Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes
Abstract
1. Introduction
2. Results and Discussion
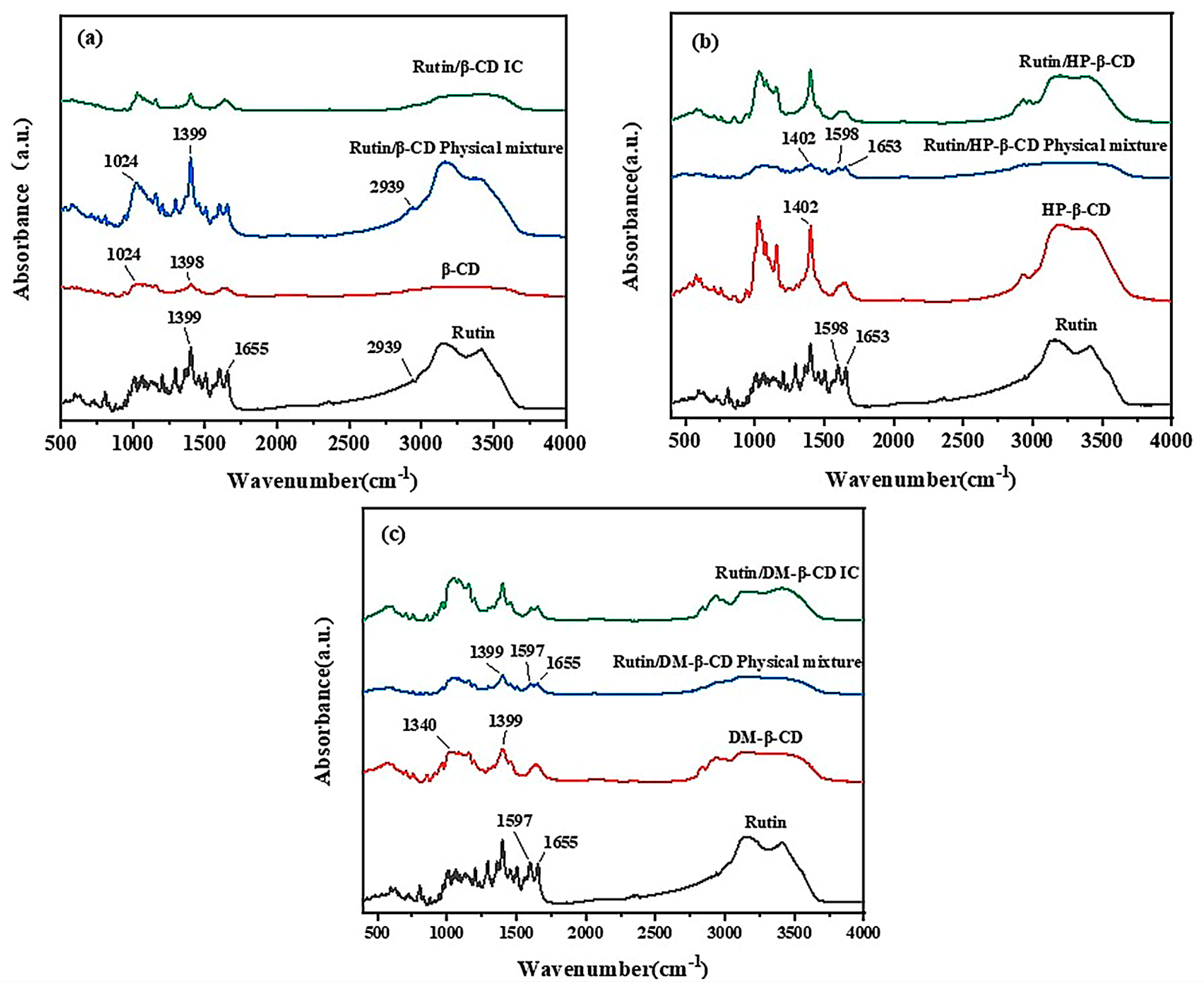
2.1. Fourier Infrared Spectroscopy Analysis
2.2. XRD Diffraction Analysis
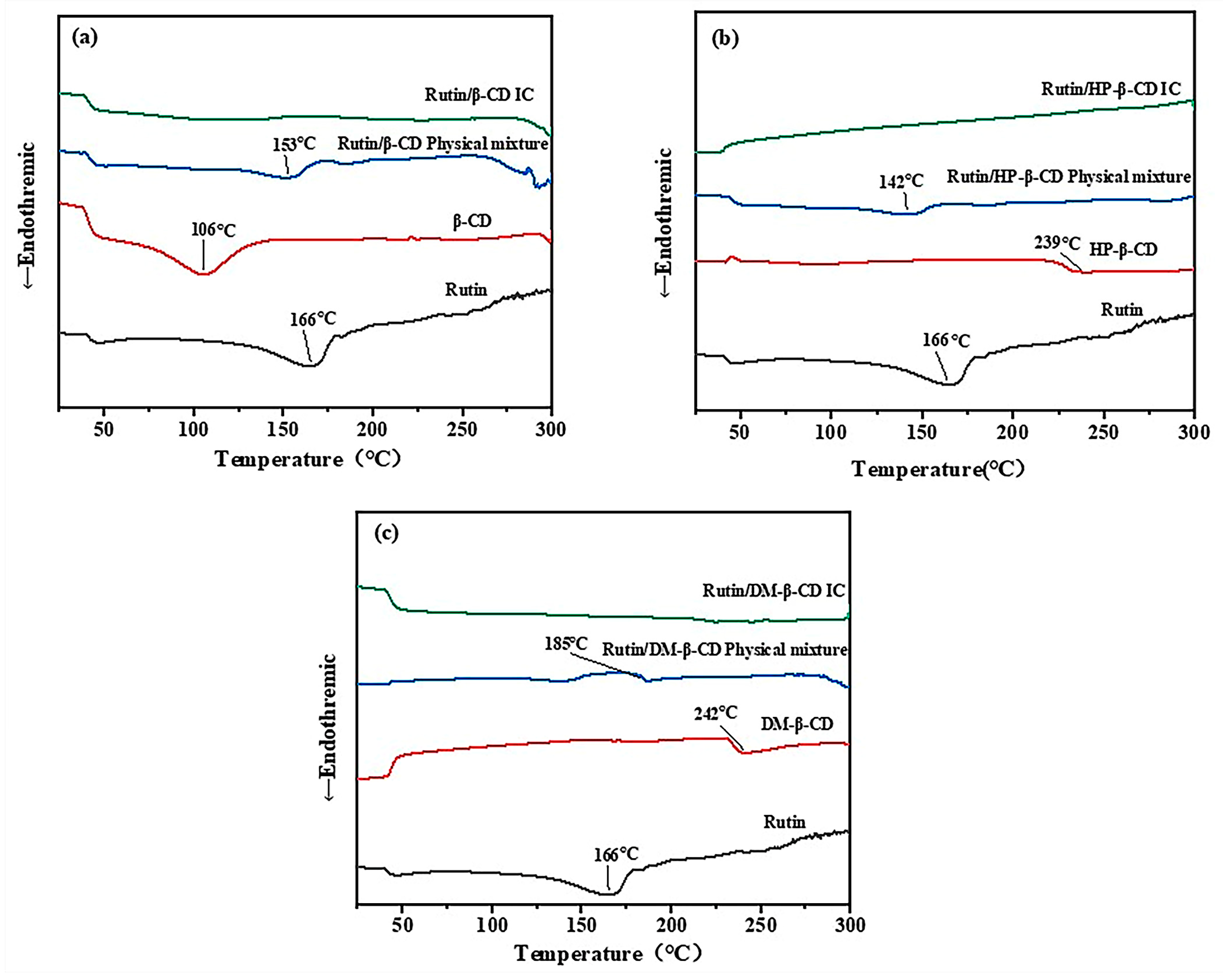
2.3. Differential Scanning Calorimetry (DSC) Analysis
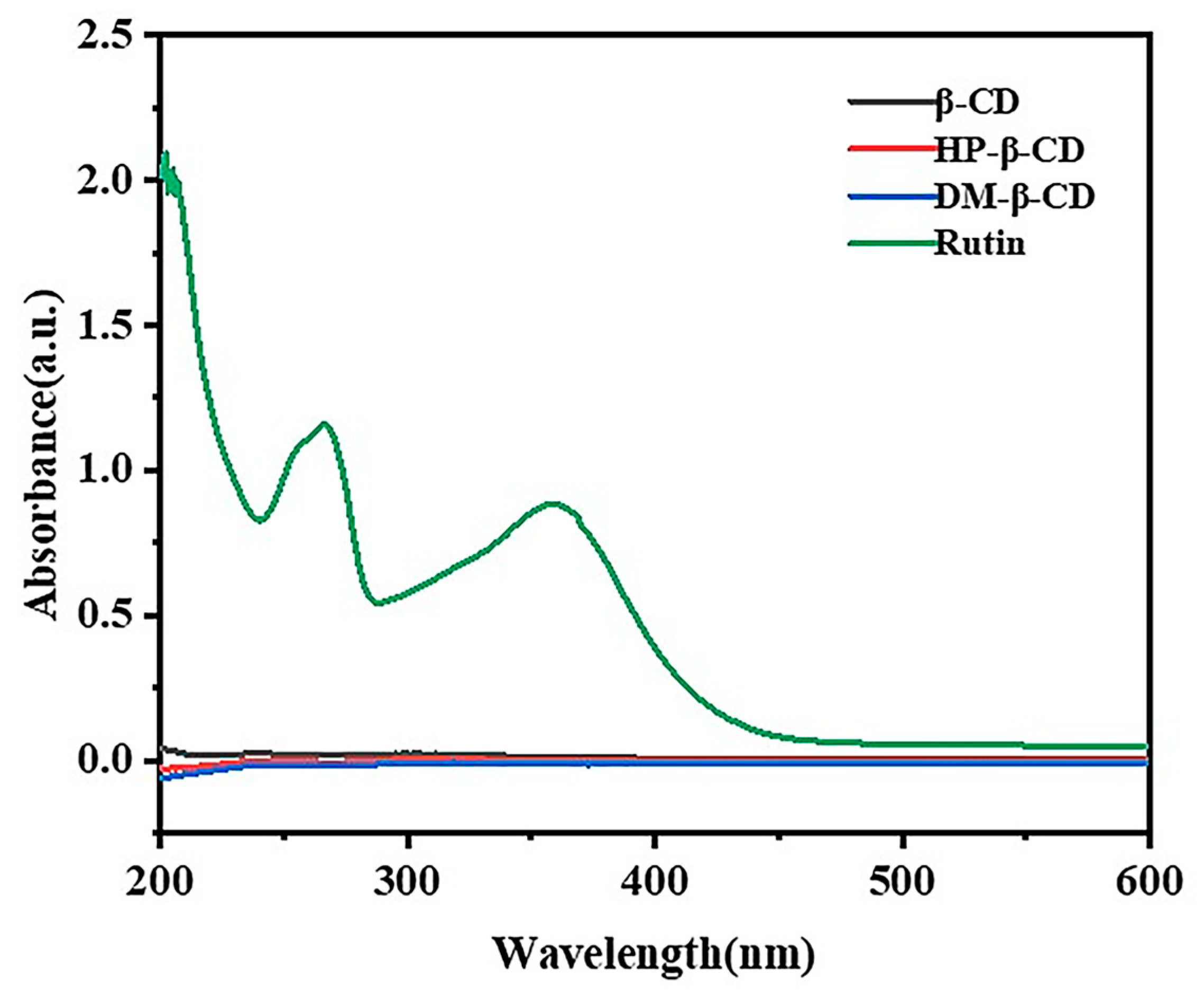
2.4. UV–Visible Spectral Analysis
2.4.1. Detection Wavelength of Rutin
2.4.2. Standard Curve of Rutin
2.4.3. Solubility Diagram of Rutin Phase
2.4.4. Inclusion Stability Constants
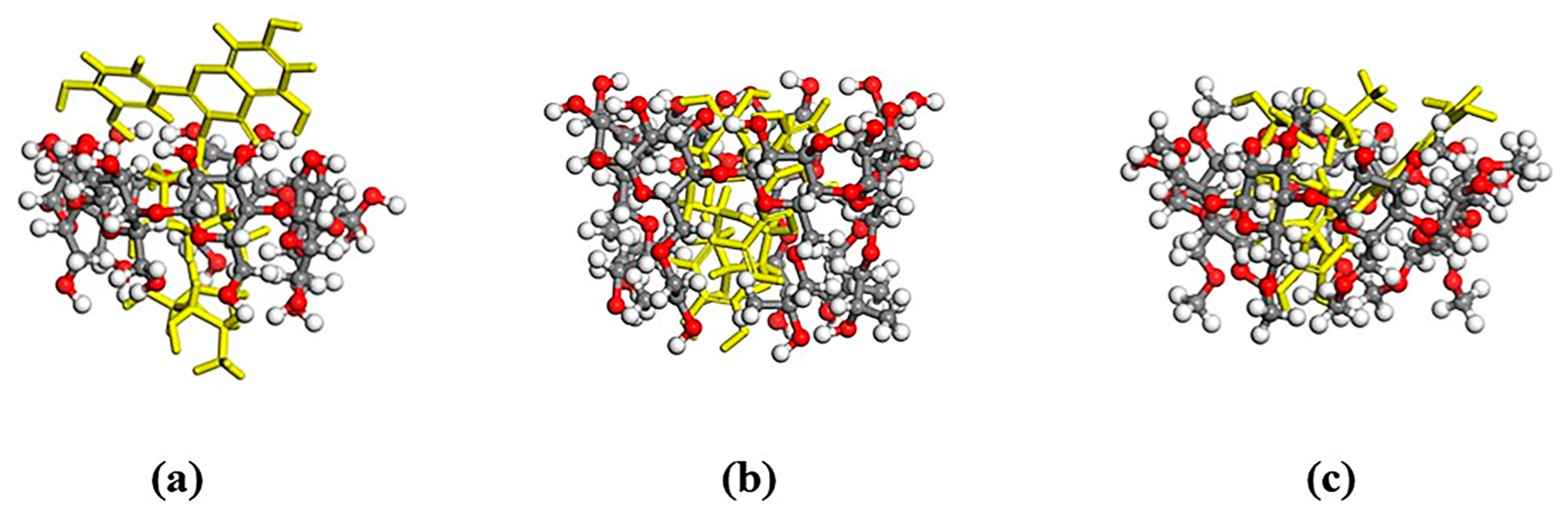
2.5. Molecular Docking
2.6. Judgment of System Equilibrium
2.7. Solubility Parameters Analysis
2.8. Binding Energy Analysis
2.9. Mean Square Displacement Analysis
2.10. Hydrogen Bond Analysis of Cyclodextrin System
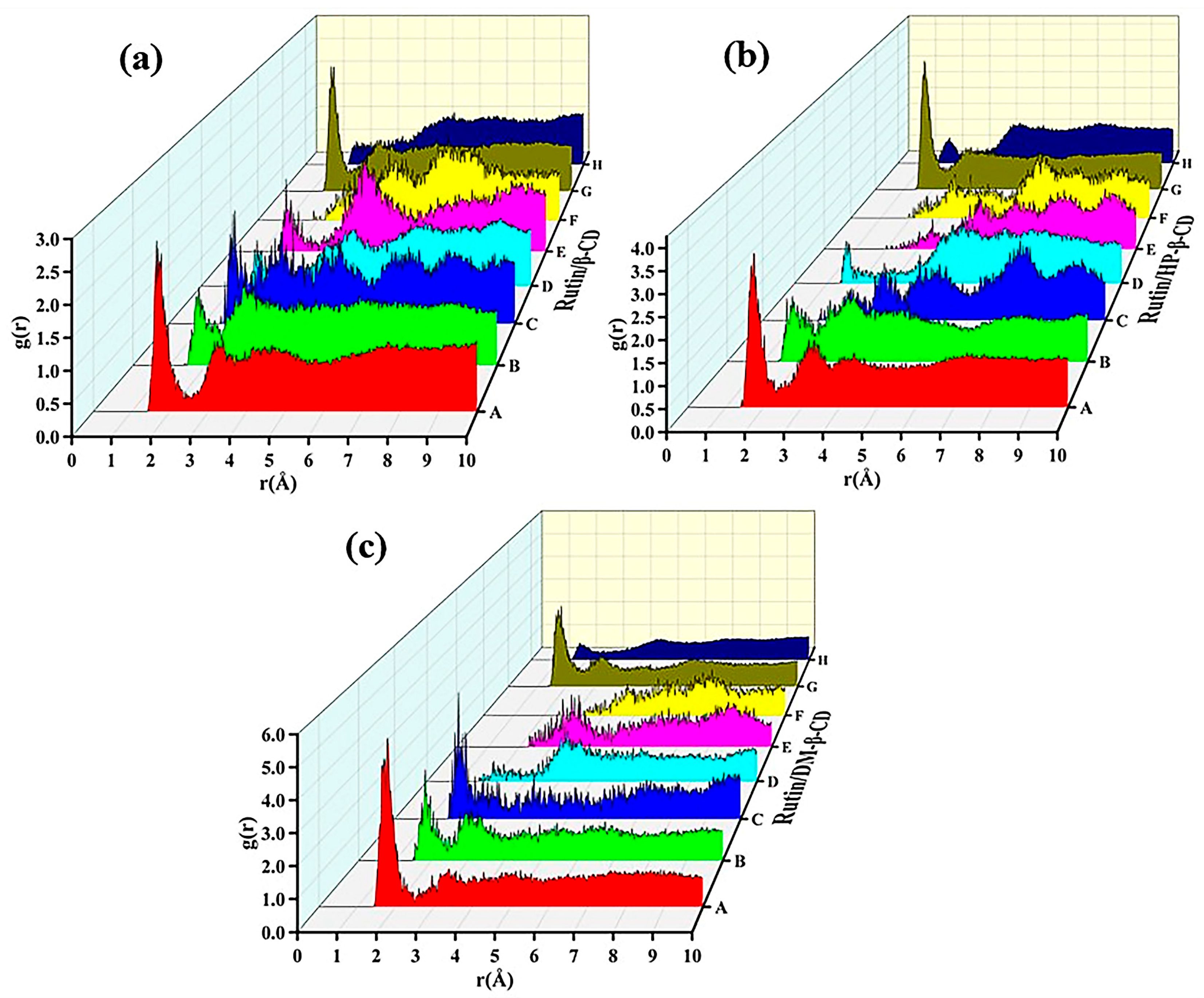
2.11. Hydrogen Bond Predictive Analysis and Radial Distribution Function (RDF) of Inclusion Complexes
3. Experimental Section
3.1. Materials and Instruments
3.2. Preparation of Rutin Cyclodextrin Inclusion Complexes (IC)
3.3. Preparation of the Rutin–Cyclodextrin Physical Mixture (PM)
3.4. Characterization
3.4.1. Fourier Infrared Spectrometry (FTIR)
3.4.2. X-ray Diffraction Analysis (XRD)
3.4.3. Differential Scanning Calorimetry (DSC)
3.4.4. Ultraviolet–Visible Spectral Analysis (UV)
4. Simulation Part
4.1. Molecular Docking
4.2. Molecular Dynamics (MD) Simulation
