Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells
Abstract
1. Introduction
2. Results
2.1. Identification of Potential Genistein Targets against Human Cervical Cancer
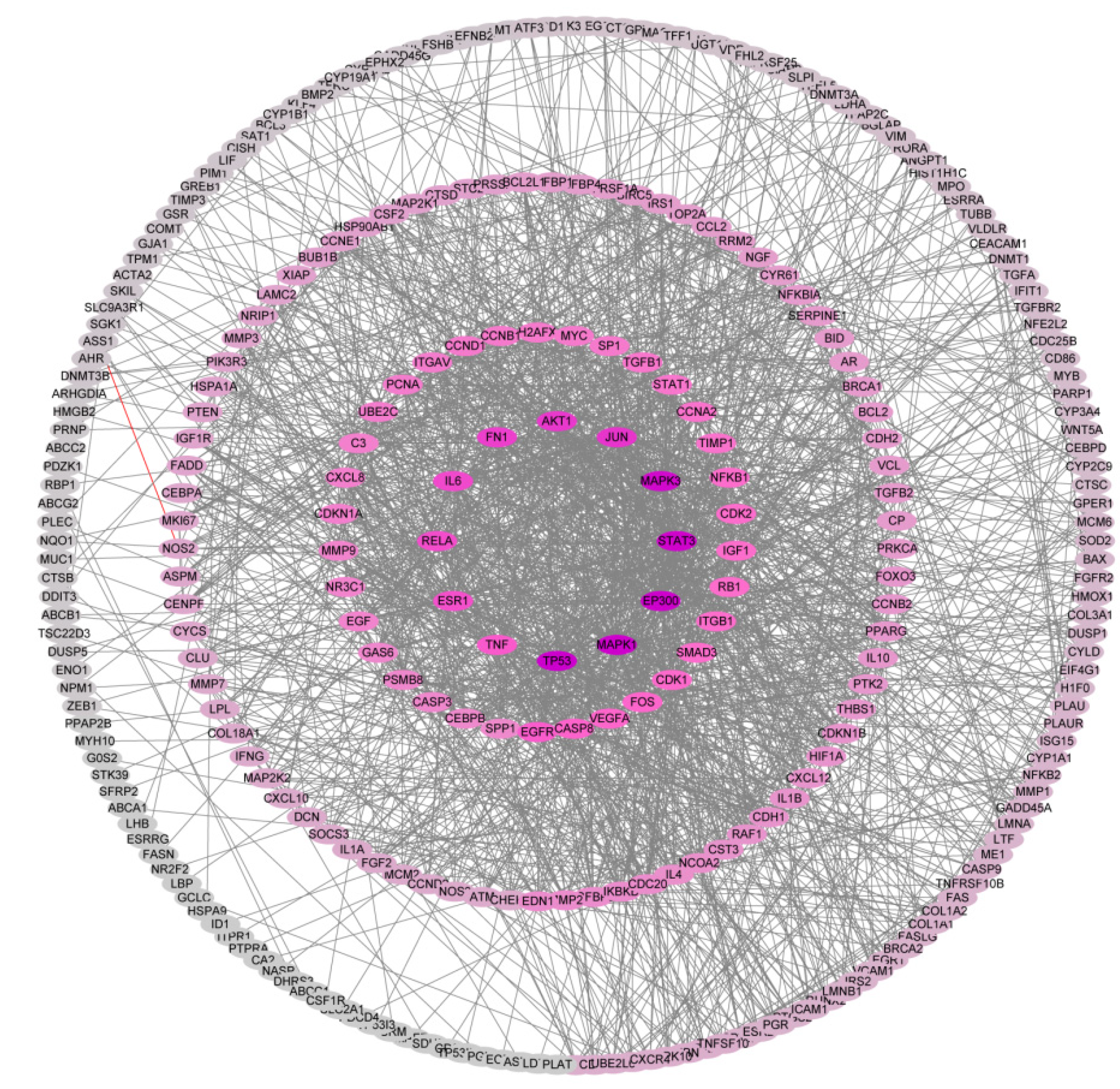
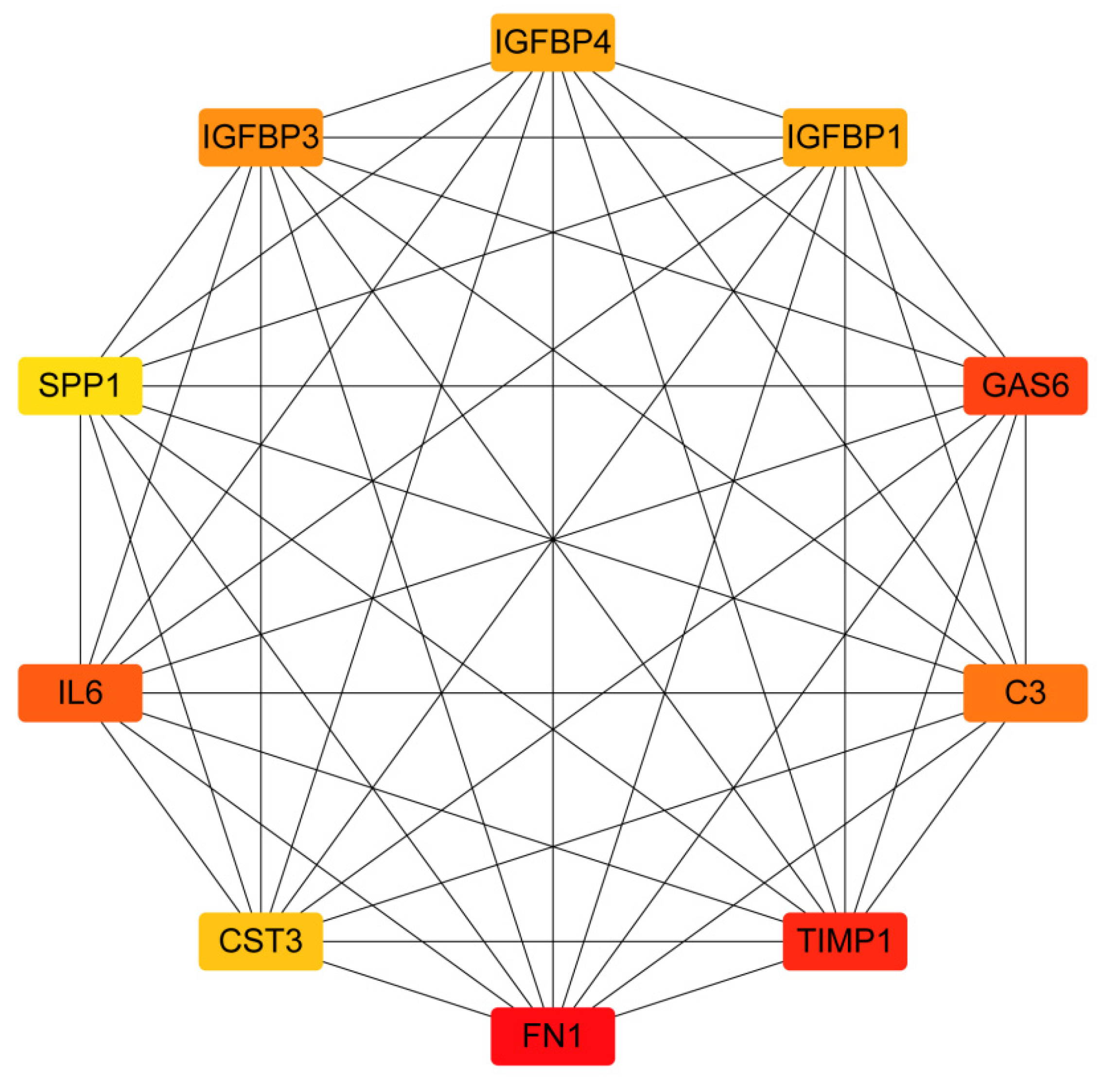
2.2. PPI Network Construction and Identification of Hub Genes
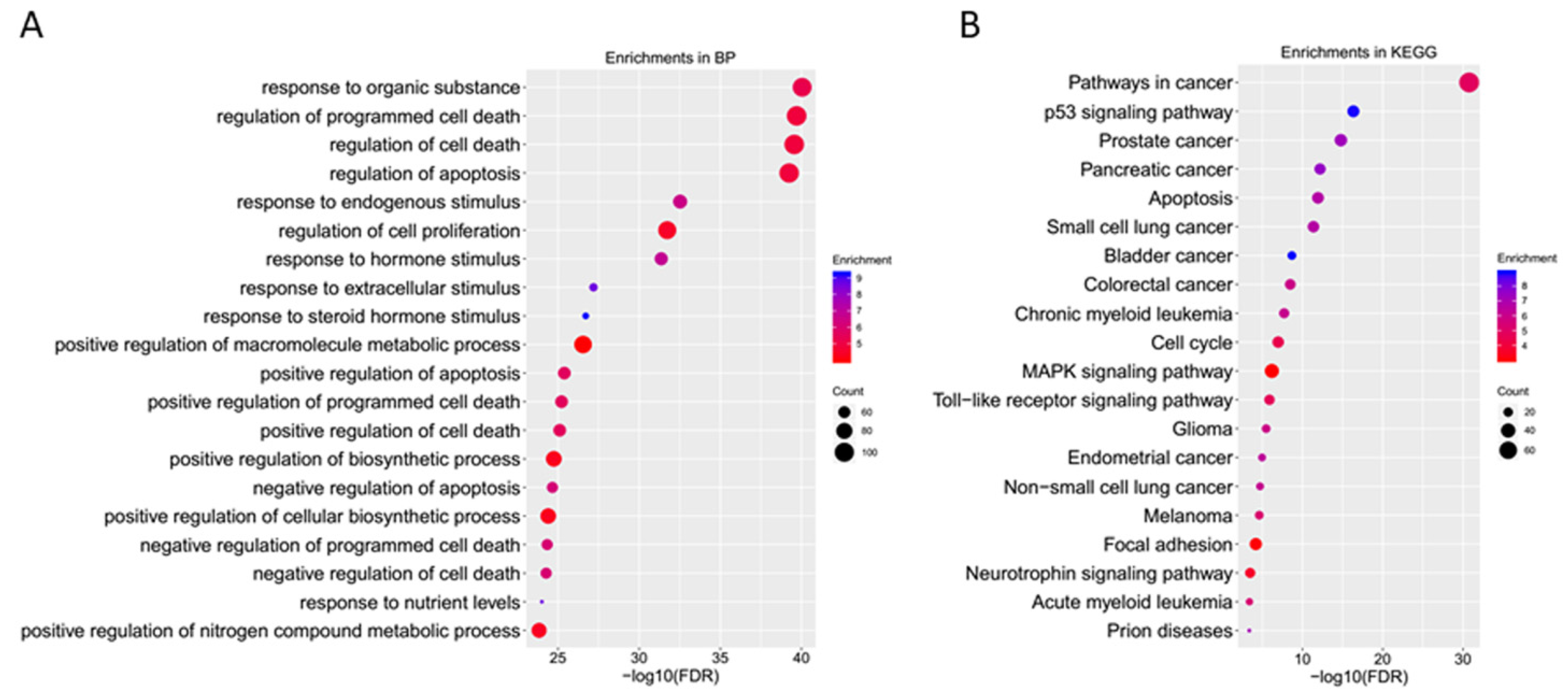
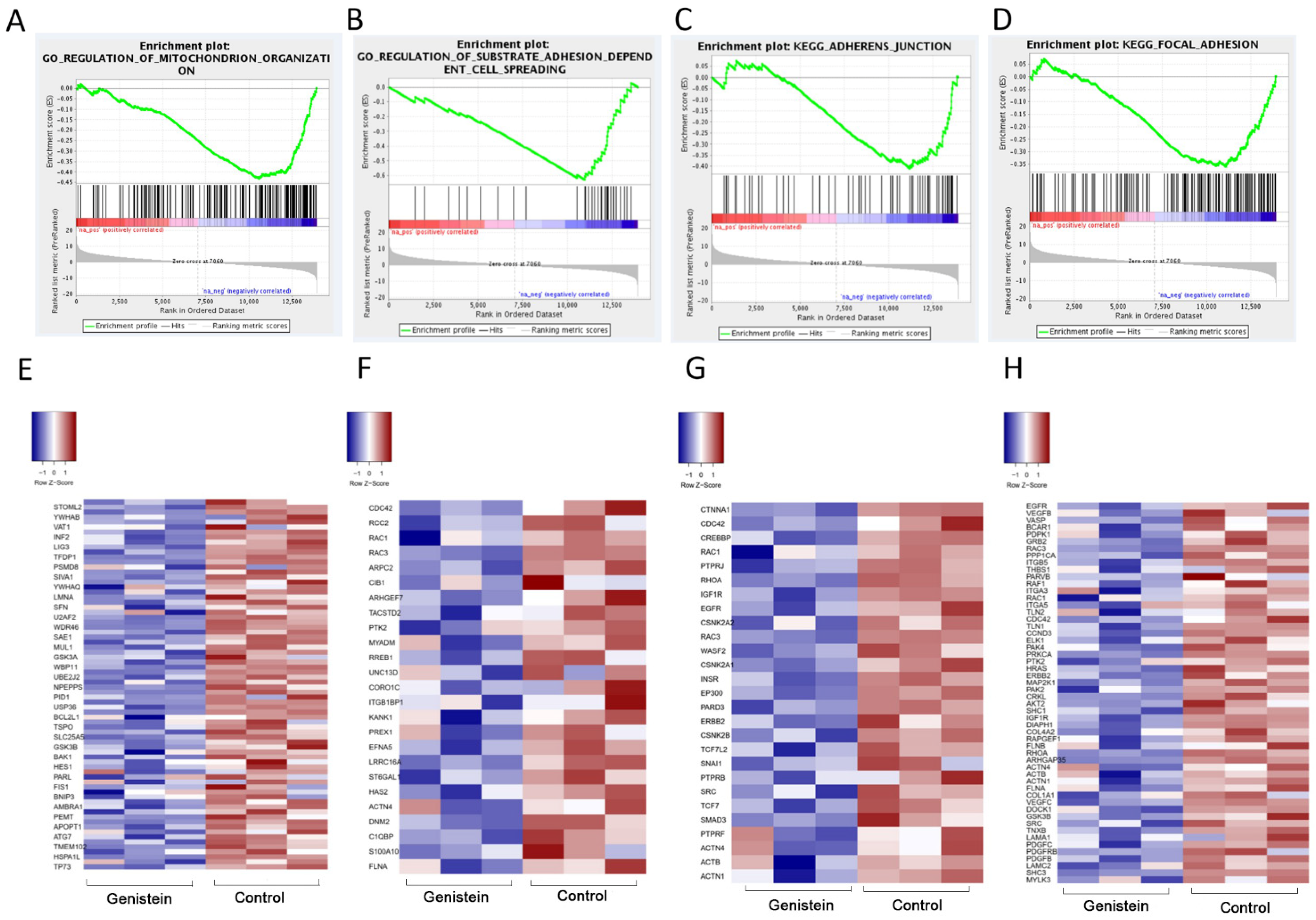
2.3. GO Enrichment Analyses of Core Targets
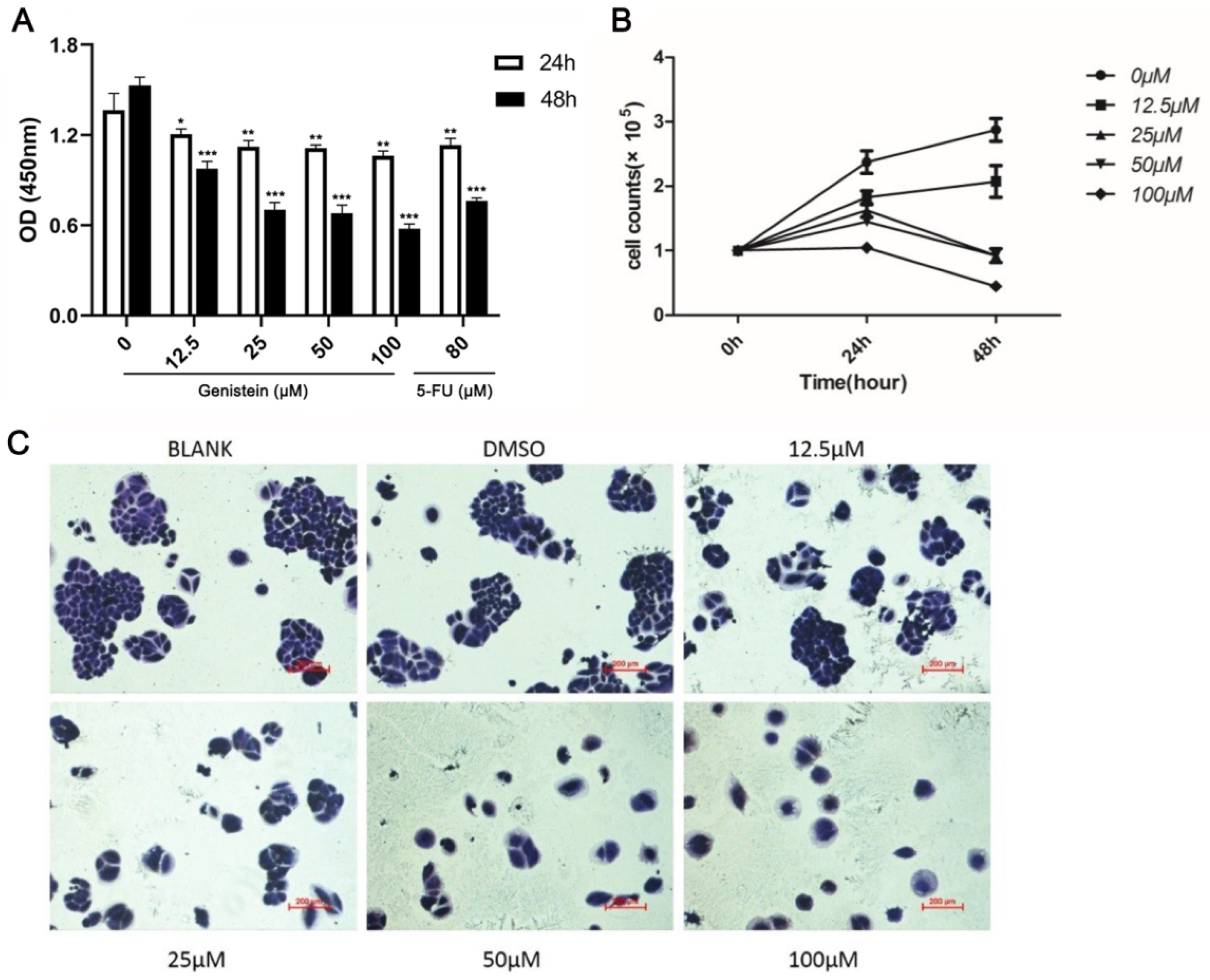
2.4. Genistein Attenuates Cell Viability and Growth in HeLa Cells
2.5. Genistein Inhibits HeLa Cell Adhesion
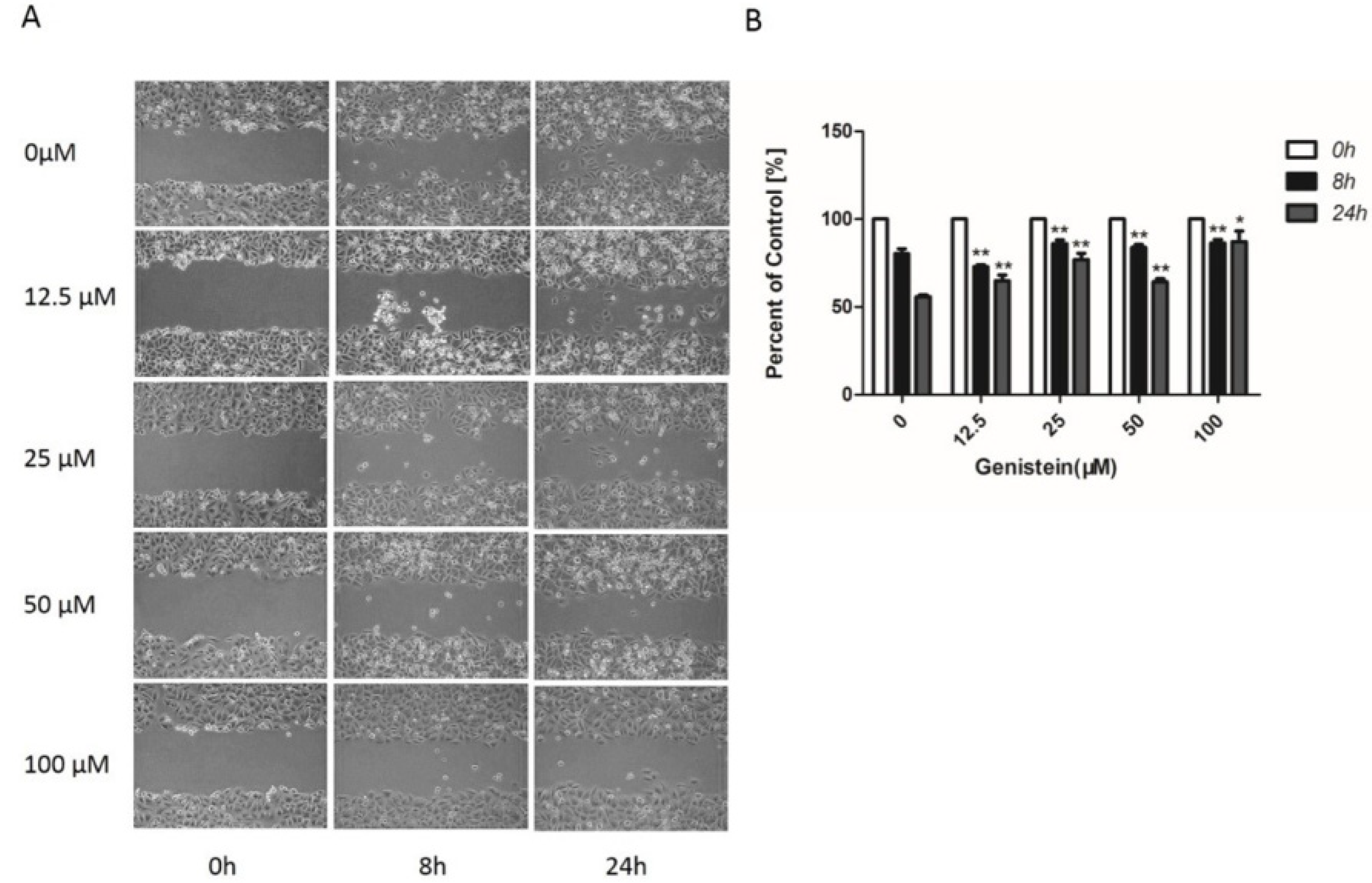
2.6. Genistein’s Inhibition of Cell Migration in the Wound−Healing Assay
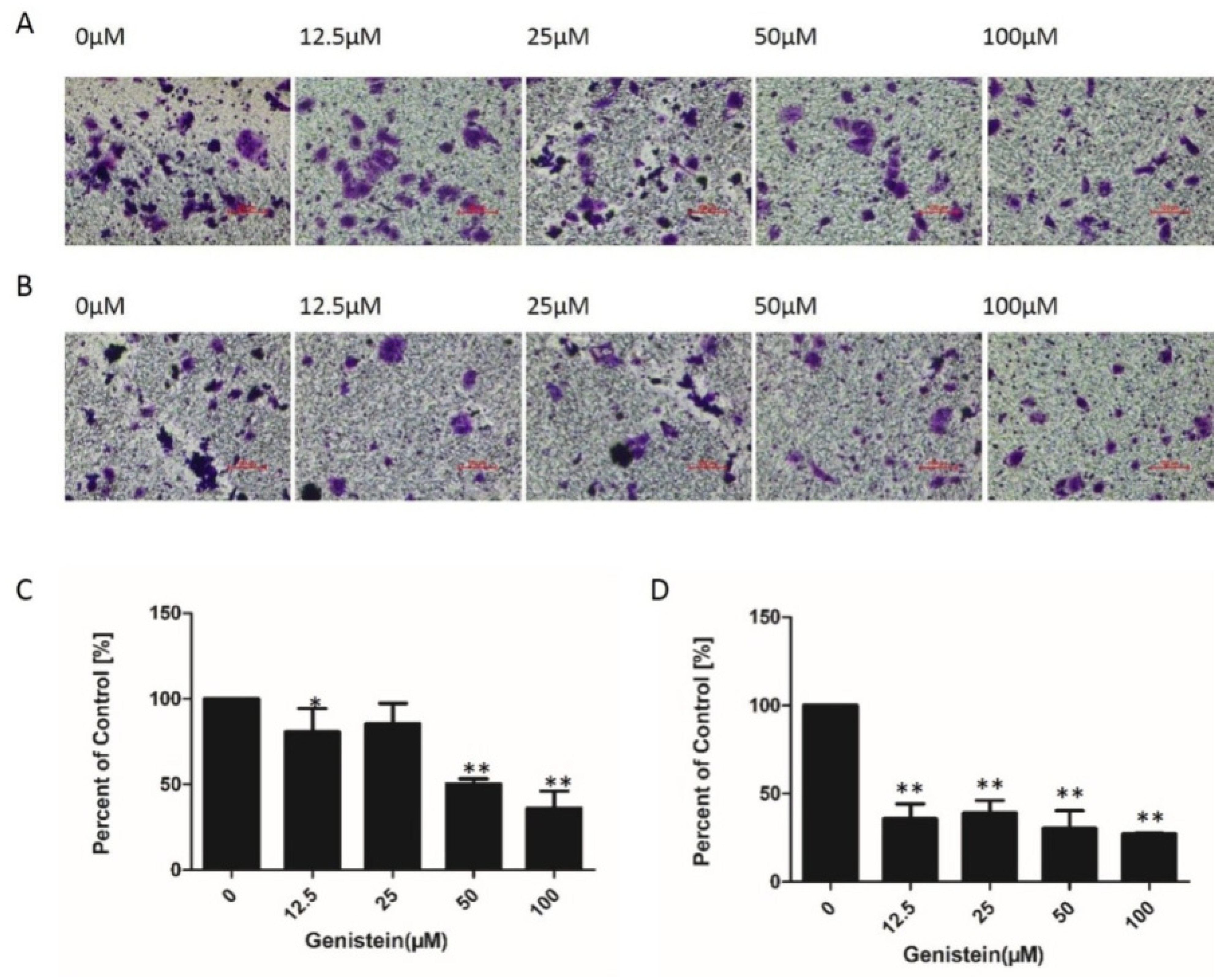
2.7. Genistein Inhibited Cell Migration and Invasion of HeLa Cells in Transwell® Assays
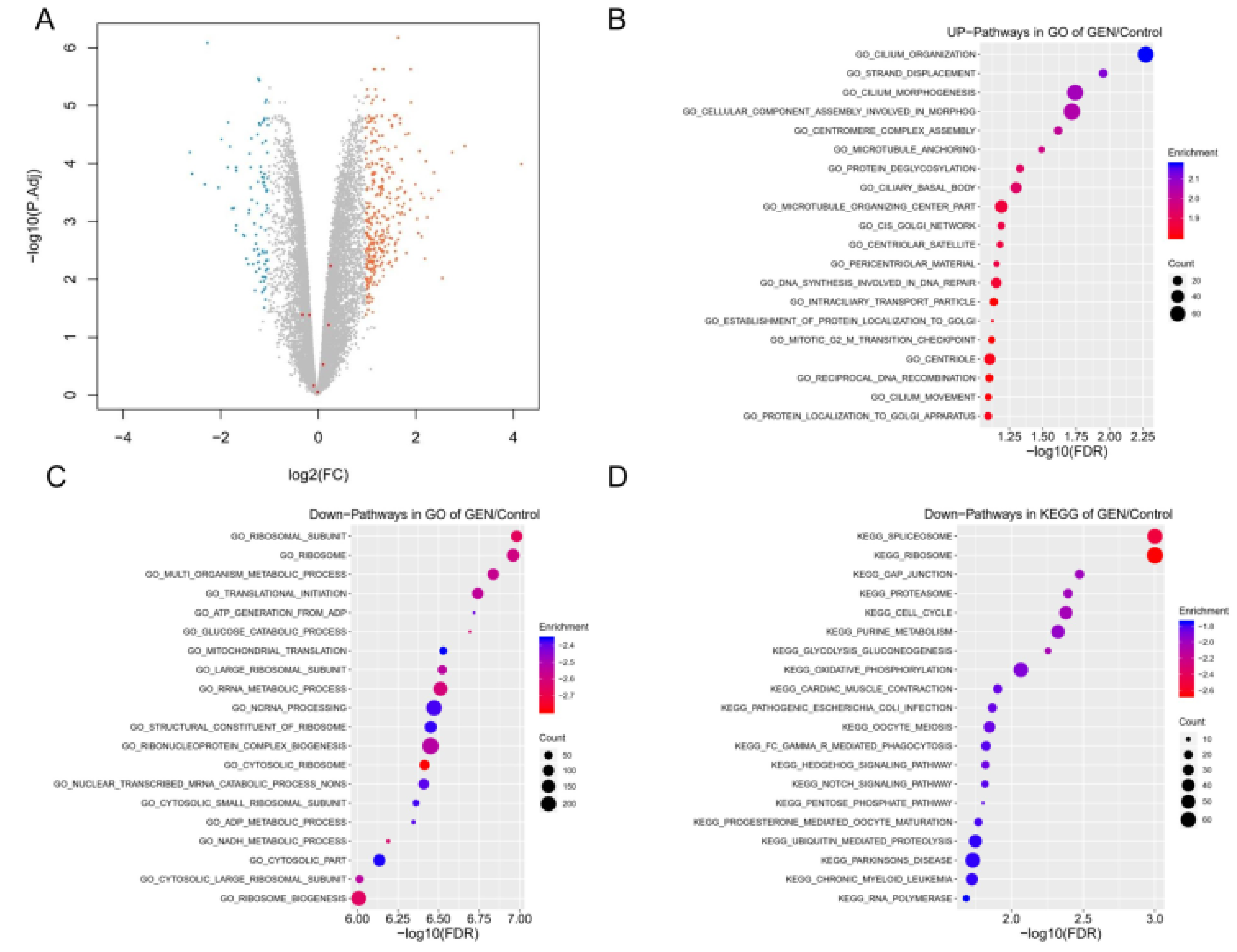
2.8. Identification of Differentially Expressed Genes (DEGs) Associated with Genistein Treatment
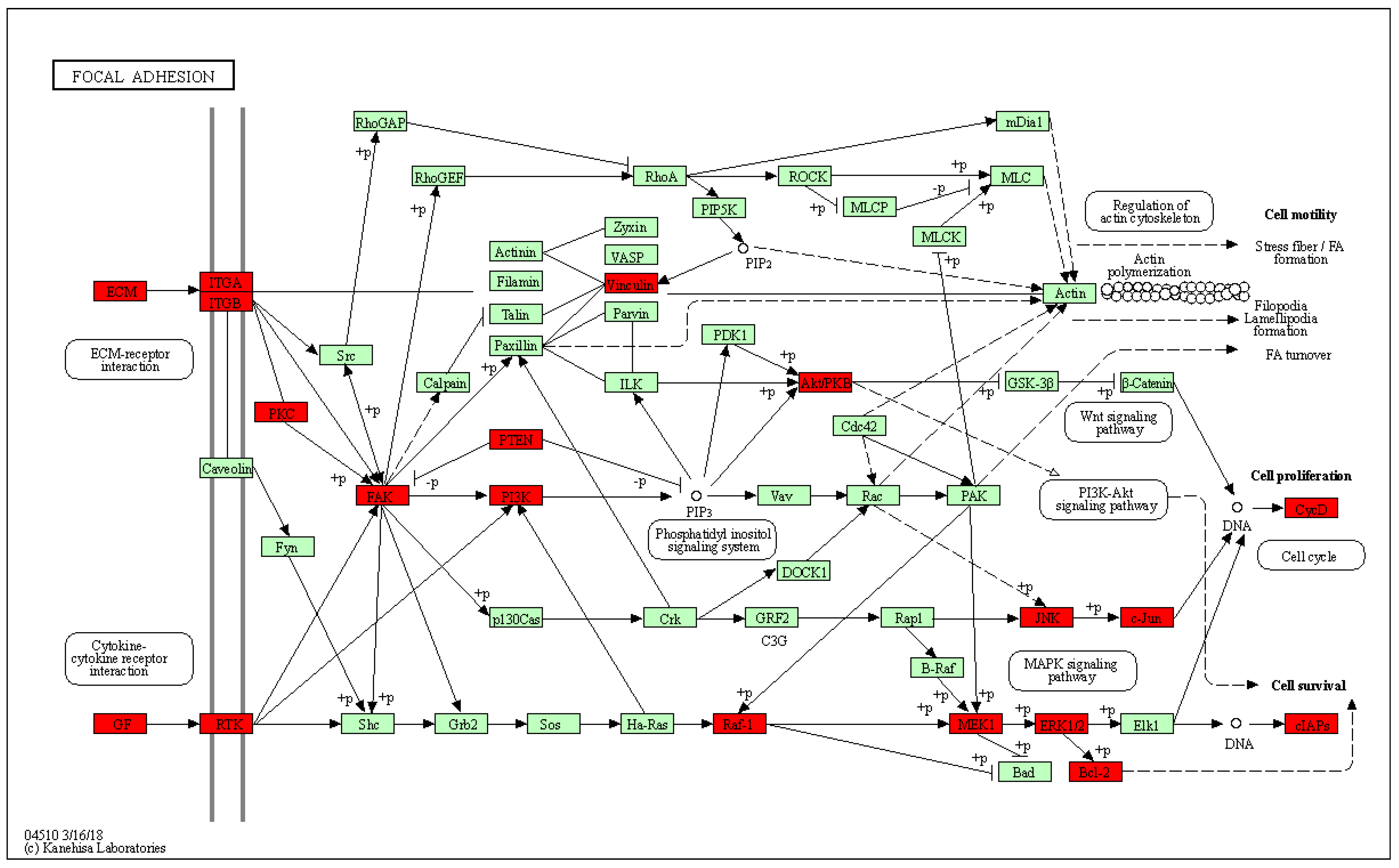
2.9. Genistein Inhibits Activation of the FAK–Paxillin Pathway
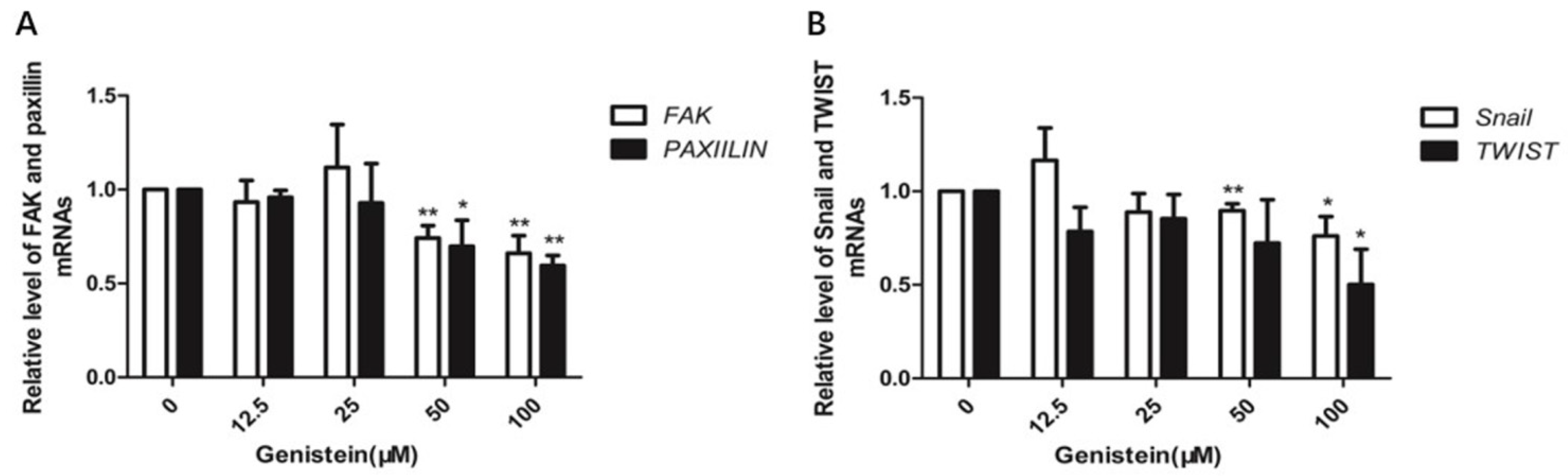
2.10. Genistein Inhibits Gene Expression of FAK, Paxillin, Snail, and Twist
3. Discussion
4. Materials and Methods
4.1. Prediction of Genistein’s Anti-Human Cervical Cancer Targets
4.2. PPI Network Analysis and Identification of Hub Genes
4.3. GO and KEGG Pathway Enrichment Analyses of Core Targets
4.4. Experimental Verification
4.4.1. Reagent Source
4.4.2. Cell Culture and CCK-8 Cell Viability Test
4.4.3. Colony Formation Assay
4.4.4. Adhesion Assay
4.4.5. Wound-Healing Mobility Assay
4.4.6. Transwell Assay
4.4.7. Western Blotting Assay
4.4.8. RNA Sequencing
4.4.9. Quantitative Real-Time RT-PCR
4.5. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.; Aimagambetova, G.; Ukybassova, T.; Kongrtay, K.; Azizan, A. Human Papillomavirus Infection and Cervical Cancer: Epidemiology, Screening, and Vaccination-Review of Current Perspectives. J. Oncol. 2019, 2019, 3257939. [Google Scholar] [CrossRef]
