7.88 (td, J = 2.1, 7.7 Hz, 2H, H-o), 11.33 (s, 1H, NH). 13C NMR (151 MHz, DMSO-d6) δ 55.42 (OCH3), 55.56 (OCH3), 108.58 (C-2), 111.49 (C-5), 121.00 (C-6), 126.36 (s, 1C), 127.26 (C-o), 129.21 (C-m), 133.05 (C-p), 139.01 (C-i), 147.45 (CH=N), 148.90 (C-3), 150.66 (C-4). HREIMS m/z [M + H]+ 321.090353 (calcd for C15H16N2O4S, [M + H]+ 321.0895).
N’-[(E)-(3,4-dimethoxyphenyl)methylidene]-4-methylbenzenesulfonohydrazide, 1g
White solid. Yield: 82%; m.p. 174–175 °C. 1H NMR (DMSO-d6) δ(ppm): 2.36 (s, 3H, CH3), 3.76 (s, 6H, OCH3), 6.95 (d, J = 8.3 Hz, 1H, H-5), 7.08 (dd, J = 1.9, 8.3 Hz, 1H, H-6), 7.12 (d, J = 1.8 Hz, 1H, H-2), 7.40 (d, J = 8.1 Hz, 2H, H-3′ and H-5′), 7.76 (d, J = 8.3 Hz, 2H, H-2′ and H-6′), 7.82 (s, 1H, CH=N), 11.21 (s, 1H, NH). 13C NMR (DMSO-d6) δ(ppm): 20.98 (CH3), 55.41 (OCH3), 55.54 (OCH3), 108.62 (C-2), 111.50 (C-5), 120.92 (C-6), 126.41 (C-1), 127.27 (C-2′ and C-6′), 129.57 (C-3′ and C5′), 136.14 (C-1′), 143.36 (C-4′), 147.18 (CH=N), 148.89 (C-3), 150.61 (C-4). HREIMS m/z [M + H]+ 335.10518 (calcd for C16H18N2O4S, [M+H]+ 335.106003).
N’-[(E)-(4-chlorophenyl)methylidene]benzenesulfonohydrazide,1h
White solid. Yield: 80%; m.p. 161–163 °C. 1H NMR (600 MHz, DMSO-d6) δ 7.45 (td, J = 2.2, 9.1 Hz, 2H, H-3 and H-5), 7.58 (td, J = 2.2, 9.1 Hz, 2H, H-2 and H-6), 7.61 (tt, J = 1.5, 7.2 Hz, 2H, H-m), 7.67 (tt, J = 1.6, 7.1 Hz, 1H, H-p), 7.88 (td, J = 1.5, 6.6 Hz, 2H, H-o), 7.91 (s, 1H, CH=N), 11.64 (s, 1H, NH). 13C NMR (151 MHz, DMSO-d6) δ 145.90 (CH=N), 127.18 (C-o), 128.44 (C-2 and C-6), 128.93 (C-3 and C-5), 129.32 (C-m), 132.56 (C-1), 133.16 (C-p), 134.60 (C-4), 138.95 (C-i). HREIMS m/z [M + H]+ 295.030252 (calcd for C19H17NO4, [M + H]+ 295.03044).
N’-[(E)-(5-methoxy-1-methyl-1H-indol-3-yl)methylidene]benzenesulfonohydrazide, 1i
Yellow solid. Yield: 83%; m.p. 190–191 °C. 1H NMR (600 MHz, DMSO-d6) δ 3.73 (s, 3H, NCH3), 3.75 (s, 3H, OCH3), 6.85 (dd, J = 2.6, 8.9 Hz, 1H, H-6), 7.35 (d, J = 8.9 Hz, 1H, H-7), 7.63 (tt, J = 1.4, 7.4 Hz, 1H, H-p), 7.64 (s, 1H, H-2), 10.93 (s, 1H, NH-indol), 7.44 (d, J = 2.5 Hz, 1H, H-4), 7.59 (tt, J = 1.7, 7.3 Hz, 2H, H-m), 7.92 (dd, J = 1.5, 7.0 Hz, 2H, H-o), 8.05 (s, 1H, CH=N). 13C NMR (151 MHz, DMSO-d6) δ 32.94 (NCH3), 55.21 (OCH3), 103.21 (C-4), 109.62 (C-3), 111.09 (C-7), 112.58 (C-6), 124.87 (C-3a), 127.34 (C-o), 154.68 (C-5), 129.09 (C-m), 132.52 (C-7a), 132.86 (C-p), 134.39 (C-2), 139.16 (C-i), 145.35 (CH=N). HREIMS m/z [M + H]+ 344.106338 (calcd for C17H17N3O3S, [M + H]+ 344.10625).
4.5. In Vitro Anticancer Activity
4.5.1. MTT Method
The cytostatic activity of the experimental compounds was investigated using an established methodology for assessing cell viability known as the Mosmann MTT method [45]. The assay is colorimetric and measures the activity of mitochondrial enzymes by reducing the yellow dye MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) to violet formazan crystals. Exponential-phased cells were harvested and seeded (100 μL/well) in 96-well plates at 1.5 × 105 density and incubated for 24 h. Cell cultures were treated and exposed to various concentrations (200–6.25 μM) of the tested compounds for 72 h, following which cell survival was quantified as percentage (%) relative to untreated control (100% cell viability).
