Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I
Abstract
1. Introduction
2. Results
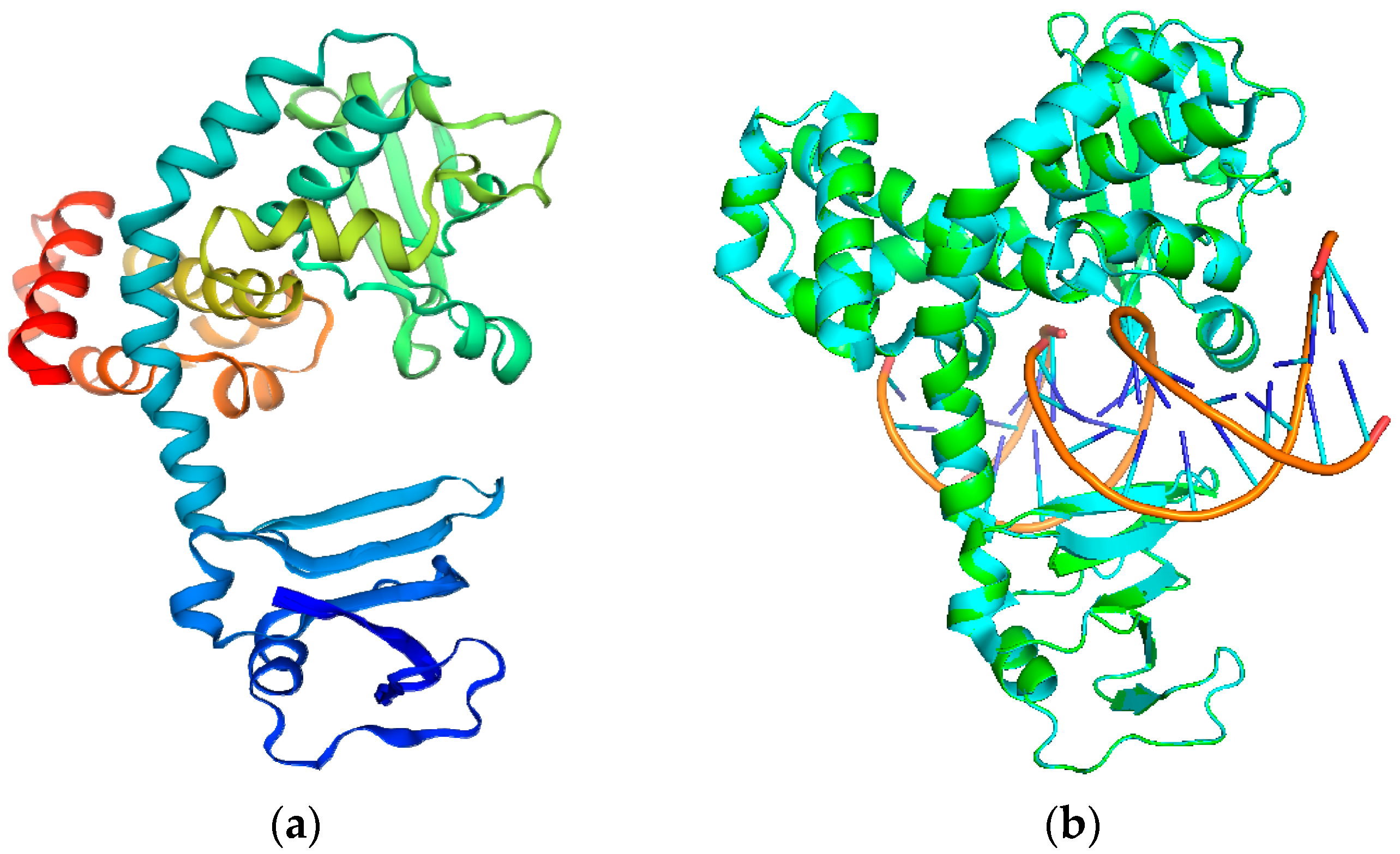
2.1. Modeling the Structure of TOP1
2.2. Quality Evaluation of MPXV TOP1 Models
2.3. Docking-Based Virtual Screening
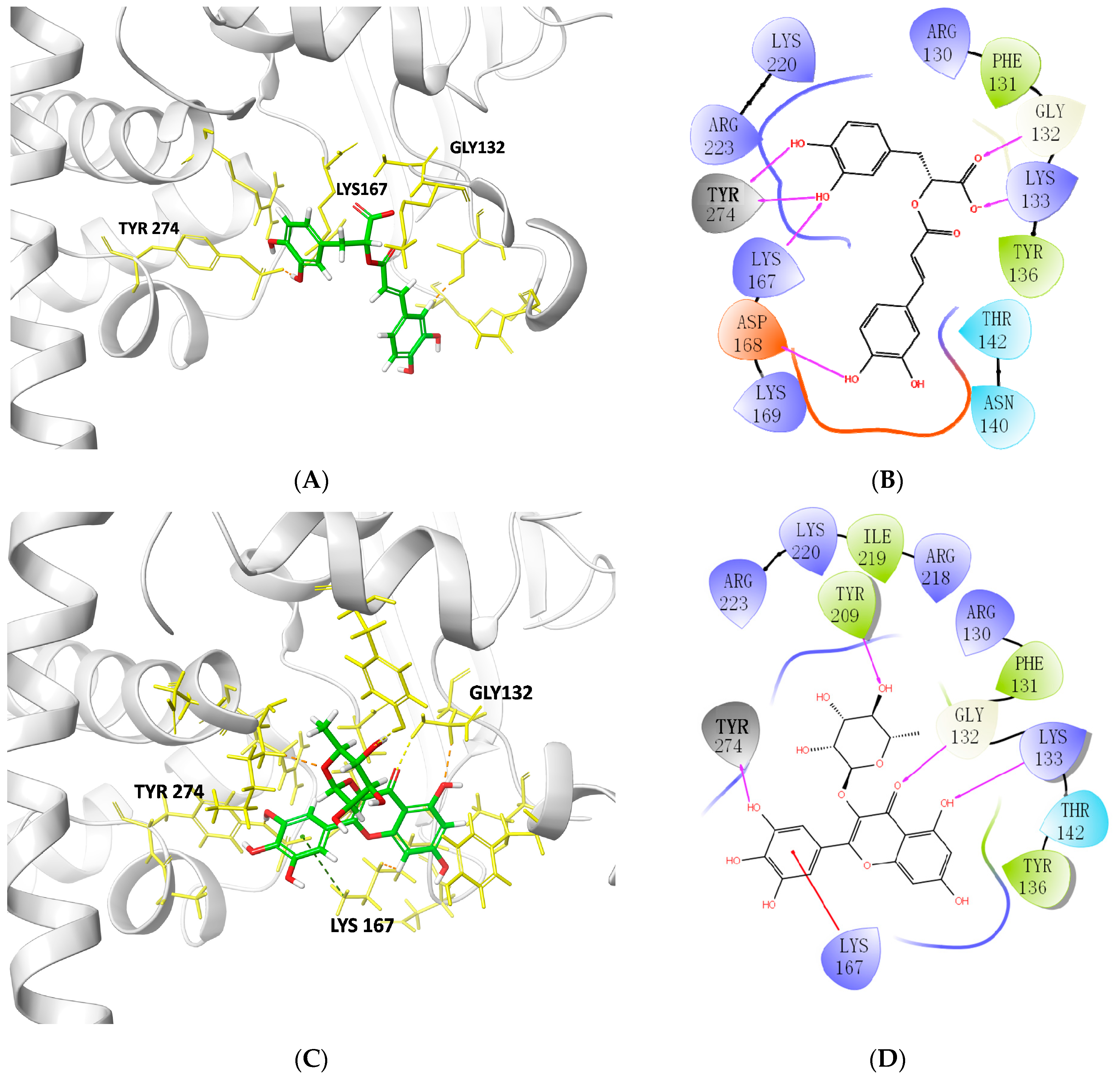
2.4. Characterization of TOP1–Ligand Interactions
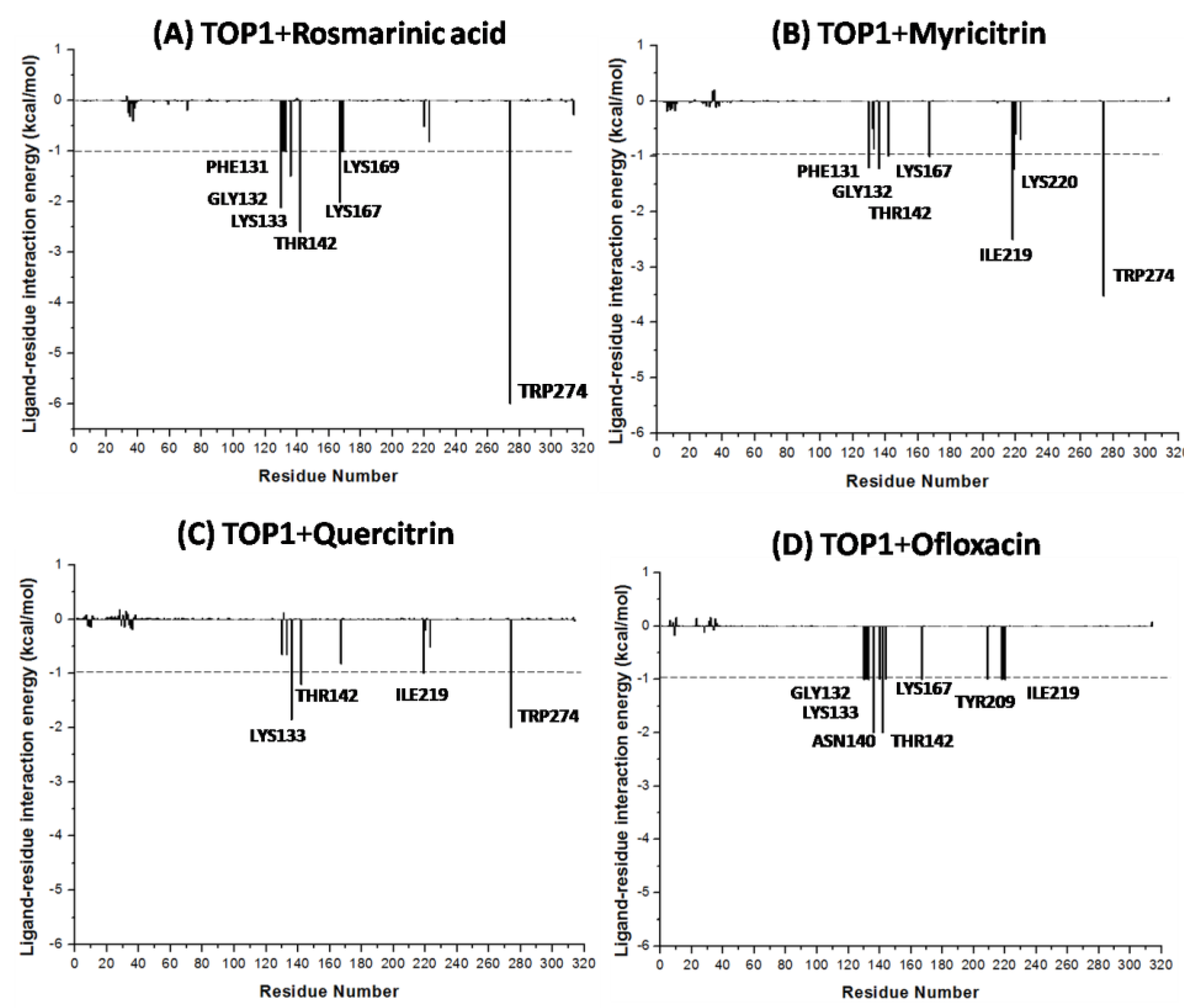
2.5. Molecular Dynamics Simulations
2.6. SPR Results
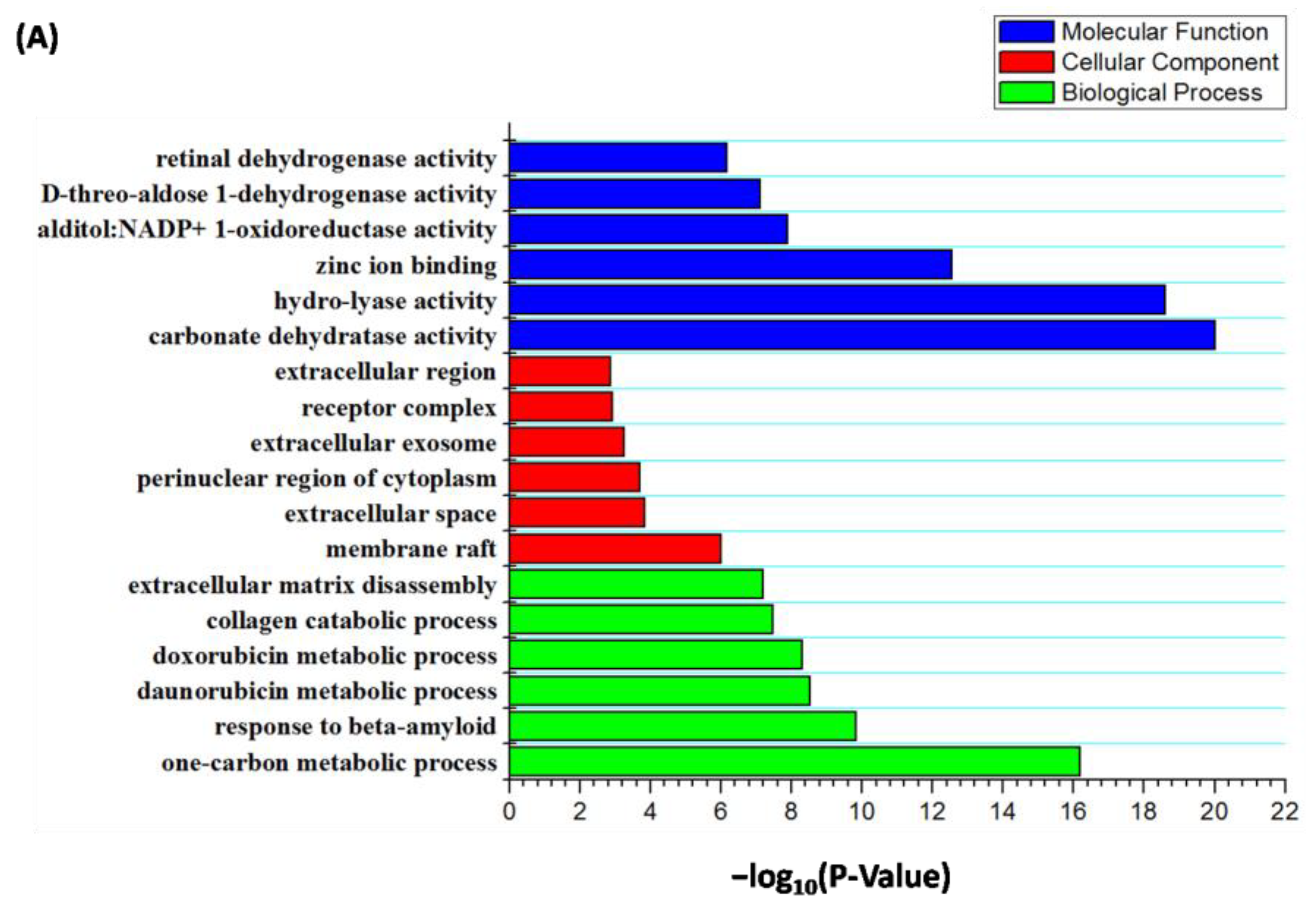
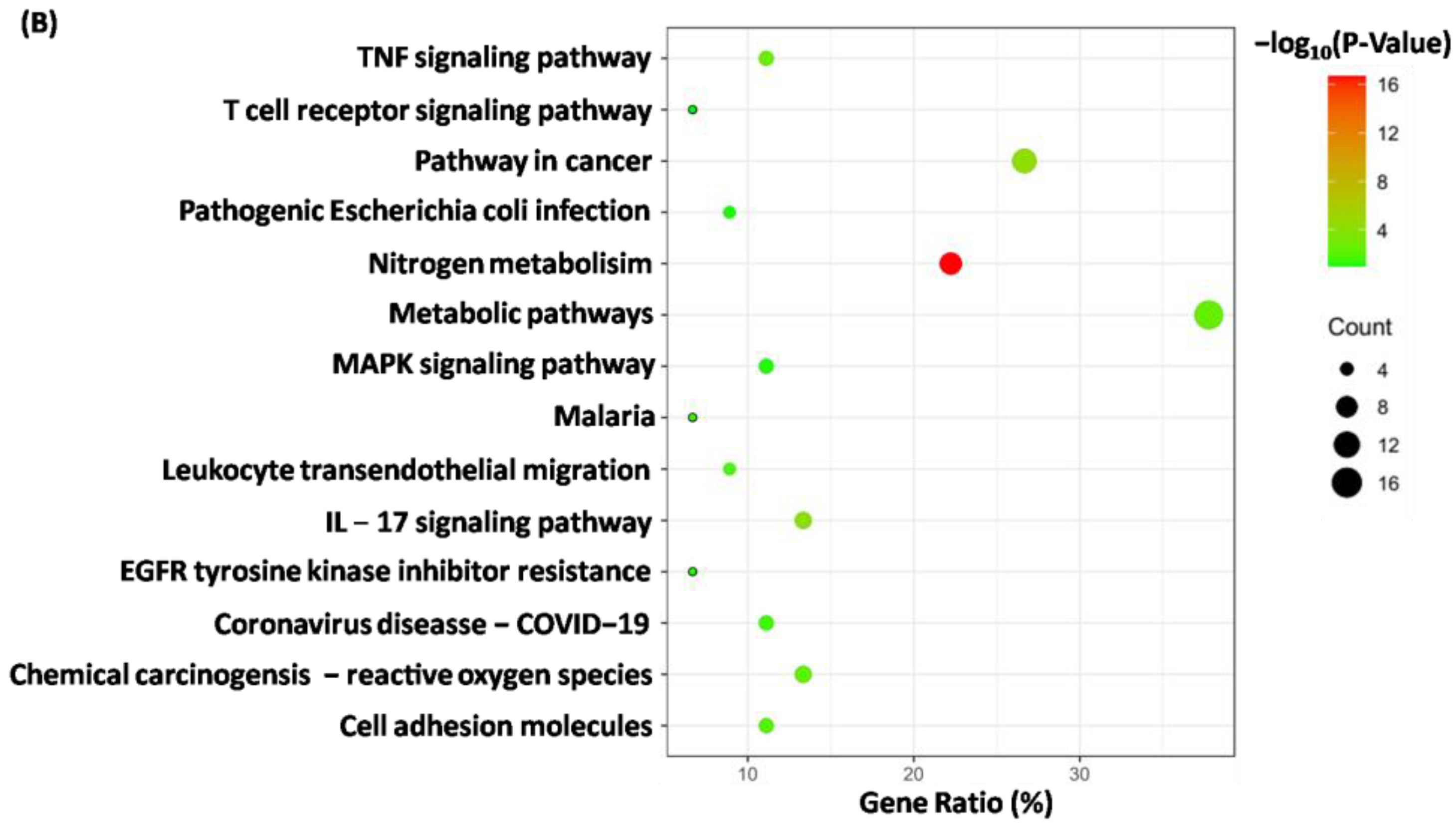
2.7. Analysis of Feasible Targets toward Rosmarinic Acid through GO and KEGG Enrichment
3. Discussion
4. Materials and Methods
4.1. Homology Modeling
4.2. Structural Validation
4.3. Prediction of Binding Sites
4.4. Preparation of Protein Targets and Ligand Libraries
4.5. Receptor Grid Generation and Virtual Screening
4.6. Characterisation of Binding Mechanism
4.7. MD Simulations of Protein–Ligand Complexes and Proteins
4.8. Surface Plasmon Resonance (SPR) Analysis
4.9. Forecasting for Mechanism of Action towards Rosmarinic Acid
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Alakunle, E.; Moens, U.; Nchinda, G.; Okeke, M. Monkeypox Virus in Nigeria: Infection Biology, Epidemiology, and Evolution. Viruses 2020, 12, 1257. [Google Scholar] [CrossRef]
