MOZART, a QSAR Multi-Target Web-Based Tool to Predict Multiple Drug–Enzyme Interactions
Abstract
1. Introduction
2. Results and Discussion
2.1. ANN Multi-Target Model
2.2. Web-Based Tool
3. Materials and Methods
3.1. Dataset
3.2. Molecular Descriptors
3.3. Artificial Neural Network Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zou, Y.; Li, C.; Brunzelle, J.S.; Nair, S.K. Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme. Biochemistry 2007, 46, 4294–4304. [Google Scholar] [CrossRef] [PubMed]
- Celis, J.E.; Gromova, I.; Gromov, P.; Moreira, J.M.; Cabezón, T.; Friis, E.; Rank, F. Molecular pathology of breast apocrine carcinomas: A protein expression signature specific for benign apocrine metaplasia. FEBS Lett. 2006, 580, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, S.M.; Wilkinson, B.L.; Golde, T.E.; Landreth, G. Statins reduce amyloid-beta production through inhibition of protein isoprenylation. J. Biol. Chem. 2007, 282, 26832–26844. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.G.; Notarnicola, M.; Cavallini, A.; Di Leo, A. 3-Hydroxy-3-methylglutaryl coenzyme A reductase activity and low-density lipoprotein receptor expression in diffuse-type and intestinal-type human gastric cancer. J. Gastroenterol. 2002, 37, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Dinavahi, S.S.; Bazewicz, C.G.; Gowda, R.; Robertson, G.P. Aldehyde Dehydrogenase Inhibitors for Cancer Therapeutics. Trends Pharm. Sci. 2019, 40, 774–789. [Google Scholar] [CrossRef]
- Ford-Hutchinson, A.F.; Ali, Z.; Seerattan, R.A.; Cooper, D.M.; Hallgrímsson, B.; Salo, P.T.; Jirik, F.R. Degenerative knee joint disease in mice lacking 3’-phosphoadenosine 5’-phosphosulfate synthetase 2 (Papss2) activity: A putative model of human PAPSS2 deficiency-associated arthrosis. Osteoarthr. Cartil. 2005, 13, 418–425. [Google Scholar] [CrossRef]
- Krayenbuehl, J.; Zamburlini, M.; Ghandour, S.; Pachoud, M.; Tanadini-Lang, S.; Tol, J.; Guckenberger, M.; Verbakel, W. Planning comparison of five automated treatment planning solutions for locally advanced head and neck cancer. Radiat. Oncol. 2018, 13, 170. [Google Scholar] [CrossRef]
- Park, B.N.; Lee, S.J.; Roh, J.H.; Lee, K.H.; An, Y.S.; Yoon, J.K. Radiolabeled Anti-Adenosine Triphosphate Synthase Monoclonal Antibody as a Theragnostic Agent Targeting Angiogenesis. Mol. Imaging 2017, 16, 1536012117737399. [Google Scholar] [CrossRef]