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gullon, B.; Lú-Chau, T.A.; Moreira, M.T.; Lema, J.M.; Eibes, G. Rutin: A review on extraction, identification and purification methods, biological activities and approaches to enhance its bioavailability. Trends Food Sci. Technol. 2017, 67, 220–235. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhou, Q.H.; Yang, Y.Q.; Wang, Y.Q. Investigation of the interaction between rutin and trypsin in solution by multi-spectroscopic method. Spectrosc. Lett. 2010, 43, 183–191. [Google Scholar] [CrossRef]
- de Sousa Fideles, L.; de Miranda, J.A.L.; de Souza Pimentel, P.V.; Ribeiro, M.E.N.P.; Ricardo, N.M.P.S.; Araújo, J.L.; Rocha, J.A.; Silva, M.F.S.; Cerqueira, G.S. Scientific and technological investigation of the protective effect of rutin on intestinal mucositis induced by 5-fluorouracil. Res. Soc. Dev. 2021, 10, e32710615625. [Google Scholar] [CrossRef]
- Arowoogun, J.; Akanni, O.O.; Adefisan, A.O.; Owumi, S.E.; Tijani, A.S.; Adaramoye, O.A. Rutin ameliorates copper sulfate-induced brain damage via antioxidative and anti-inflammatory activities in rats. J. Biochem. Mol. Toxicol. 2021, 35, e22623. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Fan, B.T.; Fassi, N.E.; Zakrzewska, K.; Jia, Z.J.; Zheng, R.L.; Panaye, A.; Couesnon, T.; Doucet, J.P. Comparative study of activities between verbascoside and rutin by docking method. QSAR Comb. Sci. 2003, 22, 18–28. [Google Scholar] [CrossRef]
- Nandana, C.N.; Christeena, M.; Bharathi, D. Synthesis and characterization of chitosan/silver nanocomposite using rutin for antibacterial, antioxidant and photocatalytic applications. J. Clust. Sci. 2022, 33, 269–279. [Google Scholar] [CrossRef]