- Reynolds, M.G.; Doty, J.B.; Mccollum, A.M.; Olson, V.A.; Nakazawa, Y. Monkeypox re-emergence in Africa: A call to expand the concept and practice of One Health. Expert Rev. Anti-infective Ther. 2019, 17, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Thèves, C.; Crubézy, E.; Biagini, P. History of Smallpox and Its Spread in Human Populations. Microbiol. Spectr. 2016, 4, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Hu, Z.; Yu, T.; Hu, H.; Zhao, Y.; Li, C.; Zhu, Q.; Wang, M.; Zhai, P.; He, L.; et al. The Antiviral Effects of Jasminin via Endogenous TNF-α and the Underlying TNF-α-Inducing Action. Molecules 2022, 27, 1598. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.Y.; Ajisegiri, W.S.; Costantino, V.; Chughtai, A.A.; MacIntyre, C.R. Reemergence of Human Monkeypox and Declining Population Immunity in the Context of Urbanization, Nigeria, 2017–2020. Emerg. Infect. Dis. 2021, 27, 1007–1014. [Google Scholar] [CrossRef]
- Reeves, P.M.; Smith, S.K.; Olson, V.A.; Thorne, S.H.; Bornmann, W.; Damon, I.K.; Kalman, D. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and SRC family tyrosine kinases. J. Virol. 2011, 85, 21–31. [Google Scholar] [CrossRef]
- Bray, M. Pathogenesis and potential antiviral therapy of complications of smallpox vaccination. Antivir. Res. 2003, 58, 101–114. [Google Scholar] [CrossRef]
- Baker, R.; Bray, M.; Huggins, J.W. Potential antiviral therapeutics for smallpox, monkeypox and other orthopoxvirus infections. Antivir. Res. 2003, 57, 13–23. [Google Scholar] [CrossRef]