- Basu Baul, T.S.; Dutta, D.; de Vos, D.; Hopfl, H.; Pooja; Singh, P. Advancement towards tin-based anticancer chemotherapeutics: Structural modification and computer modeling approach to drug-enzyme interactions. Curr. Top. Med. Chem. 2012, 12, 2810–2826. [Google Scholar] [CrossRef]
- Zeng, X.F.; Li, W.W.; Fan, H.J.; Wang, X.Y.; Ji, P.; Wang, Z.R.; Ma, S.; Li, L.L.; Ma, X.F.; Yang, S.Y. Discovery of novel fatty acid synthase (FAS) inhibitors based on the structure of ketoaceyl synthase (KS) domain. Bioorg. Med. Chem. Lett. 2011, 21, 4742–4744. [Google Scholar] [CrossRef]
- Barrett, M.P.; Gilbert, I.H. Perspectives for new drugs against trypanosomiasis and leishmaniasis. Curr. Top. Med. Chem. 2002, 2, 471–482. [Google Scholar] [CrossRef]
- He, S.; Lai, L. Molecular docking and competitive binding study discovered different binding modes of microsomal prostaglandin E synthase-1 inhibitors. J. Chem. Inf. Model. 2011, 51, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Maloney, P.P.; Fujita, T.; Muir, R.M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. [Google Scholar] [CrossRef]
- Concu, R. New Computational Approaches Aimed at the Prediction of More Selective and Active Drugs. Curr. Top. Med. Chem. 2020, 20, 1581. [Google Scholar] [CrossRef] [PubMed]
- Concu, R.; Dea-Ayuela, M.A.; Perez-Montoto, L.G.; Bolas-Fernandez, F.; Prado-Prado, F.J.; Podda, G.; Uriarte, E.; Ubeira, F.M.; Gonzalez-Diaz, H. Prediction of enzyme classes from 3D structure: A general model and examples of experimental-theoretic scoring of peptide mass fingerprints of Leishmania proteins. J. Proteome Res. 2009, 8, 4372–4382. [Google Scholar] [CrossRef]
- Concu, R.; Kleandrova, V.V.; Speck-Planche, A.; Cordeiro, M. Probing the toxicity of nanoparticles: A unified in silico machine learning model based on perturbation theory. Nanotoxicology 2017, 11, 891–906. [Google Scholar] [CrossRef]
- Cronin, M.T. Quantitative structure-Activity relationship (QSAR) analysis of the acute sublethal neurotoxicity of solvents. Toxicol. In Vitro 1996, 10, 103–110. [Google Scholar] [CrossRef]
- Gonzalez-Diaz, H.; Dea-Ayuela, M.A.; Perez-Montoto, L.G.; Prado-Prado, F.J.; Aguero-Chapin, G.; Bolas-Fernandez, F.; Vazquez-Padron, R.I.; Ubeira, F.M. QSAR for RNases and theoretic-experimental study of molecular diversity on peptide mass fingerprints of a new Leishmania infantum protein. Mol. Divers. 2010, 14, 349–369. [Google Scholar] [CrossRef]