- Oluranti, I.O.; Adeyemo, V.A.; Achile, E.O.; Fatokun, B.P.; Ojo, A.O. Rutin improves cardiac and erythrocyte membrane–bound ATPase activities in male rats exposed to cadmium chloride and lead acetate. Biol. Trace Elem. Res. 2022, 200, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Cristiano, M.C.; Barone, A.; Mancuso, A.; Torella, D.; Paolino, D. Rutin-loaded nanovesicles for improved stability and enhanced topical efficacy of natural compound. J. Funct. Biomater. 2021, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, G.; Papageorgiou, G.Z.; Bikiaris, D.N. β-cyclodextrin inclusion complexes of budesonide with enhanced bioavailability for COPD treatment. Appl. Sci. 2021, 11, 12085. [Google Scholar] [CrossRef]
- Mokhtar, M.S.; Suliman, F.O.; Elbashir, A.A. Atrazine and ametryne inclusion complexes with 2-hydroxypropyl-β/γ-cyclodextrin: Spectroscopic studies and molecular dynamics simulation. J. Mol. Struct. 2019, 1179, 161–170. [Google Scholar] [CrossRef]
- Li, J.X.; Zhang, X.S. Preparation and characterization of the inclusion complex of ofloxacin with β-CD and HP-β-CD. J. Incl. Phenom. Macro. 2011, 69, 173–179. [Google Scholar] [CrossRef]
- Li, S.J.; Yuan, L.; Chen, Y.; Zhou, W.; Wang, X.R. Studies on the inclusion complexes of daidzein with β-cyclodextrin and derivatives. Molecules 2017, 22, 2183. [Google Scholar] [CrossRef]
- Guendouzi, O.; Guendouzi, A.; Ouici, H.B.; Brahim, H.; Boumediene, M.; Elkeurti, M. A quantum chemical study of encapsulation and stabilization of gallic acid in beta-cyclodextrin as a drug delivery system. Can. J. Chem. 2020, 98, 204–214. [Google Scholar] [CrossRef]
- Viernstein, H.; Wolschann, P. Cyclodextrin inclusion complexation and pharmaceutical applications. Sci. Asia 2020, 46, 254–262. [Google Scholar] [CrossRef]
