Abstract
The inflammatory cytokine interleukin-17 (IL17) plays an important role in innate immunity by binding to its receptors (IL17Rs) to activate immune defense signals. To date, information on members of the IL17 family is still very limited in molluscan species. Here, a novel member of the IL17 family was identified and characterized from thick shell mussel Mytilus coruscus, and this gene was designated as McIL17-1 by predicting structural domains and phylogenetic analysis. McIL17-1 transcripts existed in all examined tissues with high expression levels in gills, hemocytes and digestive glands. After the stimuli of different pathogen associated molecular patterns (PAMPs) for 72 h, transcriptional expression of McIL17-1 was significantly upregulated, except for poly I:C stimulation. Cytoplasm localization of McIL17-1 was shown in HEK293T cells by fluorescence microscopy. Further, in vivo and in vitro assays were performed to evaluate the potential function of McIL17-1 played in immune response. McIL17-1 was either knocked down or overexpressed in vivo through RNA inference (RNAi) and recombinant protein injection, respectively. With the infection of living Vibrio alginolyticus, a high mortality rate was exhibited in the McIL17-1 overexpressed group compared to the control group, while a lower mortality rate was observed in the McIL17-1 knocked down group than control group. In vitro, the flow cytometric analysis showed that the apoptosis rate of McIL17-1 inhibited hemocytes was significantly lower than that of the control group after lipopolysaccharide stimulation. These results collectively suggested that the newly identified IL17 isoform is involved in the inflammatory response to bacterial infection in M. coruscus.
1. Introduction
Due to the lack of antibody-based adaptive immunity, bivalves are protected from pathogens by anti-infective effectors such as antimicrobial peptides and a range of more sophisticated defense mechanisms known as innate immunity [1]. Innate immunity represents an ancient evolutionary defense strategy shared by vertebrates and invertebrates for nonspecific recognition of pathogenic microorganisms [2]. Mussels, the largest phyla of bivalves, are frequently exposed to a wide range of microorganisms due to their filtering activity [3]. They rely entirely on the innate immune system to confront pathogen invasion, which is activated by pattern recognition receptors (PRRs) that recognize pathogen-related molecular patterns (PAMPs) or damage-related molecular patterns (DAMPs) [4,5,6].
As one of the oldest PRRs, the toll-like receptors (TLRs) have a broad pattern recognition spectrum and can recognize diverse PAMPs [7], including lipids, lipoproteins, proteins, and nucleic acids derived from a wide range of microbes such as bacteria, viruses, fungi and protozoa [8,9,10,11]. Upon recognition of PAMPs, TLRs drive downstream signaling pathways mediated by the transcription factor NF-κB and IRFs, which in turn triggers the expression of multiple pro- and anti-inflammatory genes, among which interleukin-17 (IL17) is one of the most typical pro-inflammatory factors [12,13].
IL17 is one member of the most ancient cytokine family; six IL17 family ligands (IL17A-F), as well as five receptors of them (IL17RA-E), have been identified in mammals [14]. Structurally, these cytokines generally embrace an IL17 domain at the C-terminus, where four conserved cysteine residues form intra-chain disulfide bonds to promote dimerization [15]. The unique structural characteristics of 4-cysteine enable IL17 to fulfill its function by binding or interacting with its specific receptors, meanwhile providing a basis that distinguishes IL17 sequences from other cytokines [16]. Unlike traditional inflammatory cytokines, activated T lymphocytes and other cell types relevant to the host immunity, such as the neutrophils and the mucosal epithelial cells, produce IL-17s [17,18]. Nevertheless, IL17 plays a fundamental role in innate immunity as its secretion triggers the production of a large number of chemokines, leading to the recruitment of neutrophils and macrophages, which, in turn, clear pathogens [19]. By this means, IL17 mediates the interaction between the innate and adaptive immunity, thereby coordinating an effective immune response [20].
IL17 and its mediated signaling pathway have long been thought to be unique to vertebrates. However, thirty IL17s and two IL17Rs were identified in the genome of sea urchins in 2006, thereby opening the prologue to IL17s in invertebrates [21]. According to written records, the molluscan IL17 was first found in Crassostrea gigas in 2008, and two clones encoding a protein analogous to vertebrate IL17s were obtained from C. gigas hemocyte cDNA library [22]. Further research showed that the transcriptional expression of CgIL17s was distinctly increased in hemocytes in response to PAMPs, suggesting that CgIL17s can participate in innate immune response [23]. Moreover, an IL17 receptor named CgIL17R1 was also identified from C. gigas, which binds to CgIL17 in granulocytes to mediate hemocyte proliferation during immune modulation of C. gigas [24]. This may shed some new light on the regulation of hemocyte proliferation in invertebrates. In Pinctada fucata, IL17 has proven to be capable of activating the NF-κB signal pathway against exogenous pathogens [25]. Several IL17s and -17Rs have also been identified in other mollusks, including Haliotis discus hannai and Mytilus galloprovincialis [26,27,28,29], which provided valuable clues to elucidate the functional roles of IL17s in innate immune regulation in mollusks.
The thick shell mussel Mytilus coruscus, a species of mussel in the Mollusca genus Bivalvia, is primarily found in the western Pacific waters [30]. M. coruscus has evolved over the past several years into a model organism for investigating the physiological response and immune reactivity of molluscans to environmental stress because of its enormous economic relevance and ubiquity in coastal waters [31,32]. The clarification of the underlying mechanism for innate immune response is of great academic significance. Here, we molecularly identified an IL17 homolog (McIL17-1) from M. coruscus. Following its transcriptional response to multiple PAMPs, McIL17-1 effects on mussel survival and hemocytes apoptotic rate were assessed through laboratory means. The present study is helpful in filling the theoretical gap of the IL17 in M. coruscus, while providing some additional information to elucidate the role of proinflammatory factors acting in mollusks.