- Yakovleva, L.; Shuman, S. Chemical Mutagenesis of Vaccinia DNA Topoisomerase Lysine 167 Provides Insights to the Catalysis of DNA Transesterification. Biochemistry 2013, 52, 984–991. [Google Scholar] [CrossRef]
- Sekiguchi, J.; Stivers, J.T.; Mildvan, A.S.; Shuman, S. Mechanism of Inhibition of Vaccinia DNA Topoisomerase by Novobiocin and Coumermycin. J. Biol. Chem. 1996, 271, 2313–2322. [Google Scholar] [CrossRef]
- Broni, E.; Kwofie, S.K.; Asiedu, S.O.; Miller, W.A.; Wilson, M.D. A Molecular Modeling Approach to Identify Potential Antileishmanial Compounds Against the Cell Division Cycle (cdc)-2-Related Kinase 12 (CRK12) Receptor of Leishmania donovani. Biomolecules 2021, 11, 458. [Google Scholar] [CrossRef]
- Sarkar, B.; Kulharia, M.; Mantha, A.K. Understanding human thiol dioxygenase enzymes: Structure to function, and biology to pathology. Int. J. Exp. Pathol. 2017, 98, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Hoda, S.; Gupta, L.; Shankar, J.; Gupta, A.K.; Vijayaraghavan, P. cis-9-Hexadecenal, a Natural Compound Targeting Cell Wall Organization, Critical Growth Factor, and Virulence of Aspergillus fumigatus. ACS Omega 2020, 5, 10077–10088. [Google Scholar] [CrossRef]
- Tam, B.; Sinha, S.; Wang, S.M. Combining Ramachandran plot and molecular dynamics simulation for structural-based variant classification: Using TP53 variants as model. Comput Struct Biotechnol J. 2020, 18, 4033–4039. [Google Scholar] [CrossRef]