- Curty, G.; de Carvalho, P.S.; Soares, M.A. The Role of the Cervicovaginal Microbiome on the Genesis and as a Biomarker of Premalignant Cervical Intraepithelial Neoplasia and Invasive Cervical Cancer. Int. J. Mol. Sci. 2019, 21, 222. [Google Scholar] [CrossRef] [PubMed]
- Latsuzbaia, A.; Hebette, G.; Fischer, M.; Arbyn, M.; Weyers, S.; Vielh, P.; Schmitt, F.; Mossong, J. Introduction of liquid-based cytology and human papillomavirus testing in cervical cancer screening in Luxembourg. Diagn. Cytopathol. 2017, 45, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Iwanari, O.; Arakawa, I.; Moriya, T.; Mikami, Y.; Iihara, K.; Konoro, R. Cervical Cancer Screening With Human Papillomavirus DNA and Cytology in Japan. Int. J. Gynecol. Cancer 2017, 27, 523–529. [Google Scholar] [CrossRef]
- Gupta, S. Adjuvant chemotherapy in locally advanced cervical cancer: The ceiling remains unbroken. J. Gynecol. Oncol. 2019, 30, e97. [Google Scholar] [CrossRef]
- Liontos, M.; Kyriazoglou, A.; Dimitriadis, I.; Dimopoulos, M.A.; Bamias, A. Systemic therapy in cervical cancer: 30 years in review. Crit. Rev. Oncol. Hematol. 2019, 137, 9–17. [Google Scholar] [CrossRef]
- Azmi, A.S. Adopting network pharmacology for cancer drug discovery. Curr. Drug Discov. Technol. 2013, 10, 95–105. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key Molecular Events in Cervical Cancer Development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef]
- He, C.; Lv, X.; Huang, C.; Angeletti, P.C.; Hua, G.; Dong, J.; Zhouu, J.; Wang, Z.; Ma, B.; Chen, X.; et al. A Human Papillomavirus-Independent Cervical Cancer Animal Model Reveals Unconventional Mechanisms of Cervical Carcinogenesis. Cell Rep. 2019, 26, 2636–2650.e5. [Google Scholar] [CrossRef]
- Wang, C.; Davis, J.S. At the center of cervical carcinogenesis: Synergism between high-risk HPV and the hyperactivated YAP1. Mol. Cell Oncol. 2019, 6, e1612677. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shu, C.; Huang, Y. Fibronectin promotes cervical cancer tumorigenesis through activating FAK signaling pathway. J. Cell Biochem. 2019, 120, 10988–10997. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, J.; Khan, A.; Ali, A.; Fang, L.; Yanjing, W.; Xu, Q.; Wei, D.Q. Network Pharmacology: Exploring the Resources and Methodologies. Curr. Top. Med. Chem. 2018, 18, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, S.B.; Sandoval, M.J.; Rauschemberger, M.B.; Massheimer, V.L. Beneficial role of the phytoestrogen genistein on vascular calcification. J. Nutr. Biochem. 2017, 50, 26–37. [Google Scholar] [CrossRef]
- Cui, S.; Bilitewski, U. Effect of genistein on the TLR and MAPK transduction cascades in lipopolysaccharide-stimulated macrophages. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2014, 30, 233–236. [Google Scholar]
- Cui, S.N.; Wang, J.; Wu, Q.Q.; Qian, J.; Yang, C.S.; Bo, P. Genistein inhibits the growth and regulates the migration and invasion abilities of melanoma cells via the FAK/paxillin and MAPK pathways. Oncotarget 2017, 8, 21674–21691. [Google Scholar] [CrossRef]
- Jaiswal, N.; Akhtar, J.; Singh, S.P.; Badruddeen; Ahsan, F. An Overview on Genistein and its Various Formulations. Drug Res. 2019, 69, 305–313. [Google Scholar] [CrossRef]
- Mukund, V.; Mukund, D.; Sharma, V.; Mannarapu, M.; Alam, A. Genistein: Its role in metabolic diseases and cancer. Crit. Rev. Oncol. Hematol. 2017, 119, 13–22. [Google Scholar] [CrossRef]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Sundaram, M.K.; Ansari, M.Z.; Al Mutery, A.; Ashraf, M.; Nasab, R.; Rai, S.; Rais, N.; Hussain, A. Genistein Induces Alterations of Epigenetic Modulatory Signatures in Human Cervical Cancer Cells. Anticancer Agents Med. Chem. 2018, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Yang, K.W.; Hsu, Y.T.; Chang, C.L.; Yang, Y.C. The differential inhibitory effects of genistein on the growth of cervical cancer cells in vitro. Neoplasma 2001, 48, 227–233. [Google Scholar] [PubMed]
- Yang, Y.M.; Yang, Y.; Dai, W.W.; Li, X.M.; Ma, J.Q.; Tang, L.P. Genistein-induced apoptosis is mediated by endoplasmic reticulum stress in cervical cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3292–3296. [Google Scholar] [PubMed]