2. Results
2.1. Molecular Cloning and Characterization of McIL17-1
The McIL17-1 cDNA containing the complete ORF region was cloned from M. coruscus (GenBank: GCA_011752425.2). McIL17-1 contains a 585 bp ORF region encoding 194 amino acids (Figure 1A). The predicted molecular weight is 20.35 kDa, and the isoelectric point is 9.43. SMART analysis revealed a typical IL17 domain in this protein (Figure 1B). A phylogenetic tree was constructed with vertebrate interleukin-2 cytokines (IL-2s) as outgroup. Clearly, these IL17s clustered into a distinct clade to distinguish them from IL-2s. In the IL17s cluster, McIL17-1 first bound to its corresponding molecule in another Mytilus species i.e., Mytilus galloprovincialis, and then clustered with other IL17s from M. galloprovincialis and Crassostrea gigas into one subbranch. In addition, the vertebrate IL17s converged into a distinct clade distinct from the molluscan IL17s clade (Figure 2).
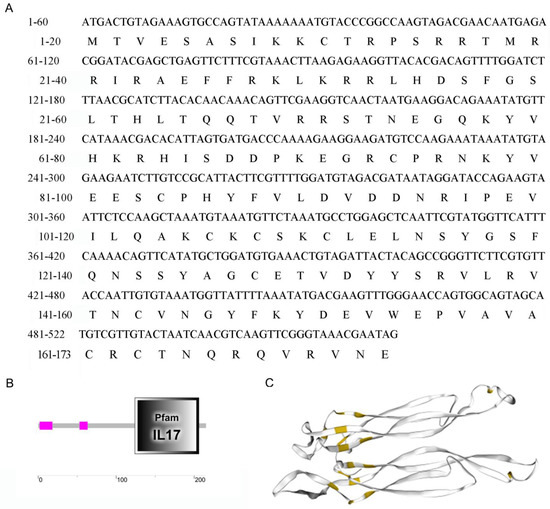
Figure 1.
Molecular characterization of McIL17-1. (A) The nucleotide sequences and the deduced amino acid sequences of IL17-1 in thick shell mussel Mytilus coruscus. It contains a 585 bp ORF region coding the protein of 194 amino acid residues; (B) Schematic diagram of IL17 structure domain; (C) Schematic diagram of cysteine nodule structure (Yellow marked: cysteines).
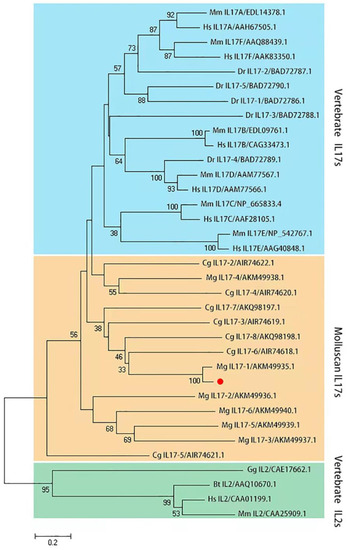
Figure 2.
Phylogenetic analysis of McIL17-1. The phylogenetic tree is constructed using MEGA-X software with 2000 replications of bootstrap in the way of the neighbor-joining method. McIL17-1 is labeled with a red dot. Species included in the phylogenetic tree are all retrieved from the GenBank database and accession numbers are also listed in the tree. Sequences from various species are abbreviated as follows: Hs, Homo sapiens; Mm, Mus musculus; Bt, Bos taurus; Dr, Danio rerio; Gg, Gallus gallus; Mg, Mytilus galloprovincialis; Cg, Crassostrea gigas.
2.2. Transcriptional Expression of McIL17-1
The tissue distribution profile of McIL17-1 transcripts was assessed by qPCR assay. As shown in Figure 3A, the expression level of McIL17-1 mRNA was the highest in gills, followed by in hemocytes and digestive glands, and lowest in adductor muscle. Additionally, the transcriptional response to multiple PAMPs was also evaluated for McIL17-1 (Figure 3B). With the stimulation of live V. alginolyticus, the transcriptional expression of McIL17-1 was significantly upregulated at 12 and 72 hps. Similarly, McIL17-1 transcripts also showed a response to other immune stimuli, such as LPS, PGN and GLU, which all peaked at 72 hps. However, poly I:C injection did not significantly change the mRNA level of McIL17-1.
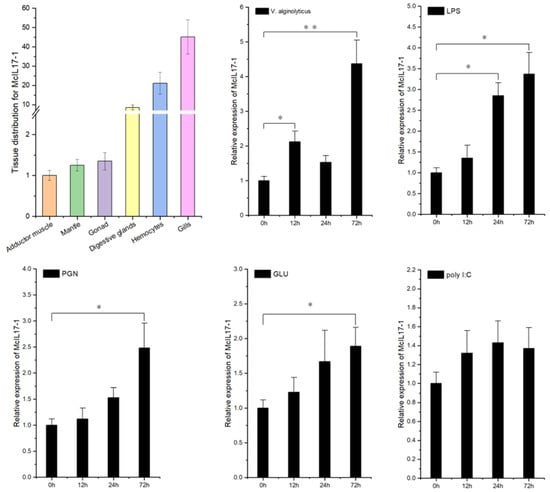
Figure 3.
Expression profile analysis of McIL17-1 transcripts in common adult tissues and after challenge with live V. alginolyticus, lipopolysaccharide (LPS) (from Escherichia coli O111: B4), peptidoglycans (PGN) (from Staphylococcus aureus), glucan (GLU) (from Saccharomyces cerevisiae), or polyinosinic-polycytidylic acid (poly I:C), respectively. The results were expressed as mean ± S.D. (n = 3). Significant difference relative to control is indicated with asterisk symbol (* p < 0.05, ** p < 0.01).
