Rhamnopyranoside-Based Fatty Acid Esters as Antimicrobials: Synthesis, Spectral Characterization, PASS, Antimicrobial, and Molecular Docking Studies
Abstract
1. Introduction
2. Results and Discussion
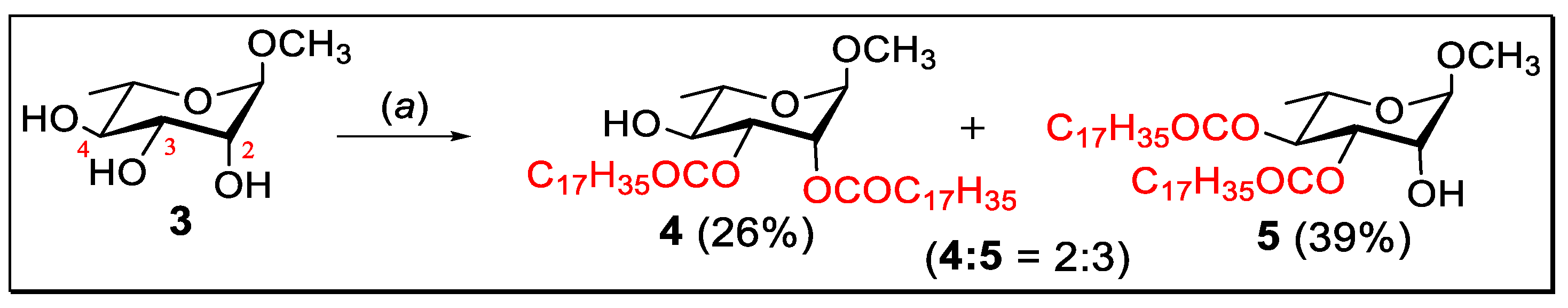
2.1. Selective Stearoylation: Synthesis of Methyl 2,3-di-O-stearoyl-α-l-rhamnopyranoside (4) and Methyl 3,4-di-O-stearoyl-α-l-rhamnopyranoside (5)
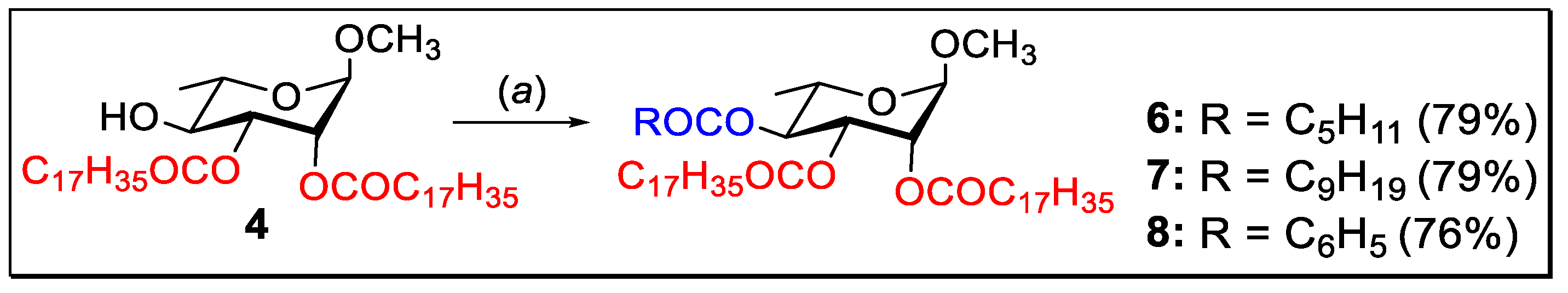
2.2. 4-O-Acylation of Di-O-stearate 4
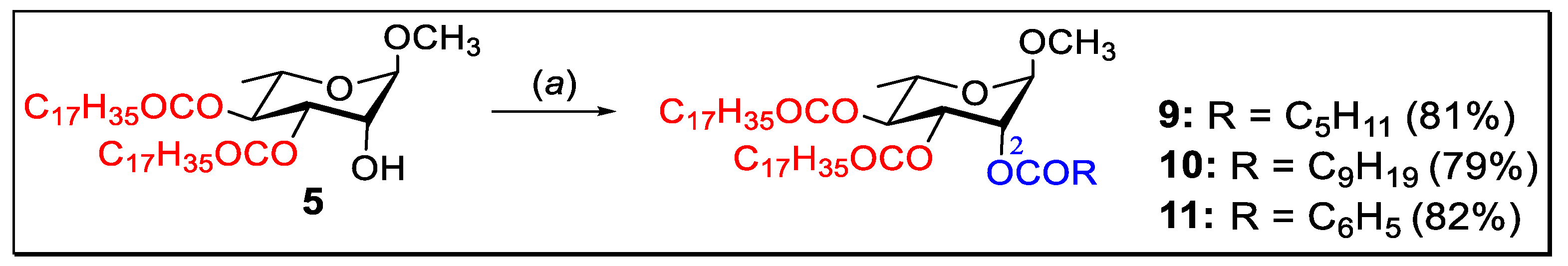
2.3. 2-O-Acylation of Di-O-stearate 5
2.4. Prediction of Activity Spectra for Substances (PASS) of 3–11
2.5. In Vitro Antimicrobial Activities of Rhamnopyranoside Esters 4–11
2.6. ADME/T Analysis
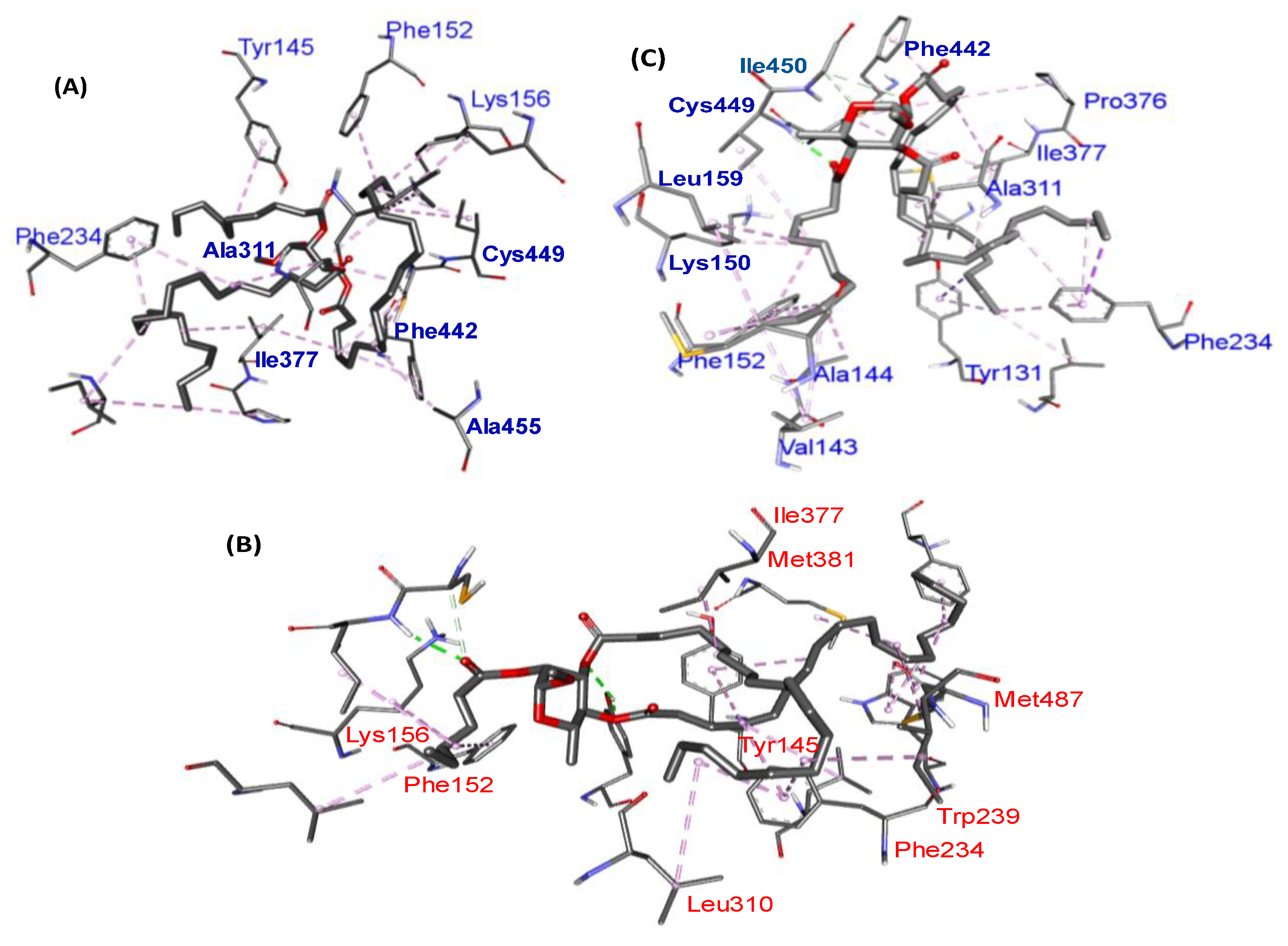
2.7. Molecular Docking Results
3. Materials and Methods
3.1. Materials and Instrumentation
3.2. Synthesis
3.3. PASS Calculation
3.4. Evaluation of In Vitro Antimicrobial Activities
3.5. ADME/T Analysis
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Teng, Y.; Stewart, S.G.; Hai, Y.-W.; Li, X.; Banwell, M.G.; Lan, P. Sucrose fatty acid esters: Synthesis, emulsifying capacities, biological activities and structure-property profiles. Crit. Rev. Food Sci. Nutr. 2021, 61, 3297–3317. [Google Scholar] [CrossRef] [PubMed]
- Dhavale, D.D.; Matin, M.M. Selective sulfonylation of 4-C-hydroxymethyl-β-L-threo-pento-1,4-furanose: Synthesis of bicyclic diazasugars. Tetrahedron 2004, 60, 4275–4281. [Google Scholar] [CrossRef]
- Zheng, Y.; Zheng, M.; Ma, Z.; Xin, B.; Guo, R.; Xu, X. Sugar Fatty Acid Esters. In Polar Lipids; Ahmad, M.U., Xu, X., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 215–243. [Google Scholar] [CrossRef]
- Matin, M.M.; Chakraborty, P.; Alam, M.S.; Islam, M.M.; Hanee, U. Novel mannopyranoside esters as sterol 14α-demethylase inhibitors: Synthesis, PASS predication, molecular docking, and pharmacokinetic studies. Carbohydr. Res. 2020, 496, 108130. [Google Scholar] [CrossRef] [PubMed]
- Snoch, W.; Wnuk, D.; Witko, T.; Staroń, J.; Bojarski, A.J.; Jarek, E.; Plou, F.J.; Guzik, M. In search of effective anticancer agents-Novel sugar esters based on polyhydroxyalkanoate monomers. Int. J. Mol. Sci. 2021, 22, 7238. [Google Scholar] [CrossRef]
- Rahman, M.A.; Matin, M.M.; Kumer, A.; Chakma, U.; Rahman, M.R. Modified D-glucofuranoses as new black fungus protease inhibitors: Computational screening, docking, dynamics, and QSAR study. Physical Chem. Res. 2022, 10, 195–209. [Google Scholar] [CrossRef]