- Dalhoff, A. Antiviral, Antifungal, and Antiparasitic Activities of Fluoroquinolones Optimized for Treatment of Bacterial Infections: A Puzzling Paradox or a Logical Consequence of Their Mode of Action? Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Kamau, E.; Grove, A. Fluoroquinolone-dependent DNA Supercoiling by Vaccinia Topoisomerase I. J. Mol. Biol. 2004, 342, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Bordoloi, M. Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr. Drug Targets 2019, 20, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Akaji, K.; Konno, H. Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease. Molecules 2020, 25, 3920. [Google Scholar] [CrossRef]
- Mohammad, T.; Shamsi, A.; Anwar, S.; Umair, M.; Hussain, A.; Rehman, M.T.; AlAjmi, M.F.; Islam, A.; Hassan, M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: Towards the development of effective COVID-19 therapy. Virus Res. 2020, 288, 198102. [Google Scholar] [CrossRef] [PubMed]
- Parashar, A.; Shukla, A.; Sharma, A.; Behl, T.; Goswami, D.; Mehta, V. Reckoning γ-Glutamyl-S-allylcysteine as a potential main protease (mpro) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Drug Dev. Ind. Pharm. 2021, 47, 699–710. [Google Scholar] [CrossRef]
- Hickman, H.D.; Reynoso, G.V.; Ngudiankama, B.F.; Rubin, E.J.; Magadán, J.G.; Cush, S.S.; Gibbs, J.; Molon, B.; Bronte, V.; Bennink, J.R.; et al. Anatomically Restricted Synergistic Antiviral Activities of Innate and Adaptive Immune Cells in the Skin. Cell Host Microbe 2013, 13, 155–168. [Google Scholar] [CrossRef]