2.3. Subcellular Localization of McIL17-1
Due to the absence of mature mussel cell lines, the cellular localization of McIL17-1 was examined in the HEK293T cells through the transfection of the constructed pEGFP-N1-McIL17-1 plasmid. Under a fluorescence microscope, the green fluorescence of GFP-labeled McIL17-1 was primarily found in the cytoplasm (Figure 4).
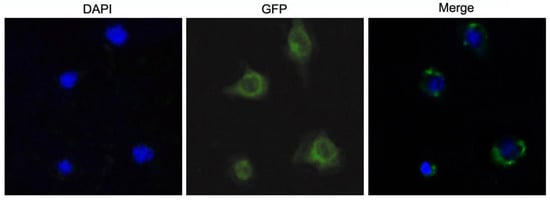
Figure 4.
Subcellular localization of McIL17-1 in HEK293T cells. The recombinant pEGFP-N1-McIL17-1 plasmid was transfected into HEK293T cell using lipofectamine 3000, the green fluorescence showed the location of proteins and the cell nucleus location was indicated by blue DAPI staining. The McIL17-1 was mainly localized in the cytoplasm of HEK293T cells.
2.4. Effects of Reduction and Overexpression of McIL17-1 on Survival Rate of Mussels
McIL17-1 was recombinantly expressed and purified by the pET-32a prokaryotic expression system. The molecular mass of rMcIL17-1, rTrx, His tag protein and cohesive amino acids combined together was compatible with the band size of roughly 40 kDa that appeared on the SDS-PAGE (Figure 5A). After injection of dsMcIL17-1, the mRNA expression level of McIL17-1 was significantly decreased in M. coruscus hemocytes (0.74 and 0.52 fold at 12 h and 24 h compared to control, respectively) (Figure 5B). Purified rMcIL17-1 was injected into healthy mussels, and a total of four groups of mussels, i.e., normal group, V. alginolyticus treated group, V. alginolyticus and dsMcIL17-1 co-treated group, V. alginolyticus and rMcIL17-1 co-treated group were employed to assess the effects of silencing and overexpression of McIL17-1 on survival rate of mussels. The results showed that the mussels of V. alginolyticus and rMcIL17-1 co-treated groups all died on the 10th day, prior to V. alginolyticus solely treated mussels that all died on the 13th day (Figure 5C). After infection with the V. alginolyticus, these McIL17-1 silenced mussels continued to die over time. However, the mortality rate was significantly lower than the other two V. alginolyticus treated groups (Figure 5C).
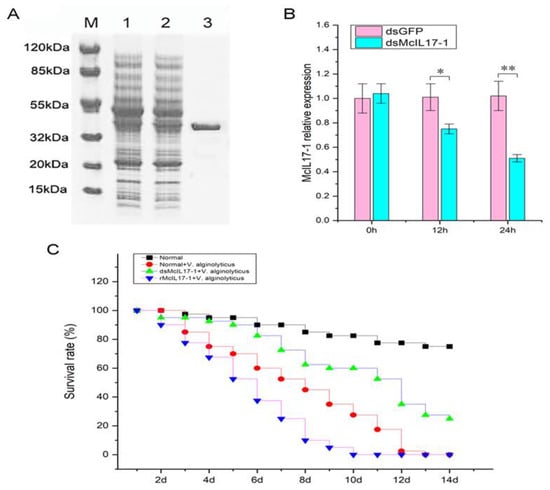
Figure 5.
Effects of silencing and overexpression of McIL17-1 on survival rate of mussels. (A) SDS-PAGE analysis of rMcIL17-1. Lane M: protein molecular standard; lane 1: negative control for rMcIL17-1(without induction); lane 2: transfected rMcIL17-1; lane3: purified rMcIL17-1. (B) McIL17-1 was silenced through dsRNA injection. (C) The survival rate (%) of mussels prior to dsMcIL17-1 and rMcIL17-1 injection following V. alginolyticus challenge. **, p < 0.01, *, p < 0.05 versus the controls.
2.5. Hemocytes Apoptotic Rate after McIL17-1 Was Inhibited
The apoptosis rate of hemocytes after LPS treatment was analyzed by flow cytometer after McIL17-1 was inhibited. For the mussels in siNC group (negative control siRNA), the apoptosis rate of hemocytes was 46.1%. However, the apoptosis rate decreased to 28.4% in siMcIL17-1 group, which was significantly lower than that in siNC group (Figure 6).
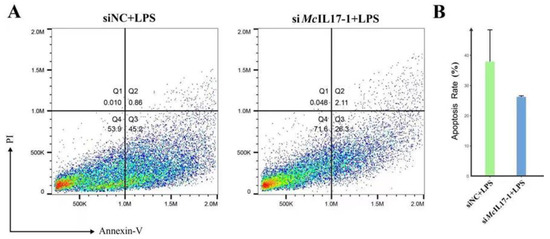
Figure 6.
Apoptotic rate in hemocytes of M. coruscus after LPS treatment and after McIL17-1 was knocked-down. The apoptotic rate of hemocytes was detected by flow cytometry with Annexin V-FITC and propidium iodide (PI) staining. (A) Apoptotic rate in RNAi mussel hemocytes. siNC + LPS: The apoptosis rate of hemocytes at 24 h after LPS challenge in siNC group; siMcIL17-1 +LPS: The apoptosis rate of hemocytes at 24 h after LPS challenge in siMcIL17-1 group. Quadrant: Q4: live cells; Q3: early apoptotic cells; Q2: late apoptotic cells; Q1: necrotic cells. (B) The changes of the apoptotic rate of hemocytes after dsRNA challenge.