- Matin, P.; Rahman, M.R.; Huda, D.; Bakri, M.K.B.; Uddin, J.; Yurkin, Y.; Burko, A.; Kuok, K.K.; Matin, M.M. Application of synthetic acyl glucopyranosides for white-rot and brown-rot fungal decay resistance in aspen and pine wood. BioResources 2022, 17, 3025–3041. [Google Scholar] [CrossRef]
- Matin, M.M.; Bhuiyan, M.M.H.; Kibria, S.M.; Hasan, M.S. Synthesis, PASS predication of antimicrobial activity and pharmacokinetic properties of hexanoyl galactopyranosides and experimental evaluation of their action against four human pathogenic bacteria and four fungal strains. Pharm. Chem. J. 2022, 56, 627–637. [Google Scholar] [CrossRef]
- Szűts, A.; Szabó-Révész, P. Sucrose esters as natural surfactants in drug delivery systems—A mini-review. Int. J. Pharm. 2012, 433, 1–9. [Google Scholar] [CrossRef]
- Verboni, M.; Lucarini, S.; Duranti, A. 6′-O-Lactose ester surfactants as an innovative opportunity in the pharmaceutical field: From synthetic methods to biological applications. Pharmaceuticals 2021, 14, 1306. [Google Scholar] [CrossRef]
- Hollenbach, R.; Delavault, A.; Gebhardt, L.; Soergel, H.; Muhle-Goll, C.; Ochsenreither, K.; Syldatk, C. Lipase-mediated mechanoenzymatic synthesis of sugar esters in dissolved unconventional and neat reaction systems. ACS Sustainable Chem. Eng. 2022, 10, 10192–10202. [Google Scholar] [CrossRef]
- Jacob, J.N.; Tazawa, M.J. Glucose–aspirin: Synthesis and in vitro anti-cancer activity studies. Bioorg. Med. Chem. Lett. 2012, 22, 3168. [Google Scholar] [CrossRef]
- El-Baz, H.A.; Elazzazy, A.M.; Saleh, T.S.; Dourou, M.; Mahyoub, J.A.; Baeshen, M.N.; Madian, H.R.; Aggelis, G. Enzymatic synthesis of glucose fatty acid esters using SCOs as acyl group-donors and their biological activities. Appl. Sci. 2021, 11, 2700. [Google Scholar] [CrossRef]
- Matin, P.; Hanee, U.; Alam, M.S.; Jeong, J.E.; Matin, M.M.; Rahman, M.R.; Mahmud, S.; Alshahrani, M.M.; Kim, B. Novel galactopyranoside esters: Synthesis, mechanism, in vitro antimicrobial evaluation and molecular docking studies. Molecules 2022, 27, 4125. [Google Scholar] [CrossRef] [PubMed]
- Kinnaert, C.; Daugaard, M.; Nami, F.; Clausen, M.H. Chemical synthesis of oligosaccharides related to the cell walls of plants and algae. Chem. Rev. 2017, 117, 11337–11405. [Google Scholar] [CrossRef]
- Hu, J.F.; Wunderlich, D.; Sattler, I.; Haertl, A.; Papastavrou, I.; Grond, S.; Grabley, S.; Feng, X.Z.; Thiericke, R. New 1-O-acyl alpha-L-rhamnopyranosides and rhamnosylated lactones from Streptomyces sp., inhibitors of 3 alpha-hydroxysteroid-dehydrogenase (3alpha-HSD). J. Antibiot. 2000, 53, 944–953. [Google Scholar]