- Yashar, C.M.; Spanos, W.J.; Taylor, D.D.; Gercel-Taylor, C. Potentiation of the radiation effect with genistein in cervical cancer cells. Gynecol. Oncol. 2005, 99, 199–205. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.Y.; Pan, J.S.; Han, S.P.; Yin, X.X.; Wang, B.; Hu, G. Combined treatment of ionizing radiation with genistein on cervical cancer HeLa cells. J. Pharmacol. Sci. 2006, 102, 129–135. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, G.; Zeng, X.; Wu, Y.; Yang, C.; Mei, L.; Wang, Z.; Huang, L. Fabrication of genistein-loaded biodegradable TPGS-b-PCL nanoparticles for improved therapeutic effects in cervical cancer cells. Int. J. Nanomed. 2015, 10, 2461–2473. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.H.; Lee, S.C.; Song, Y.S. Involvement of Both Extrinsic and Intrinsic Apoptotic Pathways in Apoptosis Induced by Genistein in Human Cervical Cancer Cells. Nat. Compd. Role Apoptotic Cell Signal. Pathw. 2009, 1171, 196–201. [Google Scholar] [CrossRef]
- Lin, Y.J.; Zhen, Y.Z.; Zhao, Y.F.; Wei, J.; Hu, G. Rhein Lysinate Induced S-Phase Arrest and Increased the Anti-Tumor Activity of 5-FU in HeLa Cells. Am. J. Chin. Med. 2011, 39, 817–825. [Google Scholar] [CrossRef]
- Lee, M.Y.; Chou, C.Y.; Tang, M.J.; Shen, M.R. Epithelial-mesenchymal transition in cervical cancer: Correlation with tumor progression, epidermal growth factor receptor overexpression, and snail up-regulation. Clin. Cancer Res. 2008, 14, 4743–4750. [Google Scholar] [CrossRef]
- Li, C.; Ao, H.; Chen, G.; Wang, F.; Li, F. The Interaction of CDH20 With beta-Catenin Inhibits Cervical Cancer Cell Migration and Invasion via TGF-beta/Smad/SNAIL Mediated EMT. Front. Oncol. 2019, 9, 1481. [Google Scholar] [CrossRef]
- Ou, J.; Guan, D.; Yang, Y. Non-contact co-culture with human vascular endothelial cells promotes epithelial-to-mesenchymal transition of cervical cancer SiHa cells by activating the NOTCH1/LOX/SNAIL pathway. Cell Mol. Biol. Lett. 2019, 24, 39. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, M.J.; Kim, Y.K.; Kim, S.M.; Park, J.Y.; Myoung, H. Combined cetuximab and genistein treatment shows additive anti-cancer effect on oral squamous cell carcinoma. Cancer Lett. 2010, 292, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, F.H.; Adsule, S.; Padhye, S.; Kulkarni, S.; Li, Y. The role of genistein and synthetic derivatives of isoflavone in cancer prevention and therapy. Mini Rev. Med. Chem. 2006, 6, 401–407. [Google Scholar] [CrossRef]
- Davis, D.A.; Sarkar, S.H.; Hussain, M.; Li, Y.W.; Sarkar, F.H. Increased therapeutic potential of an experimental anti-mitotic inhibitor SB715992 by genistein in PC-3 human prostate cancer cell line. BMC Cancer 2006, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, C.; Russo, G.L.; Orhan, I.E.; Habtemariam, S.; Daglia, M.; Sureda, A.; Nabavi, S.M. Genistein and cancer: Current status, challenges, and future directions. Adv. Nutr. 2015, 6, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Yi, Y.; Liu, F.; Wu, W.; Chen, Y.; Zhang, W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol. Lett. 2018, 16, 1003–1009. [Google Scholar] [CrossRef]
- Cao, F.Y.; Zhou, X.P.; Su, J.; Yang, X.H.; Mu, F.H.; Shen, J.Y.; Sun, W. Chemical Structure Characteristics and Bioactivity of Small Molecule FAK Inhibitors. Anticancer Agents Med. Chem. 2015, 16, 934–941. [Google Scholar] [CrossRef]
- Lv, P.C.; Jiang, A.Q.; Zhang, W.M.; Zhu, H.L. FAK inhibitors in Cancer, a patent review. Expert Opin. Ther. Pat. 2018, 28, 139–145. [Google Scholar] [CrossRef]
- Roy-Luzarraga, M.; Hodivala-Dilke, K. Molecular Pathways: Endothelial Cell FAK-A Target for Cancer Treatment. Clin. Cancer Res. 2016, 22, 3718–3724. [Google Scholar] [CrossRef]
- Alanko, J.; Ivaska, J. Endosomes: Emerging Platforms for Integrin-Mediated FAK Signalling. Trends Cell Biol. 2016, 26, 391–398. [Google Scholar] [CrossRef]