- Hazra, A.; Mondal, C.; Chakraborty, D.; Halder, A.K.; Bharitkar, Y.P.; Mondal, S.K.; Banerjee, S.; Jha, T.; Mondal, N.B. Towards the development of anticancer drugs from andrographolide: Semisynthesis, bioevaluation, QSAR analysis and pharmacokinetic studies. Curr. Top. Med. Chem. 2015, 15, 1013–1026. [Google Scholar] [CrossRef]
- Hisaki, T.; Aiba Nee Kaneko, M.; Yamaguchi, M.; Sasa, H.; Kouzuki, H. Development of QSAR models using artificial neural network analysis for risk assessment of repeated-dose, reproductive, and developmental toxicities of cosmetic ingredients. J. Toxicol. Sci. 2015, 40, 163–180. [Google Scholar] [CrossRef]
- Hitaoka, S.; Chuman, H.; Yoshizawa, K. A QSAR study on the inhibition mechanism of matrix metalloproteinase-12 by arylsulfone analogs based on molecular orbital calculations. Org. Biomol. Chem. 2015, 13, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Cvetnic, M.; Juretic Perisic, D.; Kovacic, M.; Kusic, H.; Dermadi, J.; Horvat, S.; Bolanca, T.; Marin, V.; Karamanis, P.; Loncaric Bozic, A. Prediction of biodegradability of aromatics in water using QSAR modeling. Ecotoxicol. Envrion. Saf. 2017, 139, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Cvetnic, M.; Juretic Perisic, D.; Kovacic, M.; Ukic, S.; Bolanca, T.; Rasulev, B.; Kusic, H.; Loncaric Bozic, A. Toxicity of aromatic pollutants and photooxidative intermediates in water: A QSAR study. Ecotoxicol. Envrion. Saf. 2019, 169, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, M.; Arunachalam, M. Quantitative Structure-Activity Relationship (QSAR) Studies for the Inhibition of MAOs. Comb. Chem. High Throughput Screen. 2020. [Google Scholar] [CrossRef]
- Rajathei, D.M.; Parthasarathy, S.; Selvaraj, S. Combined QSAR Model and Chemical Similarity Search for Novel HMGCoA Reductase Inhibitors for Coronary Heart Disease. Curr. Comput. Aided Drug Des. 2020, 16, 473–485. [Google Scholar] [CrossRef]
- Kumar, V.; De, P.; Ojha, P.K.; Saha, A.; Roy, K. A Multi-layered Variable Selection Strategy for QSAR Modeling of Butyrylcholinesterase Inhibitors. Curr. Top. Med. Chem. 2020, 20, 1601–1627. [Google Scholar] [CrossRef]
- Concu, R.; Gonzalez-Durruthy, M.; Cordeiro, M. Developing a Multi-target Model to Predict the Activity of Monoamine Oxidase A and B Drugs. Curr. Top. Med. Chem. 2020, 20, 1593–1600. [Google Scholar] [CrossRef]
- Son, M.; Park, C.; Rampogu, S.; Zeb, A.; Lee, K.W. Discovery of Novel Acetylcholinesterase Inhibitors as Potential Candidates for the Treatment of Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 1000. [Google Scholar] [CrossRef]