- Wu, Y.P.; Xiao, Y.; Yue, Y.X.; Zhong, K.; Zhao, Y.L.; Gao, H. A deep insight into mechanism for inclusion of 2R, 3R-dihydromyricetin with cyclodextrins and the effect of complexation on antioxidant and lipid-lowering activities. Food Hydrocoll. 2020, 103, 105718. [Google Scholar] [CrossRef]
- Nicoletti, C.D.; Faria, A.F.M.; de Sá Haddad Queiroz, M.; dos Santos Galvão, R.M.; Souza, A.L.A.; Futuro, D.O.; Ferreira, V.F. Synthesis and biological evaluation of β-lapachone and nor-β-lapachone complexes with 2-hydroxypropyl-β-cyclodextrin as trypanocidal agents. J. Bioenerg. Biomembr. 2020, 52, 185–197. [Google Scholar] [CrossRef]
- Franco, P.; Marco, I.D. Formation of rutin-β-cyclodextrin inclusion complexes by supercritical antisolvent precipitation. Polymers 2021, 13, 246. [Google Scholar] [CrossRef]
- Liu, Y.J.; Zhao, X.H.; Qiu, L.; Wu, W.W.; Li, Y.Y.; Wang, L. Characterization and preparation of inclusion complex of rutin-hydroxypropyl-β-cyclodextrin prepared by antisolvent method. Adv. Bot. Res. 2018, 7, 277–282. [Google Scholar] [CrossRef]
- Liu, J.; Qiu, L.Y.; Gao, J.Q.; Jin, Y. Preparation, characterization and in vivo evaluation of formulation of baicalein with hydroxypropyl-β-cyclodextrin. Int. J. Pharm. 2006, 312, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Omrani, Z.; Tehrani, A.D. New cyclodextrin-based supramolecular nanocapsule for codelivery of curcumin and gallic acid. Polym. Bull. 2020, 77, 2003–2019. [Google Scholar] [CrossRef]
- Sbârcea, L.; Ledei, A.; Udrescu, L.; Vărut, R.M.; Barvinschi, P.; Vlase, G.; Ledeti, I. Betulonic acid—Cyclodextrins inclusion complexes. J. Therm. Anal. Calorim. 2019, 138, 2787–2797. [Google Scholar] [CrossRef]