- Cui, S.; Wu, Q.; Wang, J.; Li, M.; Qian, J.; Li, S. Quercetin inhibits LPS-induced macrophage migration by suppressing the iNOS/FAK/paxillin pathway and modulating the cytoskeleton. Cell Adh. Migr. 2019, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kanteti, R.; Batra, S.K.; Lennon, F.E.; Salgia, R. FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget 2016, 7, 31586. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Dehart, J.P.; Murphy, J.M.; Lim, S.T. Understanding the roles of FAK in cancer: Inhibitors, genetic models, and new insights. J. Histochem. Cytochem. 2015, 63, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Timpson, P.; Horvath, L.G.; Daly, R.J. FAK signaling in human cancer as a target for therapeutics. Pharmacol. Ther. 2015, 146, 132–149. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.M.; Hsieh, M.J.; Yang, J.S.; Yang, S.F.; Chuang, Y.T.; Su, S.C.; Liang, M.-Y.; Chen, M.-K.; Lin, C.-W. Geraniin inhibits oral cancer cell migration by suppressing matrix metalloproteinase-2 activation through the FAK/Src and ERK pathways. Environ. Toxicol. 2019, 34, 1085–1093. [Google Scholar] [CrossRef]
- Herzinger, T.; Wolf, D.A. Snail puts melanoma on the fast track. Pigment. Cell Melanoma Res. 2009, 22, 150–151. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, H.; Zhao, X. Baicalin Inhibits Human Cervical Cancer Cells by Suppressing Protein Kinase C/Signal Transducer and Activator of Transcription (PKC/STAT3) Signaling Pathway. Med. Sci. Monit. 2018, 24, 1955–1961. [Google Scholar] [CrossRef]
- Bolos, V.; Gasent, J.M.; Lopez-Tarruella, S.; Grande, E. The dual kinase complex FAK-Src as a promising therapeutic target in cancer. OncoTargets Ther. 2010, 3, 83–97. [Google Scholar] [CrossRef]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef]
- Cui, S.; Wienhoefer, N.; Bilitewski, U. Genistein induces morphology change and G2/M cell cycle arrest by inducing p38 MAPK activation in macrophages. Int. Immunopharmacol. 2014, 18, 142–150. [Google Scholar] [CrossRef]
- Farruggio, S.; Cocomazzi, G.; Marotta, P.; Romito, R.; Surico, D.; Calamita, G.; Bellan, M.; Pirisi, M.; Grossini, E. Genistein and 17beta-Estradiol Protect Hepatocytes from Fatty Degeneration by Mechanisms Involving Mitochondria, Inflammasome and Kinases Activation. Cell Physiol. Biochem. 2020, 54, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Teng, J.; Zhu, Z.; Chen, J.; Huang, W.J. Genistein induces activation of the mitochondrial apoptosis pathway by inhibiting phosphorylation of Akt in colorectal cancer cells. Pharm. Biol. 2016, 54, 74–79. [Google Scholar] [CrossRef]