- Mishra, R.K.; Deibler, K.K.; Clutter, M.R.; Vagadia, P.P.; O’Connor, M.; Schiltz, G.E.; Bergan, R.; Scheidt, K.A. Modeling MEK4 Kinase Inhibitors through Perturbed Electrostatic Potential Charges. J. Chem. Inf. Model. 2019, 59, 4460–4466. [Google Scholar] [CrossRef]
- Malik, N.; Dhiman, P.; Khatkar, A. In Silico and 3D QSAR Studies of Natural Based Derivatives as Xanthine Oxidase Inhibitors. Curr. Top. Med. Chem. 2019, 19, 123–138. [Google Scholar] [CrossRef]
- Min, J.L.; Xiao, X.; Chou, K.C. iEzy-drug: A web server for identifying the interaction between enzymes and drugs in cellular networking. Biomed. Res. Int. 2013, 2013, 701317. [Google Scholar] [CrossRef] [PubMed]
- Kotera, M.; Hirakawa, M.; Tokimatsu, T.; Goto, S.; Kanehisa, M. The KEGG databases and tools facilitating omics analysis: Latest developments involving human diseases and pharmaceuticals. Methods Mol. Biol. 2012, 802, 19–39. [Google Scholar] [CrossRef]
- Quirós, M.; Gražulis, S.; Girdzijauskaitė, S.; Merkys, A.; Vaitkus, A. Using SMILES strings for the description of chemical connectivity in the Crystallography Open Database. J. Cheminform. 2018, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M. Towards a Universal SMILES representation—A standard method to generate canonical SMILES based on the InChI. J. Cheminform. 2012, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Bleakley, K.; Yamanishi, Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 2009, 25, 2397–2403. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- He, Z.; Zhang, J.; Shi, X.-H.; Hu, L.-L.; Kong, X.; Cai, Y.-D.; Chou, K.-C. Predicting Drug-Target Interaction Networks Based on Functional Groups and Biological Features. PLoS ONE 2010, 5, e9603. [Google Scholar] [CrossRef]
- Fan, Y.-N.; Xiao, X.; Min, J.-L.; Chou, K.-C. iNR-Drug: Predicting the interaction of drugs with nuclear receptors in cellular networking. Int. J. Mol. Sci. 2014, 15, 4915–4937. [Google Scholar] [CrossRef]
- Xiao, X.; Min, J.-L.; Wang, P.; Chou, K.-C. iGPCR-drug: A web server for predicting interaction between GPCRs and drugs in cellular networking. PLoS ONE 2013, 8, e72234. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2018, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2016, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Aslam, A.; Saeed, S.; Zhang, G.; Kanwal, S. Targeting highly resisted anticancer drugs through topological descriptors using VIKOR multi-criteria decision analysis. Eur. Phys. J. Plus 2022, 137, 1245. [Google Scholar] [CrossRef]