3. Discussion
Innate immunity is the main defense mechanism against pathogenic infections in marine invertebrates [33]. Since its discovery in human peripheral blood, the soluble proinflammatory cytokine IL17 family, has been widely studied as one of the key signaling molecules of innate immunity [34]. In terms of medical research, IL17 expression has long been related to transplant rejection of various solid organs such as kidney and heart [35]. In addition, IL17 also plays a role in other pathophysiological processes, including host bacterial defense, granulopoiesis, rheumatoid arthritis, tumor regulation, and asthma [36]. Consequently, any dysregulation of IL17 production and disturbance of downstream signal pathways may affect the normal physiology and function of disease pathogenesis in human. In view of its significant functional role in innate immunity, the IL17 gene family has been paid more attention in aquatic animals in the past decade [37,38,39,40,41,42]. Additionally, as genome sequencing and resequencing technologies have advanced by leaps and bounds over the past two decades, members of the IL17 gene family in mollusks have been revealed in growing numbers [24,27,28]. Most of these studies focused on the IL17 evolution and immunological function verification, providing a prelude to elucidate the immune role of IL17 family in shellfish. Nevertheless, the mechanisms underlying IL17 mediated innate immune response to bacterial infection remain unclear. Here, a novel molluscan IL17 family member (McIL17-1) was identified from M. coruscus mussels. Structural analysis of McIL17-1 showed an IL17 domain, which contains four conserved cysteine. In the phylogenetic tree, McIL17-1 clustered with MgIL17-1 and aggregated in a molluscan IL17 cluster. Based on the conservation in structural domain with its corresponding genes in other bivalves, we further explored the role of McIL17-1 in innate immune restriction.
As an essential mediator of inflammatory autoimmune diseases, IL17 is involved in innate immunology responses in many tissues [16]. Pisces represent the first group in fauna evolution to have both innate and adaptive immunity [42]. In zebrafish, tissue distribution patterns of IL17 transcripts showed that DrIL17 mRNA was expressed in the kidney, spleen, gills and intestine [43], which is consistent with previously reported results from turbot (Scophthalmus maximus) [41]. Moreover, a number of studies have labelled the constitutive expression of IL17 in molluscan species. For example, in Pacific oyster, IL17 mRNA was detected in all examined tissues, including hemocytes, heart, gills, digestive glands, gonad, mantle and muscle, and highly expressed in the gills and digestive glands tissues [23]. For freshwater pearl mussel Hyriopsis cumingii, IL17 was also found in any tissues tested, with the gills and digestive glands having the highest levels of IL17 [29]. In the present study, McIL17-1 also showed a broad spectrum of expression and significantly higher expressed in gills and hemocytes compared with that in other tissues. In mollusks, gills mediate mucosal immunity and are generally regarded as the first line of defense against infection by various pathogens [44]; hemocytes are thought to be essential immunological agents that are critical for cellular and humoral immunity in the fight against pathogen invasion [6]. Considering that McIL17-1 is highly expressed in hemocytes and gills, it is feasible to function similarly to its counterparts in other mollusks in the innate immune response.
As a potent proinflammatory cytokine, the production of IL17 in mammals is induced by infection and is thought to drive tissue inflammation and autoimmune disease [45]. To further determine the functional role of McIL17-1 in innate immunity, we investigated its transcriptional response to the challenge of V. alginolyticus, a main pathogenic bacteria of bivalve mollusks [46,47]. Following stimulation by V. alginolyticus, the mRNA expression of McIL17-1 in hemocytes increased promptly, indicating an effective response to bacterial infection. Similar results have also been observed in Crassostrea gigas and Mytilus galloprovincialis. For C. gigas, the mRNA expression of CgIL17-1 in the hemocytes increased significantly at 6 h post-V. splendidus challenge, which was 31.36-fold of that in the control group [24]. The MgIL17-1 transcripts in hemocytes also showed significant upregulation after M. galloprovincialis was injected with a mixture of heat-killed Gram+ and Gram- bacteria [28]. Sensing PAMPs is a crucial step in innate immune activation [48]. Aiming to investigate whether McIL17-1 participates in innate immune response to various exogenous pathogens, we characterized the transcriptional expression of McIL17-1 in response to four PAMPs, including LPS, PGN, GLU and poly I:C. Concomitantly, McIL17-1 was significantly induced by LPS, PGN and GLU, similar to the results of upregulated transcript levels of CgIL17s in response to PAMPs challenge in the oysters [23]. An intriguing aspect was that the McIL17-1 expression of poly I:C-injected mussels did not change significantly compared to the control group, which was different from the parallel study result of P. fucata [25]. In P. fucata, PfIL17 mRNA expression was upregulated significantly by poly I:C. In contrast, for C. gigas, poly I:C challenge raised the mRNA level of almost all CgIL17s, except CgIL17-1, indicating that CgIL17s were generally involved in fighting viral infection, but CgIL17-1 may lose this role [23]. For vertebrates, LPS and poly I:C did not affect IL17N expression in Atlantic salmon head kidney cells [49]. The responses to bacteria and viruses are generally different, which is reflected in the different IL17 responses. Considering that LPS and poly I:C are substitutes for bacteria and viruses, respectively, the susceptibility to LPS and the bluntness to poly I:C suggested that McIL17-1 may be involved in the inflammatory response to bacterial infection. In addition, the responsive reactions of McIL17-1 to various of exogenous stimuli also indicated that McIL17-1 has a broad spectrum of action to include bacteria and fungi and less for viruses.
As a cytokine associated with inflammation and autoimmunity, IL17 induces the expression of various mediators of inflammation [20]. In vertebrates, most experimental evidence suggests that members of the IL17 family play a role in coordinating local tissue inflammation, primarily through the release of proinflammatory and neutrophil-mobilizing cytokines [50]. For example, IL17s in mice have been reported to play an important role in host defense against bacterial infections by inducing CXC chemokines to recruit neutrophils and induce antimicrobial proteins at the site of infection [51]. In human, IL17A and several other family cytokines are also involved in the development of psoriatic arthritis, psoriasis and ankylosing spondylitis by inducing inflammatory cytokines and chemokines [52]. For bivalves, a series of inflammatory reactions caused by bacterial infection is the leading cause of death, especially Vibrio sp. bacteria [46]. In the present study, we further explored the possible role of McIL17-1 as an inflammatory cytokine in mediating the pathogenesis of bacterial infections by evaluating the effects of silencing and overexpression of McIL17-1 on mussel survival. The present results showed that the overexpression of McIL17-1 elevated the mortality rate. In contrast, the survival rate of the knocked-down group was significantly higher than that of V. alginolyticus alone treated group and overexpression group. These results revealed that suppression of McIL17-1 expression reduced the inflammatory response and increase the survival rate, which further reinforced the point that McIL17-1 was involved in disease promoting proinflammatory processes in response to bacterial infection.
