Design, Synthesis and Structure-Activity Relationship Studies of Nicotinamide Derivatives as Potent Antifungal Agents by Disrupting Cell Wall
Abstract
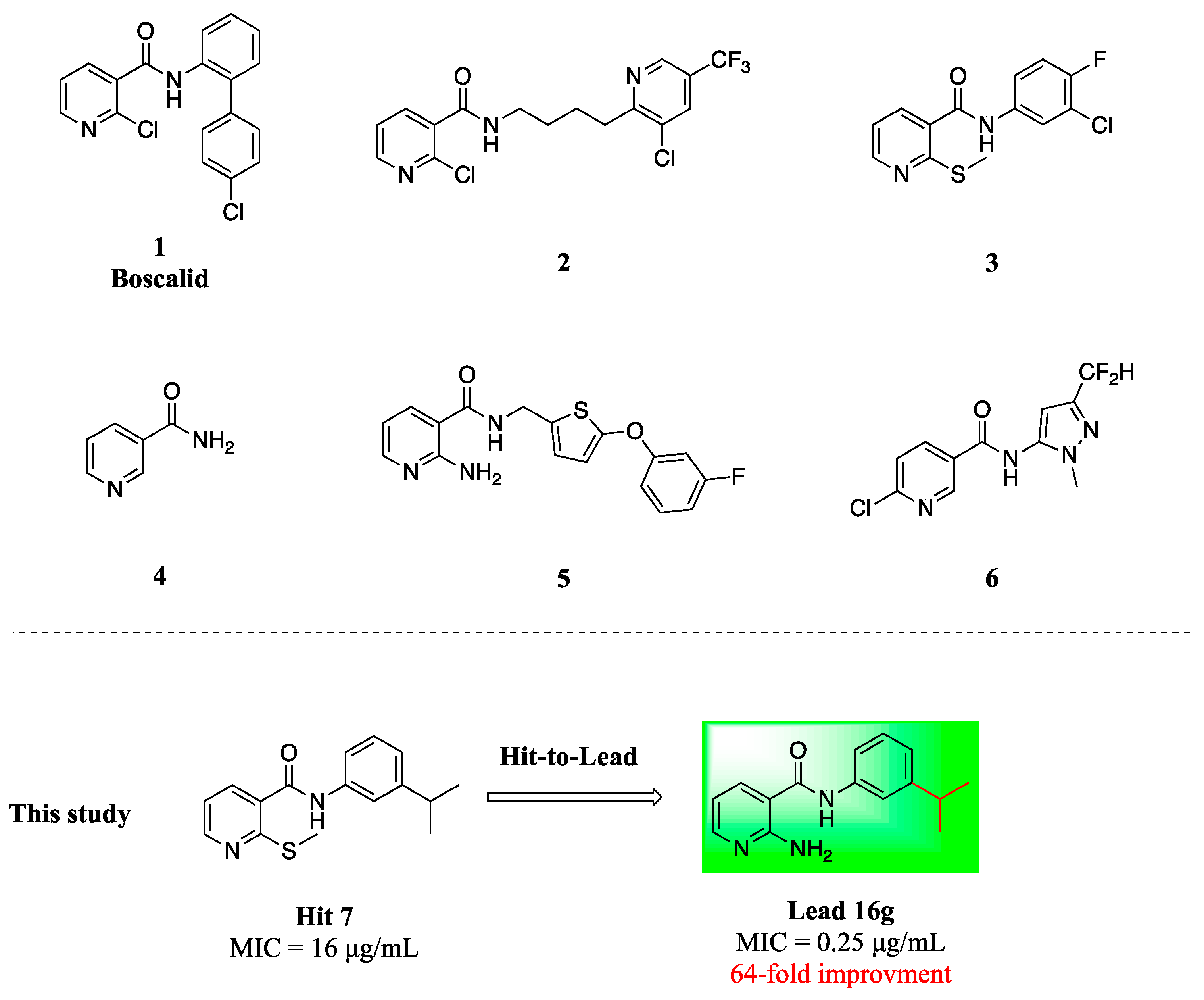
1. Introduction
2. Results and Discussion
2.1. Screening and Hit Identification
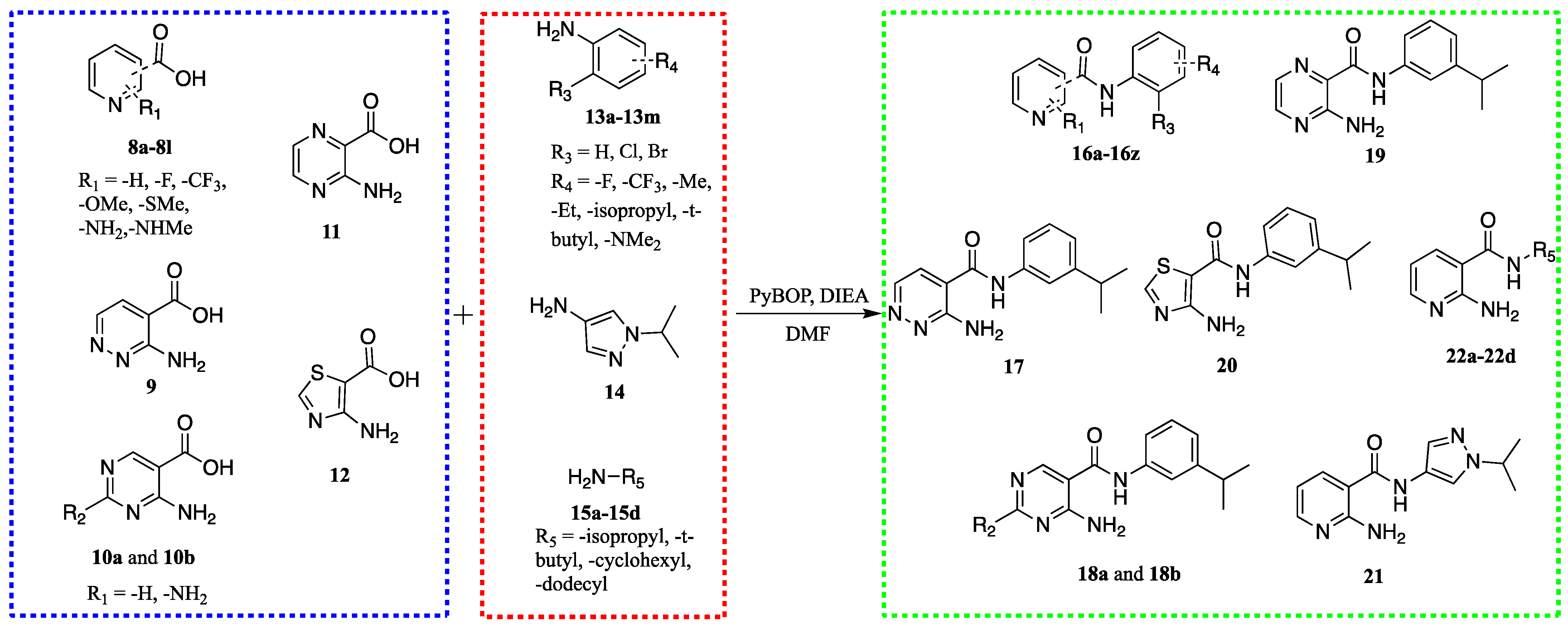