- Shigemori, H.; Komaki, H.; Yazawa, K.; Mikami, Y.; Nemoto, A.; Tanaka, Y.; Sasaki, T.; In, Y.; Ishida, T.; Kobayashi, J.; et al. A novel tricyclic metabolite with potent immunosuppressive activity from Actinomycete nocardiabrasiliensis. J. Org. Chem. 1998, 63, 6900–6904. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, Y.C. Neuroprotective phenylpropanoid esters of rhamnose isolated from roots of Scrophularia buergeriana. Phytochem. 2000, 54, 503–509. [Google Scholar] [CrossRef]
- Elmaidomy, A.H.; Mohammed, R.; Owis, A.I.; Hetta, M.H.; AboulMagd, A.M.; Siddique, A.B.; Abdelmohsen, U.R.; Rateb, M.E.; El Sayed, K.A.; Hassan, H.M. Triple-negative breast cancer suppressive activities, antioxidants and pharmacophore model of new acylated rhamnopyranoses from Premna odorata. RSC Adv. 2020, 10, 10584. [Google Scholar] [CrossRef]
- Mihoub, M.; Pichette, A.; Sylla, B.; Gauthier, C.; Legault, J. Bidesmosidic betulin saponin bearing L-rhamnopyranoside moieties induces apoptosis and inhibition of lung cancer cells growth in vitro and in vivo. PLoS ONE 2018, 13, e0193386. [Google Scholar] [CrossRef]
- Matin, M.M.; Nath, A.R.; Saad, O.; Bhuiyan, M.M.H.; Kadir, F.A.; Hamid, S.B.A.; Alhadi, A.A.; Ali, M.E.; Yehye, W.A. Synthesis, PASS-predication and in vitro antimicrobial activity of benzyl 4-O-benzoyl-α-l-rhamnopyranoside derivatives. Int. J. Mol. Sci. 2016, 17, 1412. [Google Scholar] [CrossRef] [PubMed]
- Matin, M.M.; Iqbal, M.Z. Methyl 4-O-(2-chlorobenzoyl)-α-l-rhamnopyranosides: Synthesis, characterization, and thermodynamic studies. Orbital: Electron. J. Chem. 2021, 13, 19–27. [Google Scholar] [CrossRef]
- Matin, M.M. Synthesis and antimicrobial study of some methyl 4-O-palmitoyl-α-l-rhamnopyranoside derivatives. Orbital: Electron. J. Chem. 2014, 6, 20–28. [Google Scholar]
- Matin, M.M.; Ibrahim, M.; Anisa, T.R.; Rahman, M.R. Synthesis, characterization, in silico optimization, and conformational studies of methyl 4-O-palmitoyl-α-l-rhamnopyranosides. Malaysian J. Sci. 2022, 41, 91–105. [Google Scholar] [CrossRef]
- Islam, F.; Rahman, M.R.; Matin, M.M. The effects of protecting and acyl groups on the conformation of benzyl α-l-rhamnopyranosides: An in silico study. Turkish Comp. Theor. Chem. 2021, 5, 39–50. [Google Scholar] [CrossRef]