The analysis of the subcellular localization of a protein can provide insight to its function. We performed cytoplasmic localization of McIL17-1 by fluorescence microscopy in the HEK293T cell system. The results showed that McIL17-1 was located in the cytoplasm, which was in agreement with previous studies that the CgIL17-1 and IL17s of Scophthalmus maximus were subcellular located in the cytoplasm of hemocytes from C. gigas and HEK293T cells, respectively [24,41]. IL17 functions via binding to its cognate receptor and more research is needed to confirm the binding site [20].
As a form of programmed cell death, apoptosis plays a role in both the development of immune cells and the execution of an immune response. Meanwhile, apoptosis is also an important mechanism for maintaining immune homeostasis [53]. In mice, IL17 has been proven to induce endothelial apoptosis by inducing caspase-9, caspase-3 and raising the ratio of Bax/Bcl-2 [54]. However, there has not yet been one literature report that IL17 regulates apoptosis in invertebrate cells. LPS, as a cell-wall component of gram-negative bacteria and has been increasingly recognized as a powerful stimulator of cellular immunity in previous studies [55]. In oysters, it has been reported that the apoptosis of hemocytes significantly increased after LPS treatment [56], following CgSmac was employed to activate the mitochondrial apoptosis pathway by enhancing caspase-3 activity to resist exogenous LPS invasion [57]. In the present study, the effect of McIL17-1 on hemocytes apoptosis was assessed using LPS as the inducer. The apoptosis rate of the siMcIL17-1 group decreased significantly compared with the siNC group, which suggested that McIL17-1 could promote apoptosis of hemocytes challenged by LPS in M. coruscus. Given that apoptosis plays a crucial role in pushing the resolution of acute inflammatory responses, the present results suggest that McIL17-1, a proinflammatory cytokine, mediates inflammation and is produced in response to challenge [58]. Therefore, the normal levels of McIL17-1 expression contribute to the pathogenesis of bacterial infections.
4. Materials and Methods
4.1. Animals
Healthy adult M. coruscus (about 8.0–10.5 cm in shell height) with an average weight of 60.4 ± 3.2 g were obtained from Donghe market in Zhoushan City, Zhejiang Province, China. The domestication conditions were consistent with previous studies [59]. Briefly, animals were kept in tanks with artificial sterile seawater (ASW) at 25 ± 1 °C, salinity 25%, and fed with spirulina powder.
4.2. In Silico Cloning for McIL17-1
The sequence information of McIL17-1 (GenBank: GCA_011752425.2) was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). A specific primer pair (Table 1) was designed to amplify the sequence of the McIL17-1 open reading frame (ORF) region. After DNA sequencing of the PCR products, the physicochemical characteristics of McIL17-1 were assessed by a website tool Expasy (http://www.expasy.org) and SMART (http://smart.embl-heidelberg.de/) was used to predict the conservative domains. The phylogenetic tree was constructed by the MEGA X software package with Neighbor-joining (NJ) method, and bootstrap resampling (2000 pseudo repetitions) was conducted to test the reliability of branches.

Table 1.
PCR primer pairs used in the present study.
4.3. Immune Challenge and Tissue Collection
A total of 250 mussels were randomly divided into five groups. The immune challenge assay was performed as in our previous study [60], in a word, the mussels of five treatment groups were injected in the adductor with 100 μL live Vibrio alginolyticus (3 × 107 CFU/mL), lipopolysaccharide (LPS) (L3024, Sigma, from Escherichia coli O111: B4, 50 μg/mL), peptidoglycans (PGN) (77140, Sigma, from Staphylococcus aureus, 0.5 mg/mL), glucan (GLU) (G5011, Sigma, from Saccharomyces cerevisiae, 1 mg/mL), or polyinosinic-polycytidylic acid (poly I:C), (P9582, Sigma, 1 mg/mL), respectively. Of these, V. alginolyticus was obtained from the diseased mussels as the pathogenic bacteria and the rest were purchased from Sigma-Aldrich (Shanghai). Samples of hemolymph were collected from the pericardium of mussels at 0, 12, 24 and 72 h post-stimulation (hps). Hemocytes were obtained by centrifugation at 1100× g rpm for 10 min at 4 °C. There were 3 replicates for each time point, and the hemocyte samples from 3 mussels were pooled together as one replicate to alleviate the individual variation and obtain enough cells. Each time point consisted of three replicates. Moreover, six tissues, including adductor muscle, gills, mantle, gonad, hemocytes and digestive glands, were extracted from eight untreated mussels to analyze the tissue distribution of McIL17-1.
4.4. Expression and Purification of Recombinant McIL17-1 Protein
One specific primer pair IL17-1Y1 (Table 1), incorporated with BamH I and XhoI restriction sites at its 5′ end was designed to amplify the full length of the McIL17-1 ORF sequence. A recombinant plasmid termed pET-32a-McIL17-1 was generated by subcloning the McIL17-1 ORF sequence into the pET-32a prokaryotic expression vector, following transformation into Escherichia coli cells (DE3) (Takara) to express the fusion proteins. The induction and purification of the recombinant protein (rMcIL17-1) were performed according to our previous report [61], that the isopropyl-beta-D-thiogalactopy ranoside (IPTG) and Ni-nitorilotriacetic acid (NI-NTA) used as the inducer and depurator, respectively. The purified rMcIL17-1 was analyzed using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with Coomassie brilliant blue R250. The concentration of purified recombinant protein was quantified by the BCA method [62].