2.2. Chemistry
2.3. Structure-Activity Relationship
2.4. Compounds 16g and 16j Exhibit Low Toxicity to Mammalian Cells
2.5. Compound 16g Exhibits Broad-Spectrum Antifungal Activity
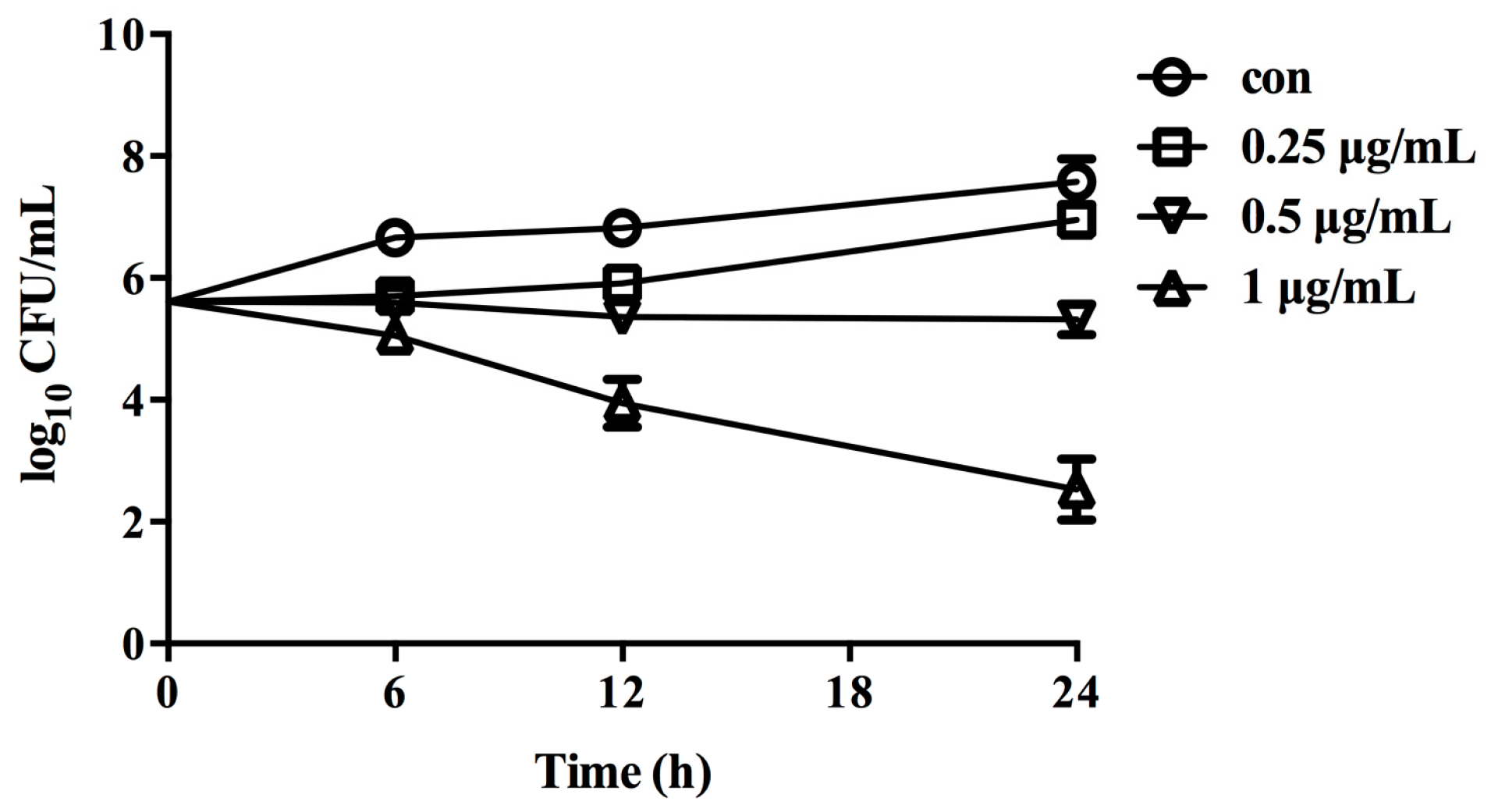
2.6. Fungicidal Activity of 16g against C. albicans
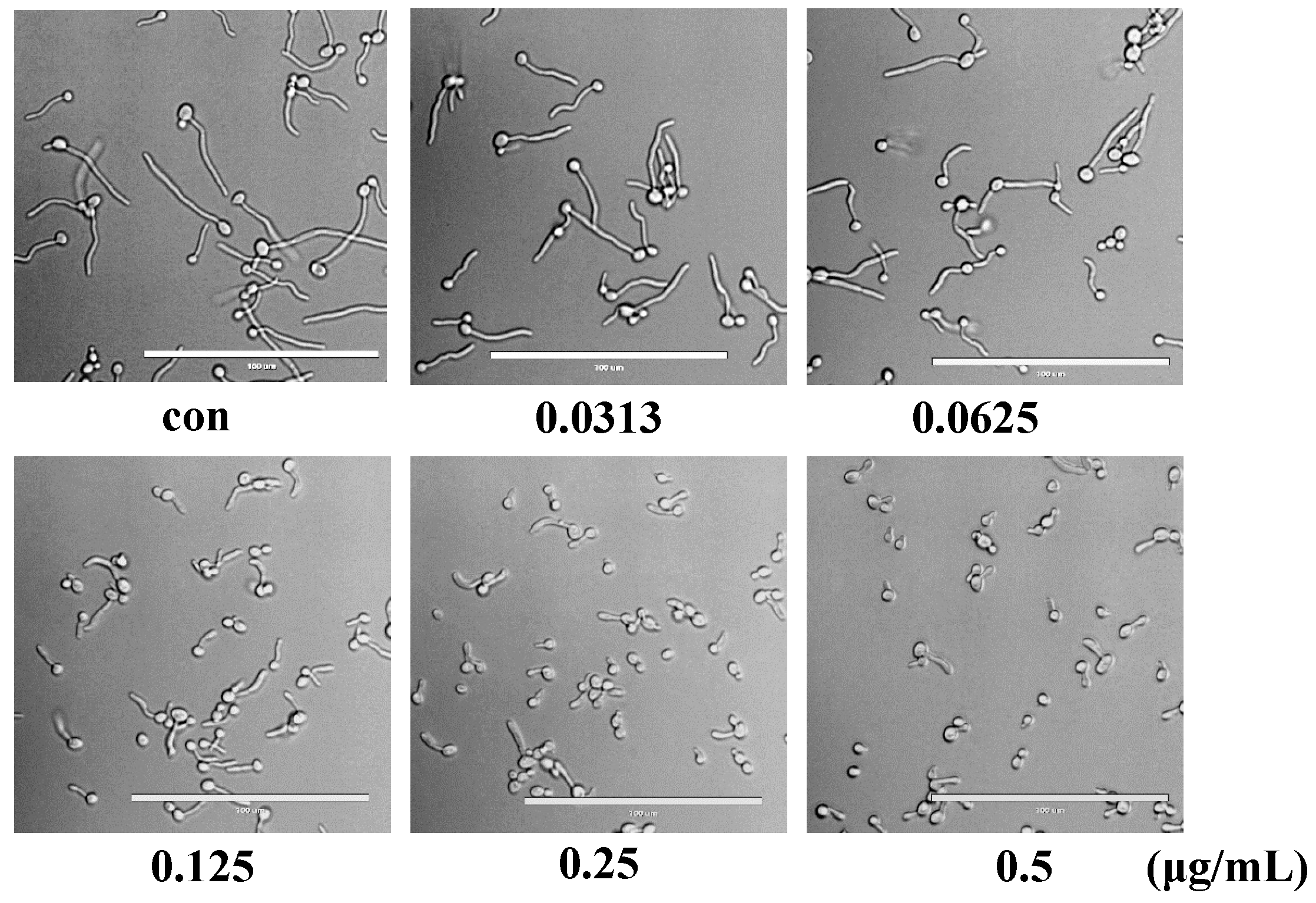