- Matin, M.M.; Islam, N.; Siddika, A.; Bhattacharjee, S.C. Regioselective synthesis of some rhamnopyranoside esters for PASS predication, and ADMET studies. J. Turkish Chem. Soc. Sect. A Chem. 2021, 8, 363–374. [Google Scholar] [CrossRef]
- Matin, M.M.; Bhuiyan, M.M.H.; Azad, A.K.M.S.; Akther, N. Design and synthesis of benzyl 4-O-lauroyl-α-l-rhamnopyranoside derivatives as antimicrobial agents. Current Chem. Lett. 2017, 6, 31–40. [Google Scholar] [CrossRef]
- An, J.; Zou, Y.Z.; Hao, X.Y.; Wang, G.C.; Li, Z.S. Antibacterial and synergy of a flavanonol rhamnoside with antibiotics against clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA). Phytomedicine 2011, 18, 990–993. [Google Scholar] [CrossRef]
- Lingaraju, M.C.; Anand, S.; Balaganur, V.; Kumari, R.R.; More, A.S.; Kumar, D.; Brijesh Bhadoria, K.; Tandan, S.K. Analgesic activity of Eugenia jambolana leave constituent: A dikaempferol rhamnopyranoside from ethyl acetate soluble fraction. Pharm. Biol. 2014, 52, 1069–1078. [Google Scholar] [CrossRef]
- Wang, C.-C.; Lee, J.-C.; Luo, S.-Y.; Kulkarni, S.S.; Huang, Y.-W.; Lee, C.-C.; Chang, K.-L.; Hung, S.-C. Regioselective one-pot protection of carbohydrates. Nature 2007, 446, 896. [Google Scholar] [CrossRef]
- Perona, A.; Hoyos, P.; Farrán, A.; Hernáiz, M.J. Current challenges and future perspectives in sustainable mechanochemical transformations of carbohydrates. Green Chem. 2020, 22, 5559–5583. [Google Scholar] [CrossRef]
- Lv, J.; Luo, T.; Zhang, Y.; Pei, Z.; Dong, H. Regio/Site-selective benzoylation of carbohydrates by catalytic amounts of FeCl3. ACS Omega 2018, 3, 17717–17723. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Ye, R.; Davidson, P.M.; Hayes, D.G.; Golden, D.A.; Zhong, Q. Sucrose monolaurate improves the efficacy of sodium hypochlorite against Escherichia coli O157:H7 on spinach. Int. J. Food Microb. 2011, 145, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Petrova, K.T.; Barros, M.T.; Calhelha, R.C.; Soković, M.; Ferreira, I.C.F.R. Antimicrobial and cytotoxic activities of short carbon chain unsaturated sucrose esters. Med. Chem. Res. 2018, 27, 980–988. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heter. Compds. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Hanee, U.; Rahman, M.R.; Matin, M.M. Synthesis, PASS, in silico ADMET, and thermodynamic studies of some galactopyranoside esters. Phys. Chem. Res. 2021, 9, 591–603. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7th ed.; CLSI Document M02-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Arastehfar, A.; Gabaldón, T.; Garcia-Rubio, R.; Jenks, J.D.; Hoenigl, M.; Salzer, H.J.F.; Ilkit, M.; Lass-Flörl, C.; Perlin, D.S. Drug-resistant fungi: An emerging challenge threatening our limited antifungal armamentarium. Antibiotics 2020, 9, 877. [Google Scholar] [CrossRef]