- Gonzalez-Diaz, H.; Herrera-Ibata, D.M.; Duardo-Sanchez, A.; Munteanu, C.R.; Orbegozo-Medina, R.A.; Pazos, A. ANN multiscale model of anti-HIV drugs activity vs AIDS prevalence in the US at county level based on information indices of molecular graphs and social networks. J. Chem. Inf. Model. 2014, 54, 744–755. [Google Scholar] [CrossRef]
- Casanola-Martin, G.M.; Le-Thi-Thu, H.; Perez-Gimenez, F.; Marrero-Ponce, Y.; Merino-Sanjuan, M.; Abad, C.; Gonzalez-Diaz, H. Multi-output model with Box-Jenkins operators of linear indices to predict multi-target inhibitors of ubiquitin-proteasome pathway. Mol. Divers. 2015, 19, 347–356. [Google Scholar] [CrossRef]
- Casanola-Martin, G.M.; Le-Thi-Thu, H.; Perez-Gimenez, F.; Marrero-Ponce, Y.; Merino-Sanjuan, M.; Abad, C.; Gonzalez-Diaz, H. Multi-output Model with Box-Jenkins Operators of Quadratic Indices for Prediction of Malaria and Cancer Inhibitors Targeting Ubiquitin- Proteasome Pathway (UPP) Proteins. Curr. Protein Pept. Sci. 2016, 17, 220–227. [Google Scholar] [CrossRef]
- Concu, R.; Cordeiro, M. Alignment-Free Method to Predict Enzyme Classes and Subclasses. Int. J. Mol. Sci. 2019, 20, 5389. [Google Scholar] [CrossRef]
- Concu, R.; MN, D.S.C.; Munteanu, C.R.; Gonzalez-Diaz, H. PTML Model of Enzyme Subclasses for Mining the Proteome of Biofuel Producing Microorganisms. J. Proteome Res. 2019, 18, 2735–2746. [Google Scholar] [CrossRef]
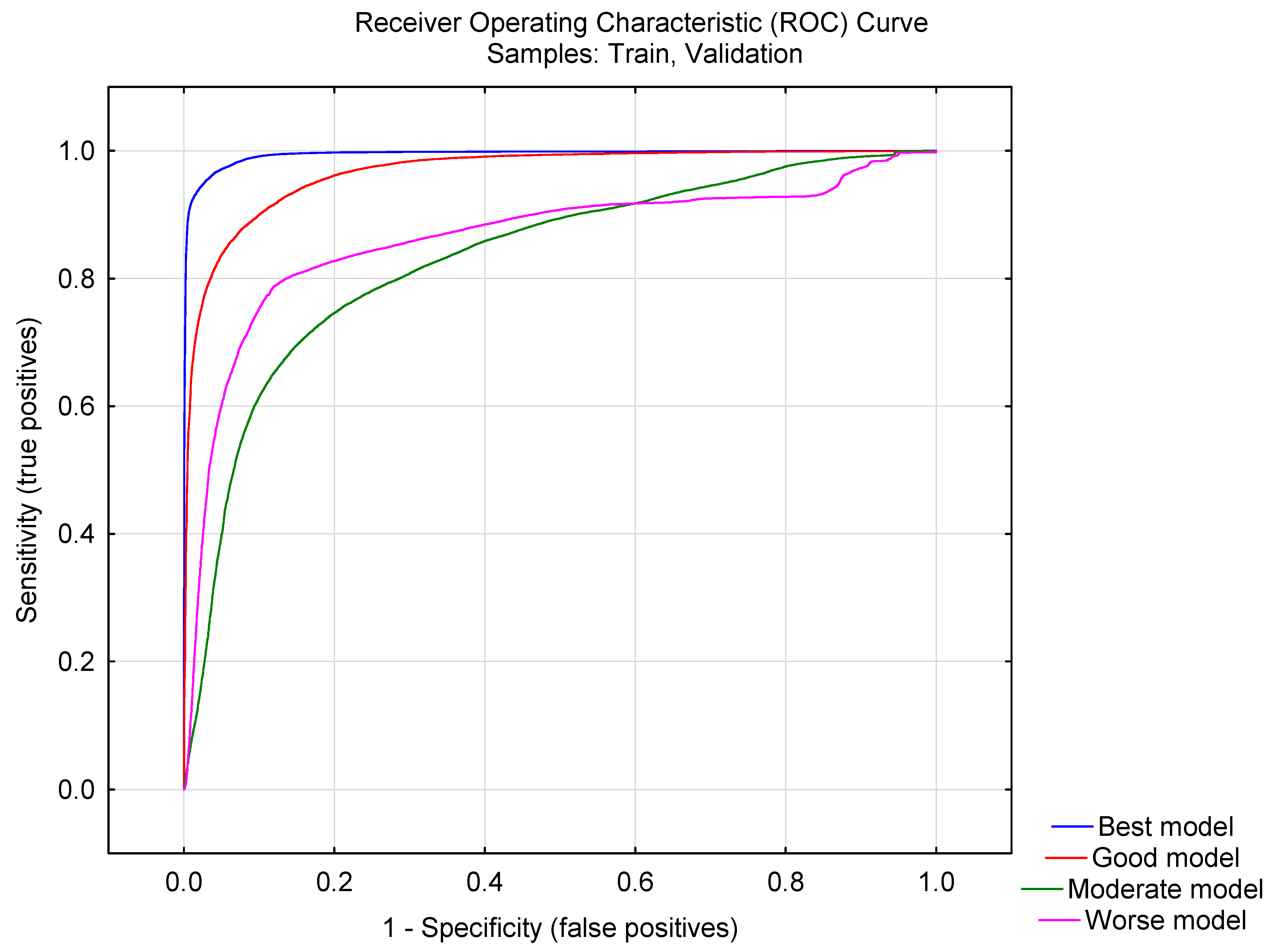

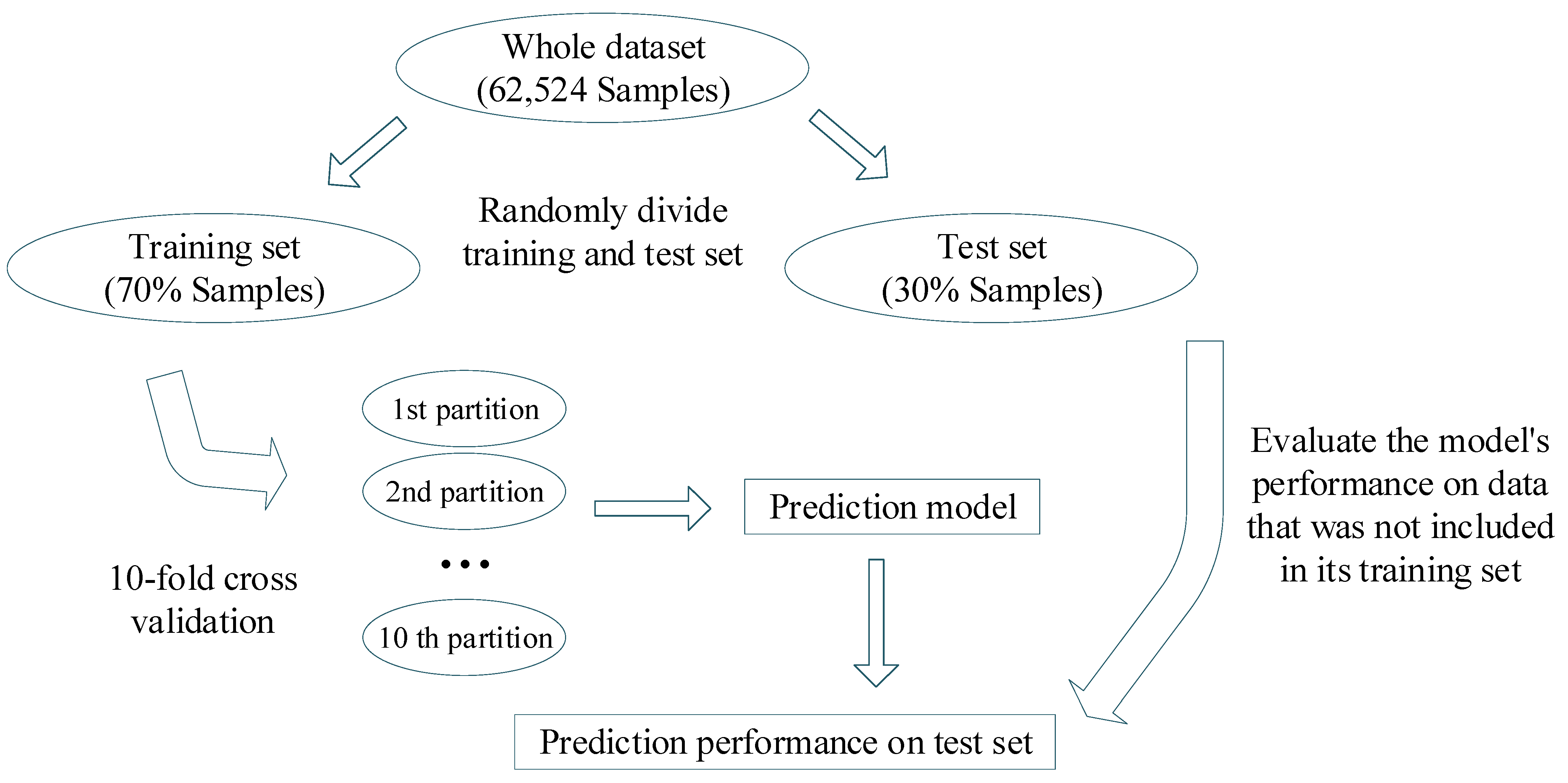
| Model Topology | Inactive * | Active * | Overall | |
|---|---|---|---|---|
| MLP 39-50-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,247 | 25,938 | 60,185 | |
| Incorrect | 1191 | 1148 | 2339 | |
| Correct (%) | 96.64 | 95.76 | 96.26 | |
| Incorrect (%) | 3.36 | 4.24 | 3.74 | |
| MLP 39-43-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,188 | 25,829 | 60,017 | |
| Incorrect | 1250 | 1257 | 2507 | |
| Correct (%) | 96.47 | 95.36 | 95.99 | |
| Incorrect (%) | 3.53 | 4.64 | 4.01 | |
| MLP 39-50-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,170 | 25,888 | 60,058 | |
| Incorrect | 1268 | 1198 | 2466 | |
| Correct (%) | 96.42 | 95.58 | 96.06 | |
| Incorrect (%) | 3.58 | 4.42 | 3.94 | |
| MLP 39-48-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,157 | 25,820 | 59,977 | |
| Incorrect | 1281 | 1266 | 2547 | |