4.5. RNA Interference
As we described earlier, the RNA interfere assay (RNAi) was performed through the dsRNA injection [61]. The dsRNA was synthesized by T7 polymerase using McIL17-1 cDNA sequence amplified by specific primer pair (Table 1, IL17-ds) as the template. Following that, the obtained substance was transferred into the mussel by adductor injection (100 μg per mussel), and another booster shot was given 24 h later. After receiving a second dsRNA injection, the mussels were given treatment with V. alginolyticus 12 h later. During the experiment, a total of 40 mussels from each group were fed with spirulina powder and seawater was changed 2 h after feeding. Meanwhile, the number of mussel deaths was recorded daily [63].
4.6. qPCR
Quantitative real-time PCR (qPCR) was performed on the 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with the SYBRR® premix ExTaq Kit (TaKaRa, Kusatsu, Japan) to assess the transcriptional expression of McIL17-1, as previously reported [61]. The qPCR reaction system was conventional, and the reaction conditions were as follows: 95 °C pre-denaturation for 10 min, 40 cycles of 95 °C denaturation for 10 s and 58 °C annealing for 20 s. Data were analyzed using the 2–ΔΔCT method with β-actin as an internal reference [64]. Three analyses were performed on all samples.
4.7. Subcellular Localization
The McIL17-1 fragment was cloned into the pEGFP-N1 vector with one specific primer pair IL17-1Y2 (Table 1) incorporated EcoR I and BamHⅠrestriction sites, followed by a DNA sequencing verification. Due to no real established cell lines available for marine bivalves, HEK293T cells were used. HEK293T cells were inoculated in sterile 6-well plates at 60–70% density. After cell adhesion was complete, the transfection reagent Lipofectamine 3000 (Invitrogen, Waltham, CA, USA) and recombinant plasmid pEGFP-N1-McIL17-1 were mixed 1:1 to form the transfection complex. The transfection complex was repeatedly blown 10 to 15 times with a pipette, then transferred to a 6-well plate, mixed gently, and placed in a cell incubator. After 48 h of culture, 4% paraformaldehyde was used for fixation. 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/mL) was used for nuclear staining after PBS cleaning. The position of McIL17-1 in cells was observed by fluorescence microscopy.
4.8. Flow Cytometric Analysis of Apoptosis
Hemocytes apoptosis was assessed and quantified according to the manual of Annexin V-FITC Apoptosis Detection Kit (Beyotime, PK, Nantong, China). Briefly, the collected hemocytes were treated with LPS (0.1 mg/mL), and either siMcIL17-1 (10 μL, 2 μM) or si-NC for 24 h. After washing with PBS, the cells were re-suspended in the L15 medium at a final concentration of 1 × 106 cells mL−1, and then they were stained with Annexin V-FITC and PI by being incubated at room temperature for 25 min in the dark. Finally, the flow cytometry instrument (Beckman CytoFLEX FCM) was employed to detect cell apoptosis, and data were analyzed using FlowJo software (New York, NY, USA).
5. Conclusions
In this work, a novel IL17 isoform was identified and characterized from the thick shell mussel Mytilus coruscus. The increased expression in immune-related tissues and effective responsiveness to PAMPs suggest that it is involved in the innate immune response. In vivo and in vitro assays further reinforce the idea that McIL17-1 may function as a proinflammatory cytokine in the immune response to bacterial infections. The present study hints at the complexity of invertebrate immunity.
Author Contributions
P.Q., W.S. and J.Z.: Conceptualization, Methodology and Writing—Reviewing and Editing. Z.D. and L.Z.: Data curation, Software, Writing—Original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers 41976111, 42176099, 42020104009 and 42076119, and the Natural Science Foundation for Distinguished Young Scholars of Zhejiang province, grant number LR22D060002.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mount, A.S.; Wheeler, A.; Paradkar, R.P.; Snider, D. Hemocyte-mediated shell mineralization in the eastern oyster. Science 2004, 304, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K. Evolution of innate immunity: Clues from invertebrates via fish to mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef]
- Saco, A.; Rey-Campos, M.; Novoa, B.; Figueras, A. Transcriptomic Response of Mussel Gills After a Vibrio splendidus Infection Demonstrates Their Role in the Immune Response. Front. Immunol. 2020, 11, 615580. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMP s and DAMP s: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.E. DAMPs, PAMPs and alarmins: All we need to know about danger. J. Leukoc. Biol. 2007, 81, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Prado-Alvarez, M.; Gestal, C.; Li, H.; Roch, P.; Novoa, B.; Figueras, A. Functional and molecular immune response of Mediterranean mussel (Mytilus galloprovincialis) haemocytes against pathogen-associated molecular patterns and bacteria. Fish Shellfish Immunol. 2009, 26, 515–523. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef]