- Reed, B.; Yakovleva, L.; Shuman, S.; Ghose, R. Characterization of DNA Binding by the Isolated N-Terminal Domain of Vaccinia Virus DNA Topoisomerase IB. Biochemistry 2017, 56, 3307–3317. [Google Scholar] [CrossRef] [PubMed]
- Merecz-Sadowska, A.; Sitarek, P.; Śliwiński, T.; Zajdel, K.; Malinowska, K.; Zielińska-Bliźniewska, H.; Kucharska, E.; Zajdel, R. In Vitro and In Silico Studies on Leonotis nepetifolia (L.) R. Br. Root Extract against Cancer Cells. Curr. Pharm. Biotechnol. 2022, 23, 1383–1395. [Google Scholar] [CrossRef]
- Langland, J.; Jacobs, B.; Wagner, C.E.; Ruiz, G.; Cahill, T.M. Antiviral activity of metal chelates of caffeic acid and similar compounds towards herpes simplex, VSV-Ebola pseudotyped and vaccinia viruses. Antivir. Res. 2018, 160, 143–150. [Google Scholar] [CrossRef]
- Jannat, K.; Paul, A.K.; Bondhon, T.A.; Hasan, A.; Nawaz, M.; Jahan, R.; Mahboob, T.; Nissapatorn, V.; Wilairatana, P.; Pereira, M.D.L.; et al. Nanotechnology Applications of Flavonoids for Viral Diseases. Pharmaceutics 2021, 13, 1895. [Google Scholar] [CrossRef]
- Mouffouk, C.; Mouffouk, S.; Mouffouk, S.; Hambaba, L.; Haba, H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CL(pro) and PL(pro)), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). Eur. J. Pharmacol. 2021, 891, 173759. [Google Scholar] [CrossRef]
- Hu, X.; Cai, X.; Song, X.; Li, C.; Zhao, J.; Luo, W.; Zhang, Q.; Ekumi, I.O.; He, Z. Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor. Futur. Virol. 2020, 15, 359–368. [Google Scholar] [CrossRef]