| Correct (%) | 96.39 | 95.33 | 95.93 | |
| Incorrect (%) | 3.61 | 4.67 | 4.07 | |
| MLP 39-49-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,118 | 25,839 | 59,957 | |
| Incorrect | 1320 | 1247 | 2567 | |
| Correct (%) | 96.28 | 95.40 | 95.89 | |
| Incorrect (%) | 3.72 | 4.60 | 4.11 | |
| MLP 39-41-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,167 | 25,839 | 60,006 | |
| Incorrect | 1271 | 1247 | 2518 | |
| Correct (%) | 96.41 | 95.40 | 95.97 | |
| Incorrect (%) | 3.59 | 4.60 | 4.03 | |
| MLP 39-48-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,242 | 25,854 | 60,096 | |
| Incorrect | 1196 | 1232 | 2428 | |
| Correct (%) | 96.63 | 95.45 | 96.12 | |
| Incorrect (%) | 3.37 | 4.55 | 3.88 | |
| MLP 39-43-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,160 | 25,842 | 60,002 | |
| Incorrect | 1278 | 1244 | 2522 | |
| Correct (%) | 96.39 | 95.41 | 95.97 | |
| Incorrect (%) | 3.61 | 4.59 | 4.03 | |
| MLP 39-49-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,148 | 25,762 | 59,910 | |
| Incorrect | 1290 | 1324 | 2614 | |
| Correct (%) | 96.36 | 95.11 | 95.82 | |
| Incorrect (%) | 3.64 | 4.89 | 4.18 | |
| MLP 39-41-2 | Total | 35,438 | 27,086 | 62,524 |
| Correct | 34,167 | 25,839 | 60,006 | |
| Incorrect | 1271 | 1247 | 2518 | |
| Correct (%) | 96.41 | 95.40 | 95.97 | |
| Incorrect (%) | 3.59 | 4.60 | 4.03 |
| Overall | |||
| Sensitivity | Specificity | Overall | |
| Total | 35,656 | 28,168 | 63,824 |
| Correct | 34,438 | 26,907 | 61,345 |
| Incorrect | 1218 | 1261 | 2479 |
| Correct (%) | 96.58 | 95.52 | 96.12 |
| Incorrect (%) | 3.42 | 4.48 | 3.88 |
| Training | |||
| Total | 25,339 | 19,338 | 44,677 |
| Correct | 24,538 | 18,537 | 43,075 |
| Incorrect | 801 | 801 | 1602 |
| Correct (%) | 96.84 | 95.86 | 96.41 |
| Incorrect (%) | 3.16 | 4.14 | 3.59 |
| Validation | |||
| Total | 10,317 | 8830 | 19,147 |
| Correct | 9900 | 8370 | 18,270 |
| Incorrect | 417 | 460 | 877 |
| Correct (%) | 95.96 | 94.79 | 95.42 |
| Incorrect (%) | 4.04 | 5.21 | 4.58 |
| EC | Enzyme Subclass Name | Total Entries | Interacting Pairs % | No Interacting Pairs % |
|---|---|---|---|---|
| 1.1 | Acting on the CH-OH group of donors | 2917 | 0.944289694 | 0.996090696 |
| 1.11 | Acting on a peroxide as an acceptor | 887 | 0.819548872 | 0.966843501 |
| 1.17 | Acting on CH or CH2 groups | 400 | 0.964912281 | 0.962099125 |
| 1.2 | Acting on the aldehyde or oxo group of donors | 27,517 | 0.989814307 | 0.82320442 |