- Sasai, M.; Yamamoto, M. Pathogen recognition receptors: Ligands and signaling pathways by Toll-like receptors. Int. Rev. Immunol. 2013, 32, 116–133. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C., Jr. Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 2000, 173, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Miyahara, Y.; Wang, H.Y. Toll-like receptors and immune regulation: Implications for cancer therapy. Oncogene 2008, 27, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Moseley, T.; Haudenschild, D.R.; Rose, L.; Reddi, A.H. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003, 14, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Iwakura, Y.; Ishigame, H.; Saijo, S.; Nakae, S. Functional specialization of interleukin-17 family members. Immunity 2011, 34, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Cua, D.J.; Tato, C.M. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010, 10, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Brocker, C.; Thompson, D.; Matsumoto, A.; Nebert, D.W.; Vasiliou, V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum. Genom. 2010, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Roy, S.; Leal, S.M., Jr.; Sun, Y.; Howell, S.J.; Cobb, B.A.; Li, X.; Pearlman, E. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORγt and dectin-2. Nat. Immunol. 2014, 15, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L. Recent advances in the IL-17 cytokine family. Curr. Opin. Immunol. 2011, 23, 613–619. [Google Scholar] [CrossRef]
- Kolls, J.K.; Lindén, A. Interleukin-17 family members and inflammation. Immunity 2004, 21, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Hibino, T.; Loza-Coll, M.; Messier, C.; Majeske, A.J.; Cohen, A.H.; Terwilliger, D.P.; Buckley, K.M.; Brockton, V.; Nair, S.V.; Berney, K. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006, 300, 349–365. [Google Scholar] [CrossRef]
- Roberts, S.; Gueguen, Y.; de Lorgeril, J.; Goetz, F. Rapid accumulation of an interleukin 17 homolog transcript in Crassostrea gigas hemocytes following bacterial exposure. Dev. Comp. Immunol. 2008, 32, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Zhang, Y.; Xiang, Z.; Tong, Y.; Qu, F.; Yu, Z. Genomic characterization and expression analysis of five novel IL-17 genes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2014, 40, 455–465. [Google Scholar] [CrossRef]
- Cao, W.; Wang, W.; Fan, S.; Li, J.; Li, Q.; Wu, S.; Wang, L.; Song, L. The receptor CgIL-17R1 expressed in granulocytes mediates the CgIL-17 induced haemocytes proliferation in Crassostrea gigas. Dev. Comp. Immunol. 2022, 131, 104376. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Huang, X.D.; Li, Q.; He, M.X. Interleukin-17 in pearl oyster (Pinctada fucata): Molecular cloning and functional characterization. Fish Shellfish Immunol. 2013, 34, 1050–1056. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, S.; Feng, C.; Zhan, W.; Zheng, Z.; Wang, Q.; Deng, Y.; Jiao, Y.; Du, X. Evolution and function analysis of interleukin-17 gene from Pinctada fucata martensii. Fish Shellfish Immunol. 2019, 88, 102–110. [Google Scholar] [CrossRef]
- Valenzuela-Munoz, V.; Gallardo-Escarate, C. Molecular cloning and expression of IRAK-4, IL-17 and I-kappaB genes in Haliotis rufescens challenged with Vibrio anguillarum . Fish Shellfish Immunol. 2014, 36, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Varotto, L.; Gerdol, M.; Pallavicini, A.; Venier, P. IL-17 signaling components in bivalves: Comparative sequence analysis and involvement in the immune responses. Dev. Comp. Immunol. 2015, 52, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, M.; Xia, N.; Yu, S.; Chen, Y.; Wang, N. Cloning and analysis of gene expression of interleukin-17 homolog in triangle-shell pearl mussel, Hyriopsis cumingii, during pearl sac formation. Fish Shellfish Immunol. 2016, 52, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Wu, Y.; Gu, Z.; Li, H.; Li, J.; Guo, B.; Liao, Z.; Yan, X. A novel molluscan TLR molecule engaged in inflammatory response through MyD88 adapter recruitment. Dev. Comp. Immunol. 2022, 131, 104373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Li, S.-Y.; He, J.-Y.; Wu, Y.-H.; Gu, Z.-Q.; Fan, M.-H.; Guo, B.-Y.; Buttino, I.; Liao, Z.; Yan, X.-J. Microalgal feeding preference of Mytilus coruscus and its effects on fatty acid composition and microbes of the digestive gland. Aquac. Rep. 2022, 23, 101024. [Google Scholar] [CrossRef]
- Yang, J.; He, J.; Liu, L.; He, M.; Zhang, X.; Buttino, I.; Guo, B.; Yan, X.; Liao, Z. Expression profiles of antimicrobial peptides in Mytilus coruscus . Aquaculture 2022, 548, 737709. [Google Scholar] [CrossRef]
- Iwanaga, S.; Lee, B.-L. Recent advances in the innate immunity of invertebrate animals. BMB Rep. 2005, 38, 128–150. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Gurney, A.L. IL-17: Prototype member of an emerging cytokine family. J. Leukoc. Biol. 2002, 71, 1–8. [Google Scholar] [CrossRef]
- Van Kooten, C.; Boonstra, J.G.; Paape, M.E.; Fossiez, F.; Banchereau, J.; Lebecque, S.; Bruijn, J.A.; De Fijter, J.; Van Es, L.A.; Daha, M.R. Interleukin-17 activates human renal epithelial cells in vitro and is expressed during renal allograft rejection. J. Am. Soc. Nephrol. 1998, 9, 1526–1534. [Google Scholar] [CrossRef]
- Benchetrit, F.; Ciree, A.; Vives, V.; Warnier, G.; Gey, A.; Sautes-Fridman, C.; Fossiez, F.; Haicheur, N.; Fridman, W.H.; Tartour, E. Interleukin-17 inhibits tumor cell growth by means of a T-cell–dependent mechanism. Blood J. Am. Soc. Hematol. 2002, 99, 2114–2121. [Google Scholar] [CrossRef]
- Tang, D.; Wu, S.; Luo, K.; Yuan, H.; Gao, W.; Zhu, D.; Zhang, W.; Xu, Q. Sequence characterization and expression pattern analysis of six kinds of IL-17 family genes in the Asian swamp eel (Monopterus albus). Fish Shellfish Immunol. 2019, 89, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, T.; Huo, D.; Yu, Z.; Ruan, Y.; Cheng, C.; Jiang, X.; Ren, C. Transcriptomic analysis of sea cucumber (Holothuria leucospilota) coelomocytes revealed the echinoderm cytokine response during immune challenge. BMC Genom. 2020, 21, 306. [Google Scholar] [CrossRef]