- Mazurek, A.H.; Szeleszczuk, L.; Gubica, T. Application of molecular dynamics simulations in the analysis of cyclodextrin complexes. Int. J. Mol. Sci. 2021, 22, 9422. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Bomzan, P.; Roy, M.N. Probing host-guest inclusion complexes of ambroxol hydrochloride with α-& β-cyclodextrins by physicochemical contrivance subsequently optimized by molecular modeling simulations. Chem. Phys. Lett. 2020, 748, 137372. [Google Scholar] [CrossRef]
- Pahari, B.; Chakraborty, S.; Sengupta, P.K. Molecular insight into the inclusion of the dietary plant flavonolfisetin and its chromophore within a chemically modified γ-cyclodextrin: Multi-Spectroscopic, molecular docking and solubility studies. Food Chem. 2018, 260, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Feng, W.; Li, C.; Tan, T.W. Investigation of the inclusions of puerarin and daidzin with beta-cyclodextrin by molecular dynamics simulation. J. Phys. Chem. B 2010, 114, 4876–4883. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Zhang, J.Q.; Ren, S.H.; Xie, X.G.; Huang, R.; Jin, Y.; Lin, J. Experimental and computational studies of naringin/cyclodextrin inclusion complexation. J. Incl. Phenom. Macrocycl. Chem. 2017, 88, 15–26. [Google Scholar] [CrossRef]
- Zhao, L.D.; Sun, Q.M.; Pu, H.Y.; Tang, P.X.; Liu, Y.Y.; Li, M.X.; Ren, X.Y.; Li, H. Experimental and computer simulation investigations of ethyl red with modified β-cyclodextrins: Inclusion mechanism and structure characterization. Chem. Phys. Lett. 2020, 754, 137725. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Liu, B.G.; Zhao, J.; Thomas, D.S.; Hook, J.M. An investigation into the supramolecular structure, solubility, stability and antioxidant activity of rutin/cyclodextrin inclusion complex. Food Chem. 2013, 136, 186–192. [Google Scholar] [CrossRef]