- Reddy, G.K.K.; Padmavathi, A.R.; Nancharaiah, Y.V. Fungal infections: Pathogenesis, antifungals and alternate treatment approaches. Curr. Res. Microb. Sci. 2022, 3, 100137. [Google Scholar] [CrossRef]
- Matin, M.M.; Bhuiyan, M.M.H.; Kabir, E.; Sanaullah, A.F.M.; Rahman, M.A.; Hossain, M.E.; Uzzaman, M. Synthesis, characterization, ADMET, PASS predication, and antimicrobial study of 6-O-lauroyl mannopyranosides. J. Mol. Struct. 2019, 1195, 189–197. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Galati, S.; Di Stefano, M.; Martinelli, E.; Macchia, M.; Martinelli, A.; Poli, G.; Tuccinardi, T. VenomPred: A Machine Learning Based Platform for Molecular Toxicity Predictions. Int. J. Mol. Sci. 2022, 23, 2105. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Wang, G.; Zhu, W. Molecular docking for drug discovery and development: A widely used approach but far from perfect. Future Med. Chem. 2016, 8, 1707–1710. [Google Scholar] [CrossRef]
- Akash, S.; Kumer, A.; Chandro, A.; Chakma, U.; Matin, M.M. Quantum calculation, docking, ADMET and molecular dynamics of ketal and non-ketal forms of D-glucofuranose against bacteria, black & white fungus, and triple-negative breast cancer. Bioint. Res. Appl. Chem. 2023, 13, 374. [Google Scholar] [CrossRef]
- Odds, F.C.; Brown, A.J.P.; Gow, N.A.R. Antifungal agents: Mechanisms of action. Trends Microbiol. 2003, 11, 272–279. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Revision D.01; Gaussian 09W; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Strushkevich, N.; Usanov, S.A.; Park, H.W. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 2010, 397, 1067–1078. [Google Scholar] [CrossRef]
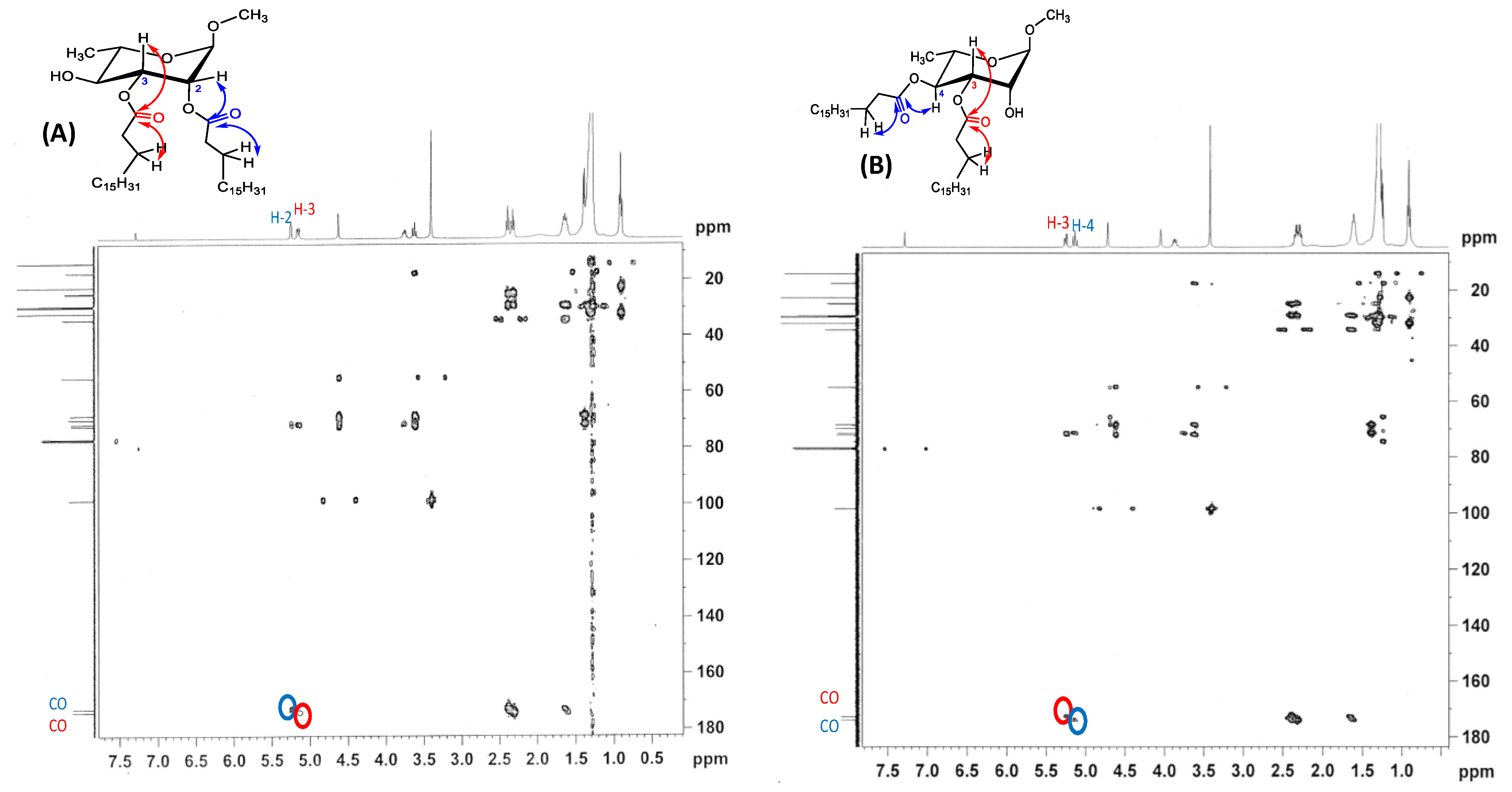
| Compound | H-1 (δ ppm) | H-2 (δ ppm) | H-3 (δ ppm) | H-4 (δ ppm) | H-5 (δ ppm) |
|---|---|---|---|---|---|
| 4 | 4.61 | 5.23 | 5.12–5.13 | 3.60 | 3.71–3.78 |
| 6 | 4.63 | 5.26 | 5.29–5.33 | 5.1 | 3.84–3.91 |
| 7 | 4.63 | 5.26 | 5.29–5.33 | 5.1 | 3.84–3.91 |
| 8 | 4.63 | 5.41 | 5.15–5.49 | 5.3 | 3.99–4.1 |
| 5 | 4.71 | 4.04 | 5.23–5.26 | 5.13 | 3.83–3.90 |
| 9 | 4.63 | 5.26 | 5.30–5.33 | 5.10 | 3.84–3.91 |
| 10 | 4.63 | 5.26 | 5.29–5.32 | 5.10 | 3.85–3.89 |
| 11 | 4.80 | 5.48 | 5.41–5.44 | 5.26 | 4.05–4.07 |