- Suri, B.; Verma, N.; Schmidtchen, A. Toll-like Receptor 3 Agonist, Polyinosinic-polycytidylic Acid, Upregulates Carbonic Anhydrase II in Human Keratinocytes. Acta Derm. Venereol. 2018, 98, 762–765. [Google Scholar] [CrossRef]
- Åkerström, S.; Mousavi-Jazi, M.; Klingström, J.; Leijon, M.; Lundkvist, A.; Mirazimi, A. Nitric Oxide Inhibits the Replication Cycle of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2005, 79, 1966–1969. [Google Scholar] [CrossRef]
- Mangmool, S.; Kunpukpong, I.; Kitphati, W.; Anantachoke, N. Antioxidant and Anticholinesterase Activities of Extracts and Phytochemicals of Syzygium antisepticum Leaves. Molecules 2021, 26, 3295. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.S.; Mar, J.M.; da Silva, L.S.; Acho, L.D.; da Silva, B.J.P.; Lima, E.S.; Campelo, P.H.; Sanches, E.A.; Bezerra, J.A.; Chaves, F.C.M.; et al. Pedra-ume caá fruit: An Amazon cherry rich in phenolic compounds with antiglycant and antioxidant properties. Food Res. Int. 2019, 123, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Madronal, M.; Caro-Leon, J.; Espinosa-Cano, E.; Aguilar, M.R.; Vazquez-Lasa, B. Chitosan–Rosmarinic acid conjugates with antioxidant, anti-inflammatory and photoprotective properties. Carbohydr. Polym. 2021, 273, 118619. [Google Scholar] [CrossRef]