4.5.2. Statistical Methods
Experimental data were processed using nonlinear regression analysis in the GraphPad Prism® software program. Semi-logarithmic “dose-response” curves were plotted and half-inhibitory concentrations (IC50) of the screened compounds were calculated for each of the tested tumor cell lines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28052058/s1, Figures S1–S28: Analytical data for the synthesized compounds; Table S1: Values of the descriptors relevant for the cytotoxic activity on MCF-7 and MDA-MB-468 of arylsulfonylhydrazones from the training set and the designed compounds.
Author Contributions
Conceptualization, V.T.A.; methodology, V.T.A. and I.D.; software, Z.Z. and I.D.; validation, Z.Z. and I.D.; investigation, V.T.A., T.T., R.M., N.V., B.P., Z.Z., and I.D.; writing—original draft preparation, I.D.; writing—review and editing, V.T.A., T.T., R.M., N.V., B.P., Z.Z. and I.D.; visualization, I.D.; supervision, I.D.; project administration, I.D.; funding acquisition, I.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Bulgarian national plan for recovery and resilience through the Bulgarian National Science Fund, grant number BG-RRP-2.004-0004-C01. The in silico calculations were performed in the Centre of Excellence for Informatics and ICT supported by the Science and Education for Smart Growth Operational Program and co-financed by the European Union through the European Structural and Investment funds (Grant No. BG05M2OP001-1.001-0003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Sample Availability
Samples of the compounds 1a–i are available from the authors.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA A Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Clemons, M.; Danson, S.; Howell, A. Tamoxifen (Nolvadex): A review. Cancer Treat. Rev. 2002, 28, 165–180. [Google Scholar] [CrossRef]
- Riemsma, R.; Forbes, C.A.; Kessels, A.; Lykopoulos, K.; Amonkar, M.M.; Rea, D.W.; Kleijnen, J. Systematic review of aromatase inhibitors in the first-line treatment for hormone sensitive advanced or metastatic breast cancer. Breast Cancer Res. Treat. 2010, 123, 9–24. [Google Scholar] [CrossRef]
- Husinka, L.; Koerner, P.H.; Miller, R.T.; Trombatt, W. Review of cyclin-dependent kinase 4/6 inhibitors in the treatment of advanced or metastatic breast cancer. J. Drug Assess. 2020, 10, 27–34. [Google Scholar] [CrossRef]
- Schlam, I.; Swain, S.M. HER2-positive breast cancer and tyrosine kinase inhibitors: The time is now. Breast Cancer 2021, 7, 56. [Google Scholar] [CrossRef]
- Hackshaw, A. Luteinizing hormone-releasing hormone (LHRH) agonists in the treatment of breast cancer. Expert Opin. Pharmacother. 2009, 10, 2633–2639. [Google Scholar] [CrossRef]
- Masoud, V.; Pagès, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef]
- MacDonald, I.; Nixon, N.A.; Khan, O.F. Triple-Negative Breast Cancer: A Review of Current Curative Intent Therapies. Curr. Oncol. 2022, 29, 4768–4778. [Google Scholar] [CrossRef]
- Henriques, B.; Mendes, F.; Martins, D. Immunotherapy in breast cancer: When, how, and what challenges. Biomedicines 2021, 9, 1687. [Google Scholar] [CrossRef]
- Şenkardeş, S.; Han, M.İ.; Kulabaş, N.; Abbak, M.; Çevik, Ö.; Küçükgüzel, İ.; Küçükgüzel, Ş.G. Synthesis, molecular docking and evaluation of novel sulfonyl hydrazones as anticancer agents and COX-2 inhibitors. Mol. Divers. 2020, 24, 673–689. [Google Scholar] [CrossRef]
- Gaur, A.; Peerzada, M.N.; Khan, N.S.; Ali, I.; Azam, A. Synthesis and anticancer evaluation of novel indole based arylsulfonylhydrazides against human breast cancer cells. ACS Omega 2022, 7, 42036–42043. [Google Scholar] [CrossRef]
- Vilar, S.; Poater, A.; Bofill, R.; Solans, X. QSAR models for the prediction of cytotoxicity of a diverse set of chemicals. Toxicol. Vitr. 2010, 24, 1611–1620. [Google Scholar]
- Jain, S.; Kumar, R.; Lal, B. In silico prediction of anticancer activity of indole derivatives using molecular docking and molecular dynamics simulation. J. Comput. Aided Mol. Des. 2013, 27, 421–429. [Google Scholar]
- Tong, W.; Li, J.; Wang, X.; Zhang, Y. QSAR study on the anticancer activity of indole derivatives. J. Mol. Graph. Model 2015, 57, 107–115. [Google Scholar]