| Drugs | Biological Activity Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Antibacterial | Antifungal | Anti-Carcinogenic | Antiviral (Herpes) | |||||
| Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | |
| 3 | 0.574 | 0.010 | 0.650 | 0.013 | 0.662 | 0.010 | 0.474 | 0.013 |
| 4 | 0.626 | 0.007 | 0.717 | 0.009 | 0.642 | 0.011 | 0.494 | 0.010 |
| 5 | 0.622 | 0.008 | 0.731 | 0.008 | 0.704 | 0.008 | 0.472 | 0.014 |
| 6 | 0.600 | 0.009 | 0.690 | 0.010 | 0.534 | 0.017 | 0.472 | 0.014 |
| 7 | 0.600 | 0.009 | 0.690 | 0.010 | 0.534 | 0.017 | 0.472 | 0.014 |
| 8 | 0.588 | 0.009 | 0.688 | 0.010 | 0.573 | 0.014 | 0.469 | 0.014 |
| 9 | 0.600 | 0.009 | 0.690 | 0.010 | 0.534 | 0.017 | 0.472 | 0.014 |
| 10 | 0.600 | 0.009 | 0.690 | 0.010 | 0.534 | 0.017 | 0.472 | 0.014 |
| 11 | 0.588 | 0.009 | 0.688 | 0.010 | 0.573 | 0.014 | 0.469 | 0.014 |
| NYS | 0.967 | 0.000 | 0.986 | 0.000 | 0.416 | 0.028 | 0.959 | 0.000 |
| FCZ | -- | -- | 0.726 | 0.008 | -- | -- | -- | -- |
| ICZ | -- | -- | 0.846 | 0.003 | -- | -- | -- | -- |
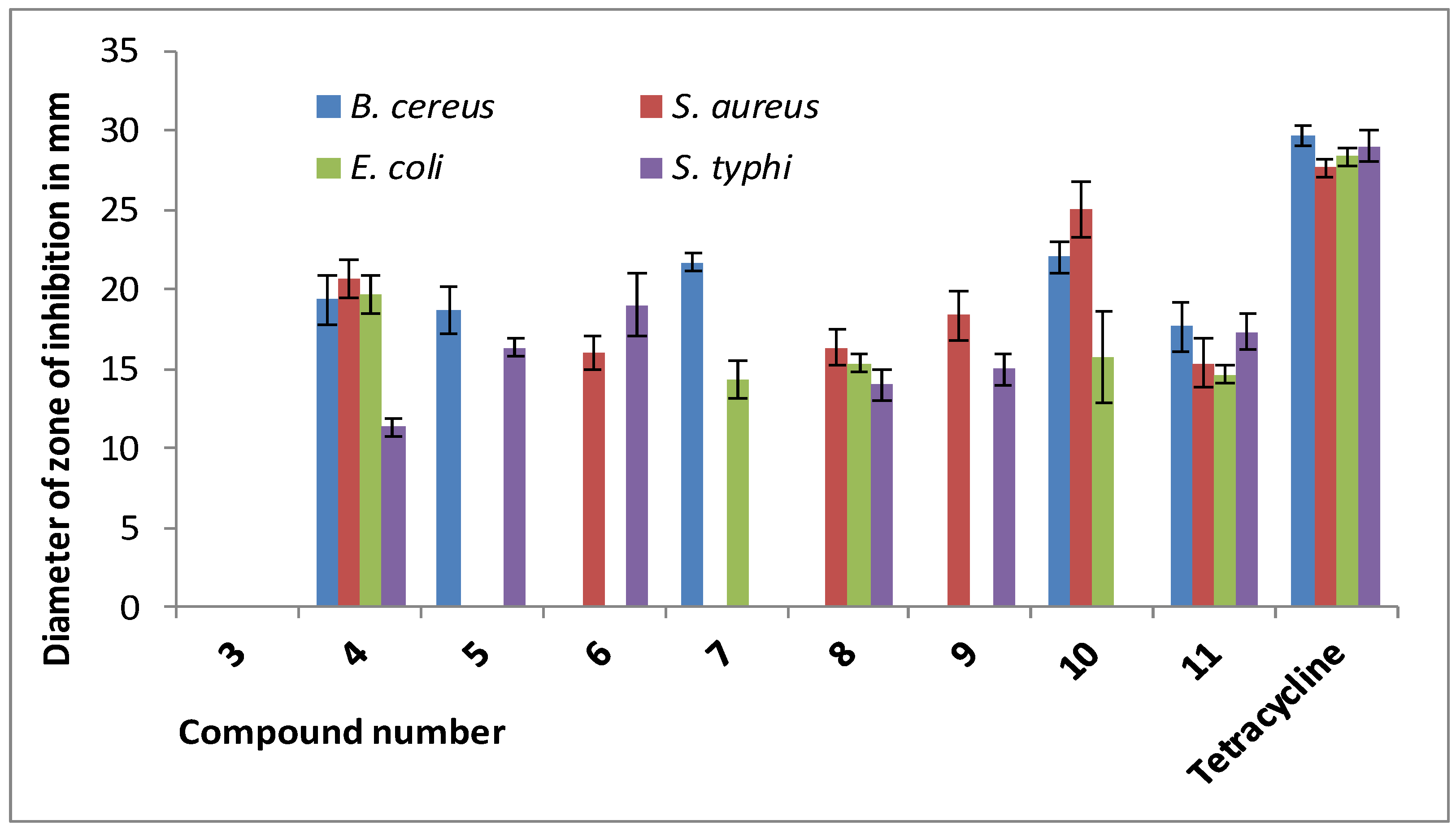
| Drugs | Diameter of Zone of Inhibition in mm (100 µg dw/disc) | |||
|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | |||
| B. cereus | S. aureus | E. coli | S. typhi | |
| 3 | NI | NI | NI | NI |
| 4 | 19.33 ± 1.52 | * 20.66 ± 1.15 | 19.66 ± 1.15 | 11.33 ± 0.57 |
| 5 | 18.66 ± 1.52 | NI | NI | 16.33 ± 0.57 |
| 6 | NI | 16.00 ± 1.00 | NI | 19.00 ± 2.00 |
| 7 | * 21.66 ± 0.57 | NI | 14.33 ± 1.15 | NI |
| 8 | NI | 16.33 ± 1.15 | 15.33 ± 0.57 | 14.00 ± 1.00 |
| 9 | NI | 18.33 ± 1.53 | NI | 15.00 ± 1.00 |
| 10 | * 22.00 ± 1.00 | * 25.00 ± 1.73 | 15.66 ± 2.88 | NI |
| 11 | 17.66 ± 1.52 | 15.33 ± 1.53 | 14.66 ± 0.57 | 17.33 ± 1.15 |
| Tetracycline ** | * 29.66 ± 0.58 | * 27.66 ± 0.58 | * 28.33 ± 0.57 | * 29.00 ± 1.00 |
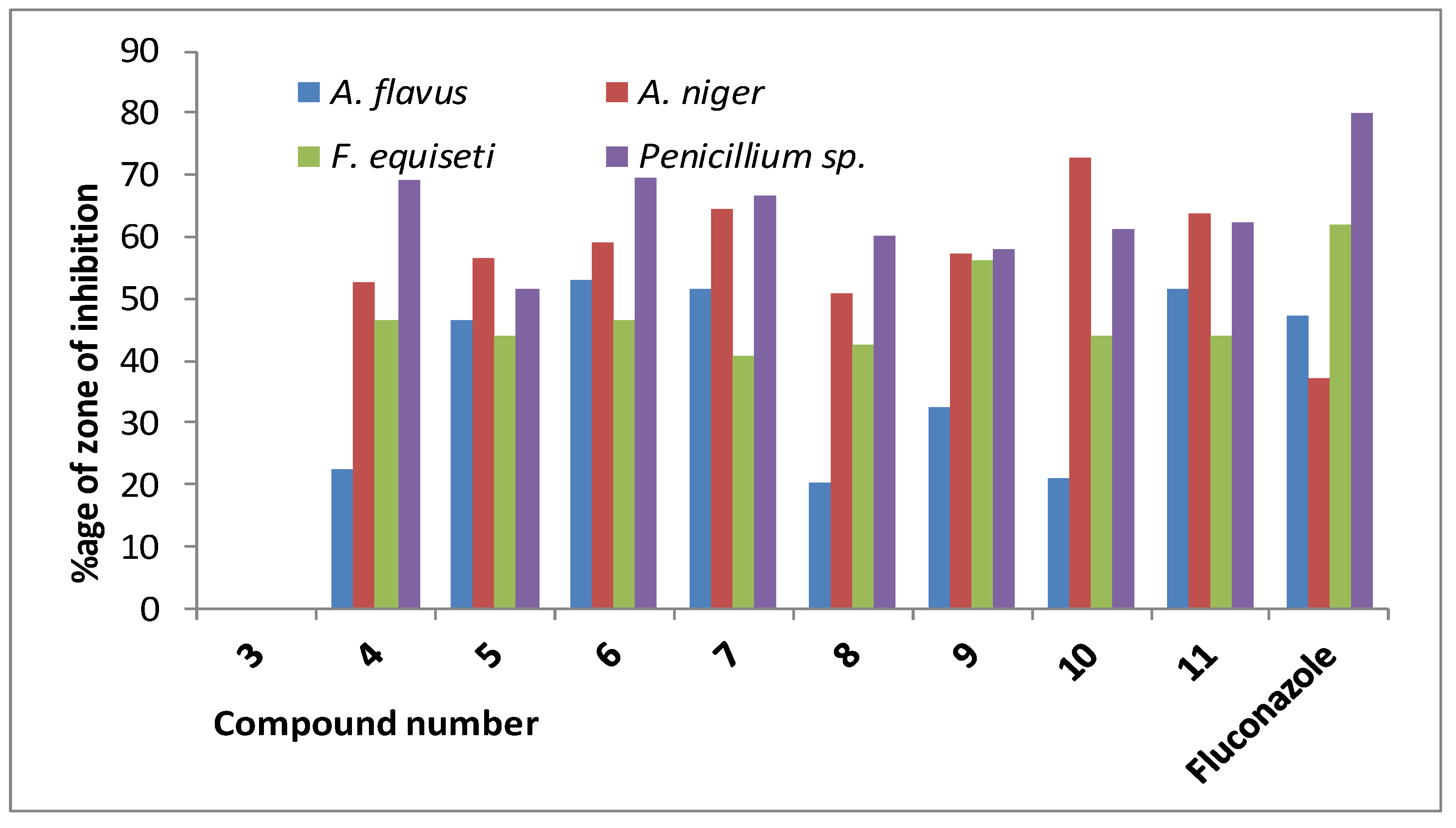
| Drug | % Inhibition of Fungal Mycelial Growth (100 µg dw/mL PDA) | |||
|---|---|---|---|---|
| A. flavus | A. niger | F.equiseti | Penicillium sp. | |
| 3 | NI | NI | NI | NI |
| 4 | 22.46 ± 1.25 | 52.61 ± 1.71 | 46.41 ± 2.00 | * 69.00 ± 1.00 |
| 5 | 46.37 ± 1.25 | 56.72 ± 1.01 | 44.04 ± 1.58 | 51.66 ± 1.53 |
| 6 | 52.89 ± 1.25 | 59.06 ± 2.67 | 46.41 ± 1.98 | * 69.33 ± 1.15 |
| 7 | 51.46 ± 1.25 | * 64.32 ± 1.01 | 40.73 ± 3.21 | * 66.66 ± 1.15 |
| 8 | 20.28 ± 3.32 | 50.87 ± 3.05 | 42.44 ± 2.02 | * 60.00 ± 2.00 |
| 9 | 32.60 ± 2.17 | 57.31 ± 2.02 | 56.11 ± 1.99 | 58.00 ± 2.00 |
| 10 | 21.01 ± 2.51 | * 72.62 ± 1.83 | 44.00 ± 1.00 | * 61.33 ± 1.15 |
| 11 | 51.45 ± 4.52 | * 63.74 ± 2.02 | 44.14 ± 1.52 | * 62.33 ± 1.53 |