- Mrityunjaya, M.; Pavithra, V.; Neelam, R.; Janhavi, P.; Halami, P.M.; Ravindra, P.V. Immune-Boosting, Antioxidant and Anti-inflammatory Food Supplements Targeting Pathogenesis of COVID-. Front. Immunol. 2020, 11, 570122. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into a Known Three-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Ikeda, S.; Yazawa, M.; Nishimura, C. Antiviral activity and inhibition of topoisomerase by ofloxacin, a new quinolone derivative. Antiviral Res. 1987, 8(3), 103–113. [Google Scholar] [CrossRef] [PubMed]
- Sk, M.F.; Roy, R.; Jonniya, N.A.; Poddar, S.; Kar, P. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J. Biomol. Struct. Dyn. 2020, 38, 1–13. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.; Liu, Z.; Ju, Y.; Xu, M.; Zhang, Y.; Wu, X.; Gu, Q.; Wang, Z.; Xu, J. Discovery of indoleamine 2,3-dioxygenase inhibitors using machine learning based virtual screening. MedChemComm 2018, 9, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]




| Molecule Name | Molecule Structure | Molecular Weight | Docking Score (kcal/mol) |
|---|---|---|---|
| Rosmarinic acid | 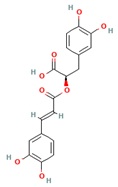 | 360.32 | −8.207 |
| Myricitrin | 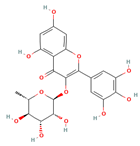 | 464.38 | −7.599 |
| Quercitrin | 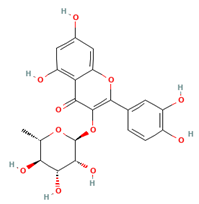 | 448.38 | −7.322 |
| Ofloxacin | 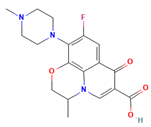 | 361.40 | −6.046 |
| Compounds | MM_PBSA (kcal/mol) | Hydrogen Bond | Hydrophobic Bond |
|---|---|---|---|