- Martínez, J.; García-Ruiz, C.; Gilarranz, M.A. QSAR modeling of anticancer activity of indole derivatives. J. Chem. Inf. Model 2012, 52, 1648–1658. [Google Scholar]
- Kier, L.B. A Shape Index from Molecular Graphs. Quant. Struct. Act. Relat. 1985, 4, 109–116. [Google Scholar] [CrossRef]
- Xie, Z.; Song, Y.; Xu, L.; Guo, Y.; Zhang, M.; Li, L.; Chen, K.; Liu, X. Rapid synthesis of N-tosylhydrazones under solvent-free conditions and their potential application against human triple-negative breast cancer. ChemistryOpen 2018, 7, 977–983. [Google Scholar] [CrossRef]
- Sidhu, J.S.; Singla, R.; Mayank, J.V. Indole derivatives as anticancer agents for breast cancer therapy: A review. Anticancer Agents Med. Chem. 2015, 16, 160–173. [Google Scholar] [CrossRef]
- Dhiman, A.; Sharma, R.; Singh, R.K. Target-based anticancer indole derivatives and insight into structure–activity relationship: A mechanistic review update (2018–2021). Acta Pharm. Sin. B 2022, 12, 3006–3027. [Google Scholar] [CrossRef]
- Congreve, M.; Carr, R.; Murray, C.; Jhoti, H. A rule of three for fragment-based lead discovery. Drug Discov. Today 2003, 8, 876–877. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Martin, Y.C. A bioavailability score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Teague, S.J.; Davis, A.M.; Leeson, P.D.; Oprea, T. The design of leadlike combinatorial libraries. Angew. Chem. Int. Ed. Engl. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Zhivkova, Z. Quantitative structure-pharmacokinetics relationship for plasma protein binding of neutral drugs. Int. J. Pharm. Pharmac. Sci. 2018, 10, 88–93. [Google Scholar] [CrossRef]
- Zhivkova, Z. Quantitative structure-pharmacokinetics modeling of the unbound clearance for neutral drugs. Int. J. Curr. Pharmac. Res. 2018, 10, 56–59. [Google Scholar] [CrossRef]
- Zhivkova, Z. Quantitative structure-pharmacokinetics relationship for the steady state volume of distribution of basic and neutral drugs. World J. Pharm. Pharmac. Sci. 2018, 7, 94–105. [Google Scholar]
- Schmidt, S.; Gonzales, D.; Derendorf, H. Significance of protein binding in pharmacokinetics and pharmacodynamics. J. Pharm. Sci. 2010, 99, 1107–1122. [Google Scholar] [CrossRef]
- Wasan, K.M.; Brocks, D.R.; Lee, S.D.; Sachs-Barrable, K.; Thornton, S.J. Impact of lipoproteins on the biological activity and disposition of hydrophobic drugs: Implications for drug discovery. Nat. Rev. Drug. Discov. 2008, 7, 84–99. [Google Scholar] [CrossRef]
- Berellini, G.; Waters, N.J.; Lombardo, S. In silico prediction of total human plasma clearance. J. Chem. Inf. Model 2012, 52, 2069–2078. [Google Scholar] [CrossRef]
- Smith, D.A.; Allerton, C.; Kalgutkar, A.; Van de Waterbeemd, H.; Walker, D.K. Renal clearance. In Pharmacokinetics and Metabolism in Drug Design, 3rd ed.; Wiley-VCH: Weinheim, Germany, 2012; pp. 103–110. [Google Scholar]
- Angelova, V.T.; Pencheva, T.; Vassilev, N.; Yovkova, E.K.; Mihaylova, R.; Petrov, B.; Valcheva, V. Development of New Antimycobacterial Sulfonyl Hydrazones and 4-Methyl-1, 2, 3-thiadiazole-Based Hydrazone Derivatives. Antibiotics 2022, 11, 562. [Google Scholar] [CrossRef]
- Horwitz, K.B.; Costlow, M.E.; McGuire, W.L. MCF-7; a human breast cancer cell line with oestrogen, androgen, progesterone, and glucocorticoid receptors. Steroids 1975, 26, 785–795. [Google Scholar] [CrossRef]
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010, 32, 35–48. [Google Scholar] [CrossRef]
- Klebe, R.J.; Ruddle, F.H. Neuroblastoma: Cell culture analysis of a differentiating stem cell system. J. Cell Biol. 1969, 43, 69A. [Google Scholar]
- Olmsted, J.B.; Carlson, K.; Klebe, R.; Ruddle, F.; Rosenbaum, J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc. Natl. Acad. Sci. USA 1970, 65, 129–136. [Google Scholar] [CrossRef]
- Peña-Morán, O.A.; Villarreal, M.L.; Álvarez-Berber, L.; Meneses-Acosta, A.; Rodríguez-López, V. Cytotoxicity, post-treatment recovery, and selectivity analysis of naturally occurring podophyllotoxins from Bursera fagaroides var. fagaroides on breast cancer cell lines. Molecules 2016, 21, 1013. [Google Scholar]
- Obach, R.S.; Lombardo, F.; Waters, N.J. Trend analysis of a database of intravenous pharmacokinetic parameters in humans for 670 drug compounds. Drug Metab. Dispos. 2008, 36, 1385–1405. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).