| 1.3 | Acting on the CH-CH group of donors | 794 | 0.985765125 | 0.990253411 |
| 1.4 | Acting on the CH-NH2 group of donors | 2222 | 0.981687014 | 0.938095238 |
| 1.5 | Acting on the CH-NH group of donors | 1291 | 0.833333333 | 0.999210734 |
| 1.8 | Acting on a sulfur group of donors | 157 | 0.914634146 | 0.866666667 |
| 2.1 | Transferring one-carbon groups | 633 | 0.983451537 | 0.980952381 |
| 2.3 | Acyltransferases | 328 | 0.962962963 | 0.991902834 |
| 2.5 | Transferring alkyl or aryl groups, other than methyl groups | 183 | 0.842105263 | 1 |
| 2.6 | Transferring nitrogenous groups | 67 | 1 | 0.8 |
| 2.7 | Transferring phosphorus-containing groups | 2036 | 0.982942431 | 0.989981785 |
| 3.1 | Acting on ester bonds | 3639 | 0.979591837 | 0.999435188 |
| 3.2 | Glycosylases | 12,598 | 0.82436189 | 0.928249045 |
| 3.3 | Acting on ether bonds | 602 | 0.935897436 | 0.992366412 |
| 3.4 | Acting on peptide bonds (peptidases) | 723 | 0.918367347 | 0.992 |
| 3.5 | Acting on carbon-nitrogen bonds, other than peptide bonds | 58 | 0.6875 | 0.976190476 |
| 4.2 | Carbon-oxygen lyases | 3678 | 0.897035881 | 0.985841291 |
| 4.6 | Phosphorus-oxygen lyases | 105 | 0.966666667 | 1 |
| 5.3 | Intramolecular isomerases | 120 | 0.983870968 | 0.965517241 |
| 5.6 | Isomerases altering the macromolecular conformation | 1530 | 0.961145194 | 0.986551393 |
| 7.2 | Catalysing the translocation of inorganic cations | 283 | 0.986013986 | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Concu, R.; Cordeiro, M.N.D.S.; Pérez-Pérez, M.; Fdez-Riverola, F. MOZART, a QSAR Multi-Target Web-Based Tool to Predict Multiple Drug–Enzyme Interactions. Molecules 2023, 28, 1182. https://doi.org/10.3390/molecules28031182
Concu R, Cordeiro MNDS, Pérez-Pérez M, Fdez-Riverola F. MOZART, a QSAR Multi-Target Web-Based Tool to Predict Multiple Drug–Enzyme Interactions. Molecules. 2023; 28(3):1182. https://doi.org/10.3390/molecules28031182
Chicago/Turabian StyleConcu, Riccardo, Maria Natália Dias Soeiro Cordeiro, Martín Pérez-Pérez, and Florentino Fdez-Riverola. 2023. "MOZART, a QSAR Multi-Target Web-Based Tool to Predict Multiple Drug–Enzyme Interactions" Molecules 28, no. 3: 1182. https://doi.org/10.3390/molecules28031182
APA StyleConcu, R., Cordeiro, M. N. D. S., Pérez-Pérez, M., & Fdez-Riverola, F. (2023). MOZART, a QSAR Multi-Target Web-Based Tool to Predict Multiple Drug–Enzyme Interactions. Molecules, 28(3), 1182. https://doi.org/10.3390/molecules28031182