- Zhang, Q.; Bao, J.; Yang, J. Genistein-triggered anticancer activity against liver cancer cell line HepG2 involves ROS generation, mitochondrial apoptosis, G2/M cell cycle arrest and inhibition of cell migration. Arch. Med. Sci. 2019, 15, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Murphy, C.G.; Saraceni-Richards, C.A.; Rosenstein, M.C.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database: A knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009, 37, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, C.J.; Rosenstein, M.C.; Davis, A.P.; Colby, G.T.; Forrest, J.N.; Boyer, J.L. The Comparative Toxicogenomics Database: A cross-species resource for building chemical-gene interaction networks. Toxicol. Sci. 2006, 92, 587–595. [Google Scholar] [CrossRef]
- Mattingly, C.J.; Colby, G.T.; Rosenstein, M.C.; Forrest, J.N.; Boyer, J.L. Promoting comparative molecular studies in environmental health research: An overview of the comparative toxicogenomics database (CTD). Pharm. J. 2004, 4, 5–8. [Google Scholar] [CrossRef]
- Mattingly, C.J.; Rosenstein, M.C.; Colby, G.T.; Forrest, J.N.; Boyer, J.L. The Comparative Toxicogenomics Database (CTD): A resource for comparative toxicological studies. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2006, 305a, 689–692. [Google Scholar] [CrossRef]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Lien, J.C.; Huang, Y.P.; Liao, C.L.; Lin, J.J.; Fan, M.J.; Ko, Y.-C.; Hsiao, Y.-P.; Lu, H.-F.; Chung, J.-G. Casticin Inhibits A375.S2 Human Melanoma Cell Migration/Invasion through Downregulating NF-kappaB and Matrix Metalloproteinase-2 and -1. Molecules 2016, 21, 384. [Google Scholar] [CrossRef]