| Rosmarinic acid | −16.18 | TYR274, LYS167, GLY132, LYS133, ASP168 | ARG223, LYS220 ARG130, PHE131 TYR136, THR142 ASN140, LYS169 |
| Myricitrin | −13.87 | TYR209, TYR274, LYS167, GLY132, LYS133 | ARG218, ILE219 LYS220, ARG223 ARG130, PHE131 TYR136, THR142 |
| Quercitrin | −9.40 | TYR209, TYR274, LYS167, LYS133, GLY132 | ILE219,LYS220, ARG223,ARG130, PHE131, TYR136, THR142 |
| Ofloxacin | −9.64 | PHE131, GLY132, ASN140 | ARG67, ARG130, LYS133, TYR136, THR142, LYS167, ASP168, ILE219, LYS220 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; An, S.; Chu, J.; Liang, B.; Liao, Y.; Jiang, J.; Lin, Y.; Ye, L.; Liang, H. Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I. Molecules 2023, 28, 1444. https://doi.org/10.3390/molecules28031444
Hu X, An S, Chu J, Liang B, Liao Y, Jiang J, Lin Y, Ye L, Liang H. Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I. Molecules. 2023; 28(3):1444. https://doi.org/10.3390/molecules28031444
Chicago/Turabian StyleHu, Xiaopeng, Sanqi An, Jiemei Chu, Bingyu Liang, Yanyan Liao, Junjun Jiang, Yao Lin, Li Ye, and Hao Liang. 2023. "Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I" Molecules 28, no. 3: 1444. https://doi.org/10.3390/molecules28031444
APA StyleHu, X., An, S., Chu, J., Liang, B., Liao, Y., Jiang, J., Lin, Y., Ye, L., & Liang, H. (2023). Potential Inhibitors of Monkeypox Virus Revealed by Molecular Modeling Approach to Viral DNA Topoisomerase I. Molecules, 28(3), 1444. https://doi.org/10.3390/molecules28031444