- Vijaya, K.V.; Kondaiah, A.; Ratna, J.V.; Annapurna, A. Preparation and characterization of quercetin and rutin cyclodextrin inclusion complexes. Drug Dev. Ind. Pharm. 2007, 33, 245–253. [Google Scholar] [CrossRef]
- Ding, H.Y.; Chao, J.B.; Zhang, G.M.; Shuang, S.M.; Pan, J.H. Preparation and spectral investigation on inclusion complex of β-cyclodextrin with rutin. Spectrochim. Acta A Mol. Biomol. 2003, 59, 3421–3429. [Google Scholar] [CrossRef]
- Savic, I.M.; Savic-Gajic, I.M.; Nikolic, V.D.; Nikolic, L.B.; Radovanovic, B.C.; Milenkovic-Andjelkovic, A. Enhencemnet of solubility and photostability of rutin by complexation with β-cyclodextrin and (2-hydroxypropyl)-β-cyclodextrin. J. Incl. Phenom. Macrocycl. Chem. 2016, 86, 33–43. [Google Scholar] [CrossRef]
- Liu, J.Y.; Zhao, S.D.; Zhao, X.Y.; Lu, Y.; Song, M.; Wu, S.Z. Molecular simulation and experimental study on the inclusion of rutin with β-cyclodextrin and its derivative. Int. J. Pharm. Sci. Res. 2022, 1254, 132359. [Google Scholar] [CrossRef]
- Song, M.; Yue, X.L.; Wang, X.J.; Cao, F.Y.; Li, Y.N.; Su, C.H.; Qin, Q. Effect of hindered phenol AO-80 on the damping properties for nitrile-butadiene rubber/phenolic resin: Molecular simulation and experimental study. Macromol. Mater. Eng. 2020, 305, 1–11. [Google Scholar] [CrossRef]