- Li, Z.; Fan, T.; Liu, X.; Liu, X.; Wang, W.; Wang, Q.; You, L.; Wang, L.; Wei, X.; Yang, J. Characterization and functional study on Octopus ocellatus interleukin-17. J. Ocean Univ. China 2019, 18, 1443–1450. [Google Scholar] [CrossRef]
- Lv, Z.; Guo, M.; Zhao, X.; Shao, Y.; Zhang, W.; Li, C. IL-17/IL-17 Receptor Pathway–Mediated Inflammatory Response in Apostichopus japonicus Supports the Conserved Functions of Cytokines in Invertebrates. J. Immunol. 2022, 208, 464–479. [Google Scholar] [CrossRef]
- Xue, T.; Liu, Y.; Cao, M.; Zhang, X.; Fu, Q.; Yang, N.; Li, C. Genome-wide identification of interleukin-17 (IL-17)/interleukin-17 receptor (IL-17R) in turbot (Scophthalmus maximus) and expression pattern analysis after Vibrio anguillarum infection. Dev. Comp. Immunol. 2021, 121, 104070. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-Y.; Nie, L.; Zhu, G.; Xiang, L.-X.; Shao, J.-Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Gunimaladevi, I.; Savan, R.; Sakai, M. Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immunol. 2006, 21, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wickamarachchi, W.D.; Whang, I.; Oh, M.; Umasuthan, N.; De Zoysa, M.; Oh, C.; Kang, D.H.; Lee, J. Immune response-related gene expression profile of a novel molluscan IkappaB protein member from Manila clam (Ruditapes philippinarum). Mol. Biol. Rep. 2013, 40, 1519–1527. [Google Scholar] [CrossRef]
- Chen, C.; Itakura, E.; Nelson, G.M.; Sheng, M.; Laurent, P.; Fenk, L.A.; Butcher, R.A.; Hegde, R.S.; de Bono, M. IL-17 is a neuromodulator of Caenorhabditis elegans sensory responses. Nature 2017, 542, 43–48. [Google Scholar] [CrossRef]
- Dubert, J.; Barja, J.L.; Romalde, J.L. New insights into pathogenic Vibrios affecting bivalves in hatcheries: Present and future prospects. Front. Microbiol. 2017, 8, 762. [Google Scholar] [CrossRef]
- Elston, R.A.; Hasegawa, H.; Humphrey, K.L.; Polyak, I.K.; Häse, C.C. Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: Severity, environmental drivers, geographic extent and management. Dis. Aquat. Org. 2008, 82, 119–134. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Kumari, J.; Larsen, A.N.; Bogwald, J.; Dalmo, R.A. Interleukin-17D in Atlantic salmon (Salmo salar): Molecular characterization, 3D modelling and promoter analysis. Fish Shellfish Immunol. 2009, 27, 647–659. [Google Scholar] [CrossRef]
- Ishigame, H.; Kakuta, S.; Nagai, T.; Kadoki, M.; Nambu, A.; Komiyama, Y.; Fujikado, N.; Tanahashi, Y.; Akitsu, A.; Kotaki, H.; et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 2009, 30, 108–119. [Google Scholar] [CrossRef]
- Huang, W.; Na, L.; Fidel, P.L.; Schwarzenberger, P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J. Infect. Dis. 2004, 190, 624–631. [Google Scholar] [CrossRef]
- Chung, S.-H.; Ye, X.-Q.; Iwakura, Y. Interleukin-17 family members in health and disease. Int. Immunol. 2021, 33, 723–729. [Google Scholar] [CrossRef]
- Nagata, S.; Tanaka, M. Programmed cell death and the immune system. Nat. Rev. Immunol. 2017, 17, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Li, Y.Y.; Wang, J.M.; Manthari, R.K.; Wang, J.D. Fluoride induces apoptosis and autophagy through the IL-17 signaling pathway in mice hepatocytes. Arch. Toxicol. 2018, 92, 3277–3289. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-S.; Lyu, S.-J.; Xu, J.-H.; Lu, B.-J.; Zhao, J.; Li, S.; Li, Y.-Q.; Chen, Y.-Y. Effect of lipopolysaccharide on the hemocyte apoptosis of Eriocheir sinensis . J. Zhejiang Univ.-Sci. B 2015, 16, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zho, Z.; Wang, L.L.; Yang, C.Y.; Jianga, S.; Song, L.S. The immunomodulation of a novel tumor necrosis factor (CgTNF-1) in oyster Crassostrea gigas. Dev. Comp. Immunol. 2014, 45, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Song, X.; Xu, J.; Jia, Z.; Yang, B.; Jia, Y.; Qiu, L.; Wang, L.; Song, L. The modulation of Smac/DIABLO on mitochondrial apoptosis induced by LPS in Crassostrea gigas. Fish Shellfish Immunol. 2019, 84, 587–598. [Google Scholar] [CrossRef]
- Savill, J. Apoptosis in resolution of inflammation. J. Leukoc. Biol. 1997, 61, 375–380. [Google Scholar] [CrossRef]
- Qi, P.; Huang, H.; Guo, B.; Liao, Z.; Liu, H.; Tang, Z.; He, Y. A novel interleukin-1 receptor-associated kinase-4 from thick shell mussel Mytilus coruscus is involved in inflammatory response. Fish Shellfish Immunol. 2019, 84, 213–222. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, Z.; Xu, Z.; Tang, Z.; Liu, L.; Lu, Z.; Qi, P. A novel invertebrate toll-like receptor is involved in TLR mediated signal pathway of thick shell mussel Mytilus coruscus . Dev. Comp. Immunol. 2019, 97, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Tang, Z. The Nrf2 molecule trigger antioxidant defense against acute benzo (a) pyrene exposure in the thick shell mussel Mytilus coruscus . Aquat. Toxicol. 2020, 226, 105554. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.e.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Viarengo, A.; Burlando, B.; Cavaletto, M.; Marchi, B.; Ponzano, E.; Blasco, J. Role of metallothionein against oxidative stress in the mussel Mytilus galloprovincialis . Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1999, 277, R1612–R1619. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).