- Wu, S.H.; Hsiao, Y.T.; Kuo, C.L.; Yu, F.S.; Hsu, S.C.; Wu, P.P.; Chen, J.C.; Hsia, T.C.; Liu, H.C.; Hsu, W.H.; et al. Bufalin Inhibits NCI-H460 Human Lung Cancer Cell Metastasis In Vitro by Inhibiting MAPKs, MMPs, and NF-kappa B Pathways. Am. J. Chin. Med. 2015, 43, 1247–1264. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Hu, C.; Jiang, J.; Jin, S.; Wei, Q.; Wei, X.; Yu, D.; Shi, F. Effects of High-Dose Genistein on the Hypothalamic RNA Profile and Intestinal Health of Female Chicks. J. Agric. Food Chem. 2019, 67, 13737–13750. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Zhou, J.; Yang, C.; Wang, G.; Tan, Y.; Wu, Y.; Zhang, S.; Yi, K.; Kang, C. The CRISPR-Cas13a Gene-Editing System Induces Collateral Cleavage of RNA in Glioma Cells. Adv. Sci. 2019, 6, 1901299. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.-L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]



| Name | Source | ID | |
|---|---|---|---|
| 1 | Paxillin | Rabbit, pAb | #2542 Cell signaling |
| 2 | Anti-rabbit IgG, HRP-linked antibody | Goat | #7074 Cell signaling |
| 3 | Phospho-Paxillin (Tyr118) | Rabbit, pAb | #2541 Cell signaling |
| 4 | FAK | Rabbit, pAb | #13430 Cell signaling |
| 5 | Phospho-FAK (Tyr925) | Rabbit, pAb | #9330 Cell signaling |
| 6 | Vimentin (D21H3) | Rabbit, mAb | #9782 Cell signaling |
| 7 | β-Catenin (D10A8) | Rabbit, mAb | #9782 Cell signaling |
| 8 | GAPDH-HRP | Mouse mAb | #: ab011 Multi Science |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Wang, J.; Li, M.; Wu, Q.; Cui, S. Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells. Molecules 2023, 28, 1919. https://doi.org/10.3390/molecules28041919
Chen T, Wang J, Li M, Wu Q, Cui S. Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells. Molecules. 2023; 28(4):1919. https://doi.org/10.3390/molecules28041919
Chicago/Turabian StyleChen, Tingting, Juan Wang, Min Li, Qingqing Wu, and Shuna Cui. 2023. "Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells" Molecules 28, no. 4: 1919. https://doi.org/10.3390/molecules28041919
APA StyleChen, T., Wang, J., Li, M., Wu, Q., & Cui, S. (2023). Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells. Molecules, 28(4), 1919. https://doi.org/10.3390/molecules28041919