- Luo, K.Q.; Ye, X.; Zhang, H.; Liu, J.Y.; Luo, Y.L.; Wu, S.Z. Vulcanization and antioxidation effects of accelerator modified antioxidant in styrene-butadiene rubber: Experimental and computational studies. Polym. Degrad. Stab. 2020, 177, 109181. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, X.J.; Xiao, J.J.; Zhu, W.H.; Sun, H.; Xiao, H.M. Molecular dynamics simulations of AP/HMX composite with a modified force field. J. Hazard. Mater. 2009, 167, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.; Hempenstall, J.; Tucker, I.; Rades, T. Selection of excipients for melt extrusion with two poorly water-soluble drugs by solubility parameter calculation and thermal analysis. Int. J. Pharm. 2001, 226, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, Y.H.; Li, Y.G.; Li, X.L.; Zhao, S.Y.; Yang, L.J.; Liu, X.Y.; Zhang, J.Q. Molecular dynamics simulations and theoretical calculations of cyclodextrin-polydatin inclusion complexes. J. Mol. Struct. 2021, 1230, 129840. [Google Scholar] [CrossRef]
- Jin, G.Y.; Chen, H. Facile preparation and extensive characterization of naproxen-β-cyclodextrin inclusion complex. J. Anal. Test. 2017, 1, 19. [Google Scholar] [CrossRef]
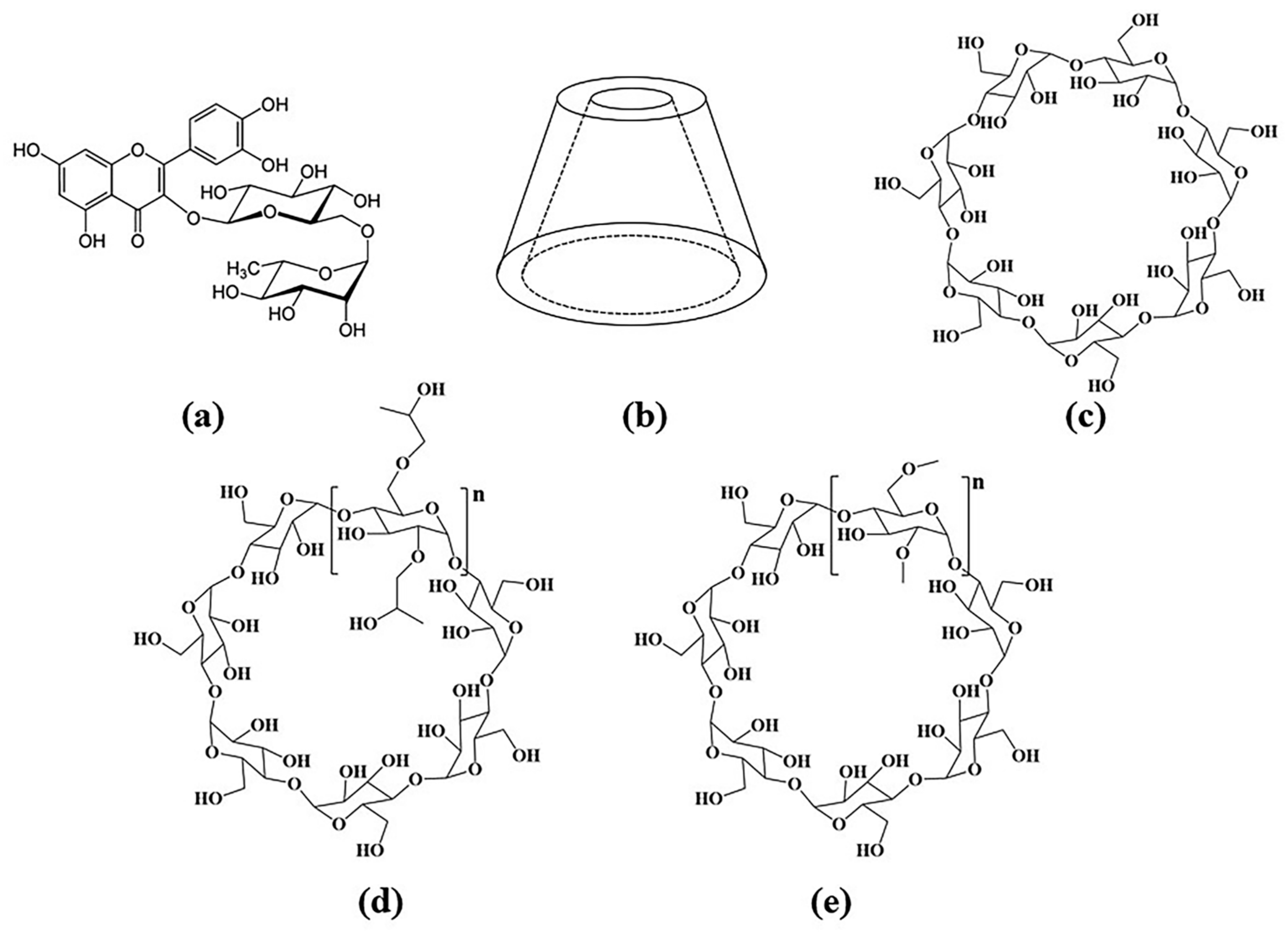

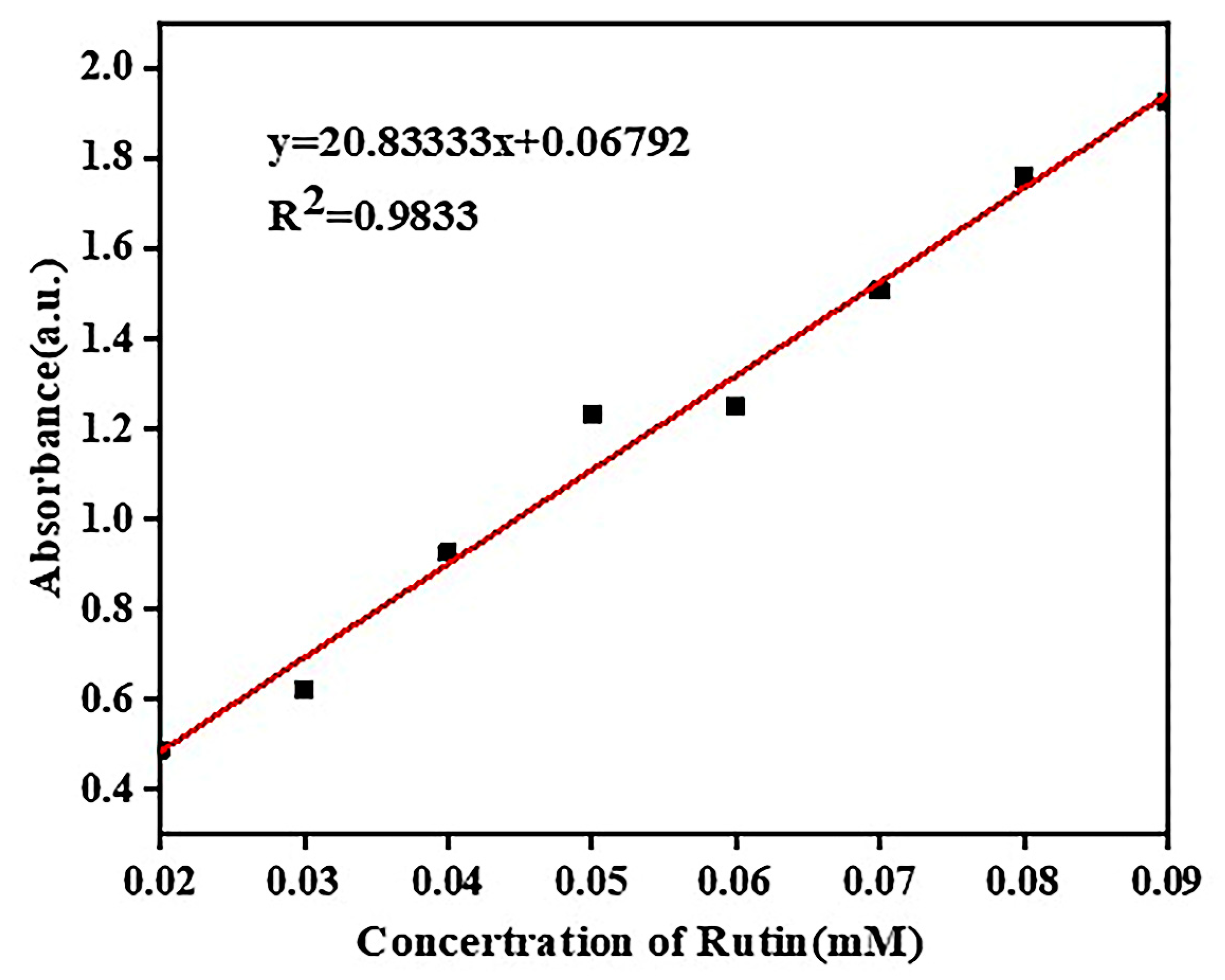
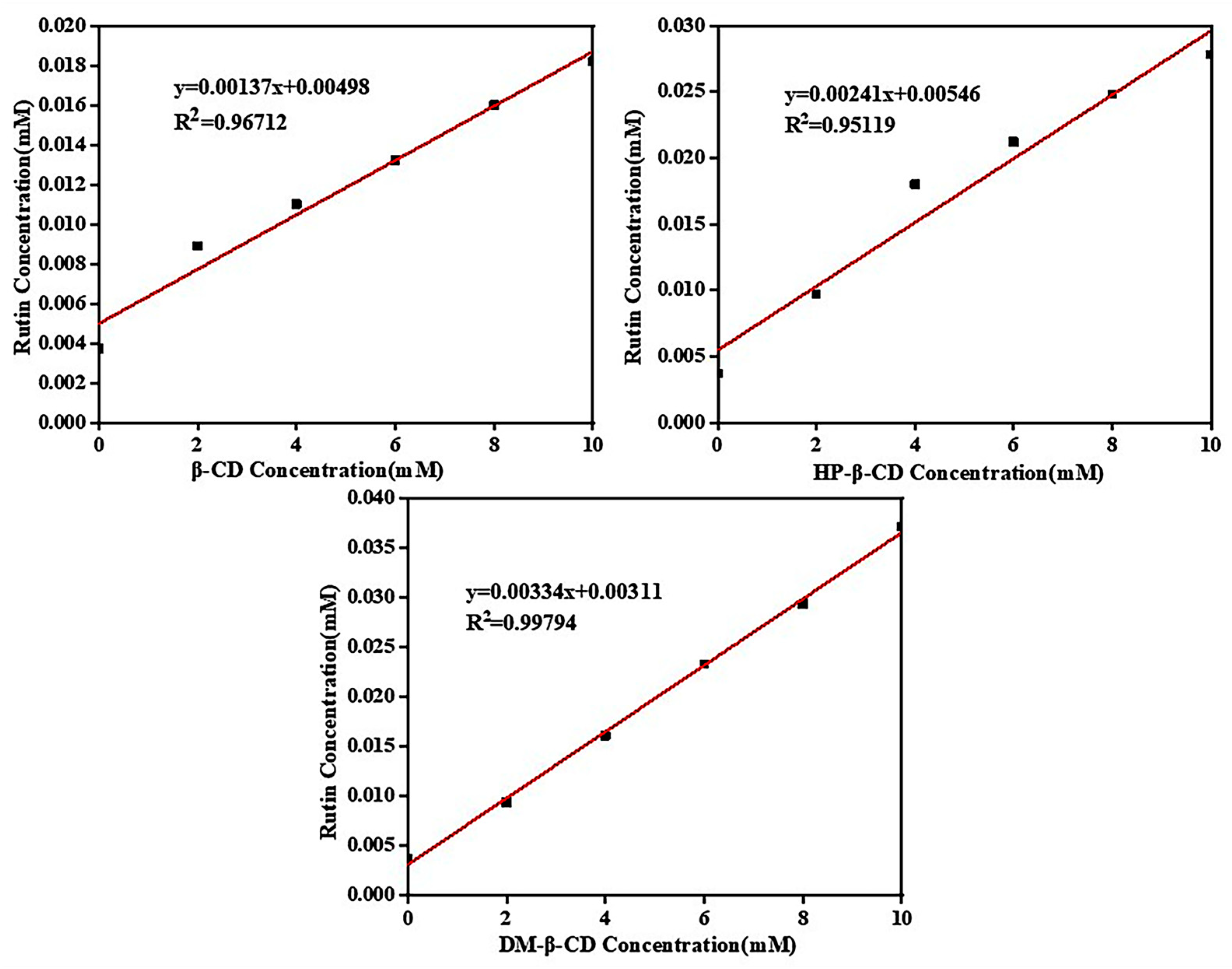




| Concentration (mM/L) | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.08 | 0.09 |
|---|---|---|---|---|---|---|---|---|
| Absorbance | 0.489 | 0.622 | 0.926 | 1.232 | 1.250 | 1.507 | 1.758 | 1.926 |
| Concentration | 0 mM/L | 2 mM/L | 4 mM/L | 6 mM/L | 8 mM/L | 10 mM/L |
|---|---|---|---|---|---|---|
| β-CD | 0.145 | 0.253 | 0.298 | 0.343 | 0.401 | 0.447 |
| HP-β-CD | 0.145 | 0.270 | 0.443 | 0.509 | 0.584 | 0.648 |
| DM-β-CD | 0.145 | 0.264 | 0.403 | 0.554 | 0.678 | 0.843 |