| Fluconazole ** | 47.03 ± 0.25 | 37.10 ± 0.17 | * 62.00 ± 0.50 | * 79.99 ± 1.53 |
| Compound | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|
| Physicochemical properties | |||||||||
| MW a (g/mol) | 178.18 | 711.11 | 711.11 | 809.25 | 865.36 | 815.21 | 809.25 | 865.36 | 815.21 |
| Rotatable bonds | 1 | 37 | 37 | 43 | 47 | 40 | 43 | 47 | 40 |
| HBA | 19 | 89 | 89 | 100 | 108 | 94 | 100 | 108 | 94 |
| HBD | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TPSA (Å2) | 79.15 | 91.29 | 91.29 | 97.36 | 97.36 | 97.36 | 97.36 | 97.36 | 97.36 |
| Absorption | |||||||||
| Lipophilicity (iLogP) | 0.77 | 9.81 | 9.32 | 9.68 | 10.26 | 10.41 | 9.51 | 11.08 | 10.05 |
| Water solubility | −0.243 | −3.690 | −3.839 | −3.219 | −3.022 | −3.240 | −3.222 | −3.022 | −3.235 |
| GI absorption (%) | 70.741 | 84.865 | 85.291 | 100 | 100 | 100 | 100 | 100 | 100 |
| Distribution | |||||||||
| BBB3 permeant (log BB) | −0.716 | −1.541 | −1.541 | −1.854 | −1.932 | −1.766 | −1.845 | −1.932 | −1.766 |
| Metabolism | |||||||||
| CYP2D6 substrate | No | No | No | No | No | No | No | No | No |
| CYP3A4 substrate | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Excretion | |||||||||
| Total Clearance (log mL/min/kg) | 0.624 | 2.062 | 1.996 | 2.003 | 2.080 | 1.815 | 2.003 | 2.080 | 1.815 |
| Toxicity | |||||||||
| Hepatotoxicity | 0.83 | 0.79 | 0.79 | 0.77 | 0.77 | 0.77 | 0.77 | 0.77 | 0.77 |
| Carcinogenicity | 0.65 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 |
| Immunotoxicity | 0.87 | 0.93 | 0.95 | 0.89 | 0.89 | 0.99 | 0.89 | 0.89 | 0.99 |
| Mutagenicity | 0.69 | 0.77 | 0.77 | 0.75 | 0.75 | 0.81 | 0.75 | 0.75 | 0.81 |
| Cytotoxicity | 0.81 | 0.81 | 0.81 | 0.79 | 0.79 | 0.78 | 0.79 | 0.79 | 0.78 |
| Predicted LD50 (mg/kg) | 23,000 | 8000 | 8000 | 8000 | 8000 | 4000 | 8000 | 8000 | 4000 |
| Toxicity class | 6 | 6 | 6 | 6 | 6 | 5 | 6 | 6 | 5 |
| Drug-likeness | |||||||||
| Lipinski’s rule | F = 4; V = 0 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 | F = 2; V = 2 |
| Veber’s rule | F = 2; V = 0 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 | F = 1; V = 1 |
| Ghosh’s rule | F = 2; V = 2 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 | F = 0; V = 4 |
 and
and  color = inactive;
color = inactive;  color = active; F = follow; V = violate.
color = active; F = follow; V = violate.| Compound/ Drug | 3LD6 (kcal/mol) | Conventional H-Bond | Hydrophobic Interaction | |
|---|---|---|---|---|
| Alkyl | Pi-alkyl | |||