| δA (cal/cm3)1/2 | δB (cal/cm3)1/2 | A–B (cal/cm3)1/2 |
|---|---|---|
| δrutin = 12.650 | δβ-CD = 9.658 | 2.992 < 3.4 |
| δrutin = 12.650 | δHP-β-CD = 10.677 | 1.973 < 3.4 |
| δrutin = 12.650 | δDM-β-CD = 12.422 | 0.228 < 1.0 |
| Complex | Binding Energy (Kcal/mol) |
|---|---|
| rutin/β-CD | 111.49 |
| rutin/HP-β-CD | 114.95 |
| rutin/DM-β-CD | 519.82 |
| Cyclodextrin | Number of Cell Hydrogen Bonds | Hydrogen Bond Concentration (mol/cm3) |
|---|---|---|
| β-CD | 143 | 1.22 × 10−3 |
| HP-β-CD | 144 | 7.92 × 10−3 |
| DM-β-CD | 42 | 2.96 × 10−2 |
| Rutin Atoms | Electronegativity | Cyclodextrin Atoms | Electronegativity |
|---|---|---|---|
| O1 | −0.57 | O7 | −0.57 |
| O2 | −0.452 | O8 | −0.32 |
| O3 | −0.419 | H2 | 0.41 |
| O4 | −0.32 | ||
| O5 | −0.1815 | ||
| O6 | −0.0685 | ||
| H1 | 0.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.; Song, M.; Ma, M.; Song, J.; Cao, F.; Qin, Q. Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes. Molecules 2023, 28, 955. https://doi.org/10.3390/molecules28030955
Chang C, Song M, Ma M, Song J, Cao F, Qin Q. Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes. Molecules. 2023; 28(3):955. https://doi.org/10.3390/molecules28030955
Chicago/Turabian StyleChang, Chaokang, Meng Song, Mingxing Ma, Jihong Song, Fengyi Cao, and Qi Qin. 2023. "Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes" Molecules 28, no. 3: 955. https://doi.org/10.3390/molecules28030955
APA StyleChang, C., Song, M., Ma, M., Song, J., Cao, F., & Qin, Q. (2023). Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes. Molecules, 28(3), 955. https://doi.org/10.3390/molecules28030955