| 3 | −4.8 | LEU310 | - | - |
| 4 | −5.1 | ALA224; ALA228; ALA237; ILE488 | ALA224; ALA228; ALA237; ILE488 | PHE195 |
| 5 | −5.6 | - | ALA74; VAL101; ALA237; ILE379 | PHE77; PHE105; HIS236; TRP239; TRP239; TRP239 |
| 6 | 5.7 | - | MET100; VAL101; ALA237; LEU240; LEU241; PRO242; ILE379 | HIS489; TRP244; HIS236; TRP239 |
| 7 | −6.9 | ILE450 | VAL143; ALA144; LYS156; LEU159; ALA311; PRO376; ILE377; CYS449; ILE450 | TYR131; PHE152; PHE234; PHE442 |
| 8 | −6.6 | HIS236 | ALA237; MET378; ILE379; MET381 | PHE98; PHE105; TYR107; HIS236; TRP239; HIS489 |
| 9 | −7.5 | TYR145; TYR145; ILE450 | LYS156; LEU310; ILE377; MET381; MET487 | TYR131; PHE152; PHE234; TRP239 |
| 10 | −7.6 | - | LYS156; ALA311; ILE377; CYS449; ALA455 | TYR145; PHE152; PHE234; PHE442 |
| 11 | −5.2 | - | ILE488; ILE488; PRO494 | - |
| KCZ | −10.3 | - | - | PHE105; TYR131; TYR145; TRP239 |
| FCZ | −7.3 | ILE379; MET487; HIS489; ILE379 | - | ILE379 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanaullah, A.F.M.; Devi, P.; Hossain, T.; Sultan, S.B.; Badhon, M.M.U.; Hossain, M.E.; Uddin, J.; Patwary, M.A.M.; Kazi, M.; Matin, M.M. Rhamnopyranoside-Based Fatty Acid Esters as Antimicrobials: Synthesis, Spectral Characterization, PASS, Antimicrobial, and Molecular Docking Studies. Molecules 2023, 28, 986. https://doi.org/10.3390/molecules28030986
Sanaullah AFM, Devi P, Hossain T, Sultan SB, Badhon MMU, Hossain ME, Uddin J, Patwary MAM, Kazi M, Matin MM. Rhamnopyranoside-Based Fatty Acid Esters as Antimicrobials: Synthesis, Spectral Characterization, PASS, Antimicrobial, and Molecular Docking Studies. Molecules. 2023; 28(3):986. https://doi.org/10.3390/molecules28030986
Chicago/Turabian StyleSanaullah, Abul Fazal Muhammad, Puja Devi, Takbir Hossain, Sulaiman Bin Sultan, Mohammad Mohib Ullah Badhon, Md. Emdad Hossain, Jamal Uddin, Md. Abdul Majed Patwary, Mohsin Kazi, and Mohammed Mahbubul Matin. 2023. "Rhamnopyranoside-Based Fatty Acid Esters as Antimicrobials: Synthesis, Spectral Characterization, PASS, Antimicrobial, and Molecular Docking Studies" Molecules 28, no. 3: 986. https://doi.org/10.3390/molecules28030986
APA StyleSanaullah, A. F. M., Devi, P., Hossain, T., Sultan, S. B., Badhon, M. M. U., Hossain, M. E., Uddin, J., Patwary, M. A. M., Kazi, M., & Matin, M. M. (2023). Rhamnopyranoside-Based Fatty Acid Esters as Antimicrobials: Synthesis, Spectral Characterization, PASS, Antimicrobial, and Molecular Docking Studies. Molecules, 28(3), 986. https://doi.org/10.3390/molecules28030986