2.7. Compound 16g Inhibits the Hyphae Formation of C. albicans
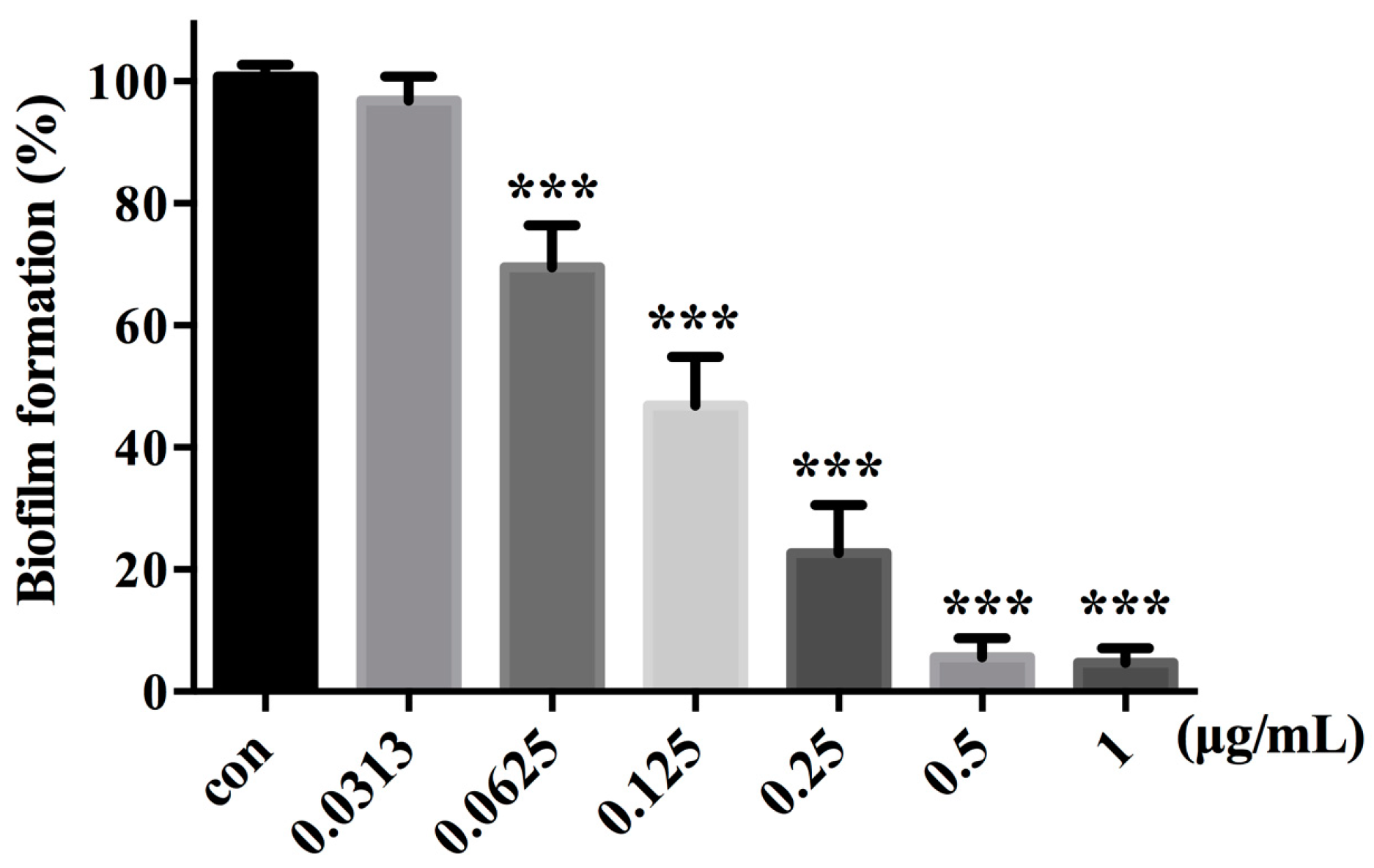
2.8. Compound 16g Inhibits the Biofilm Formation in C. albicans
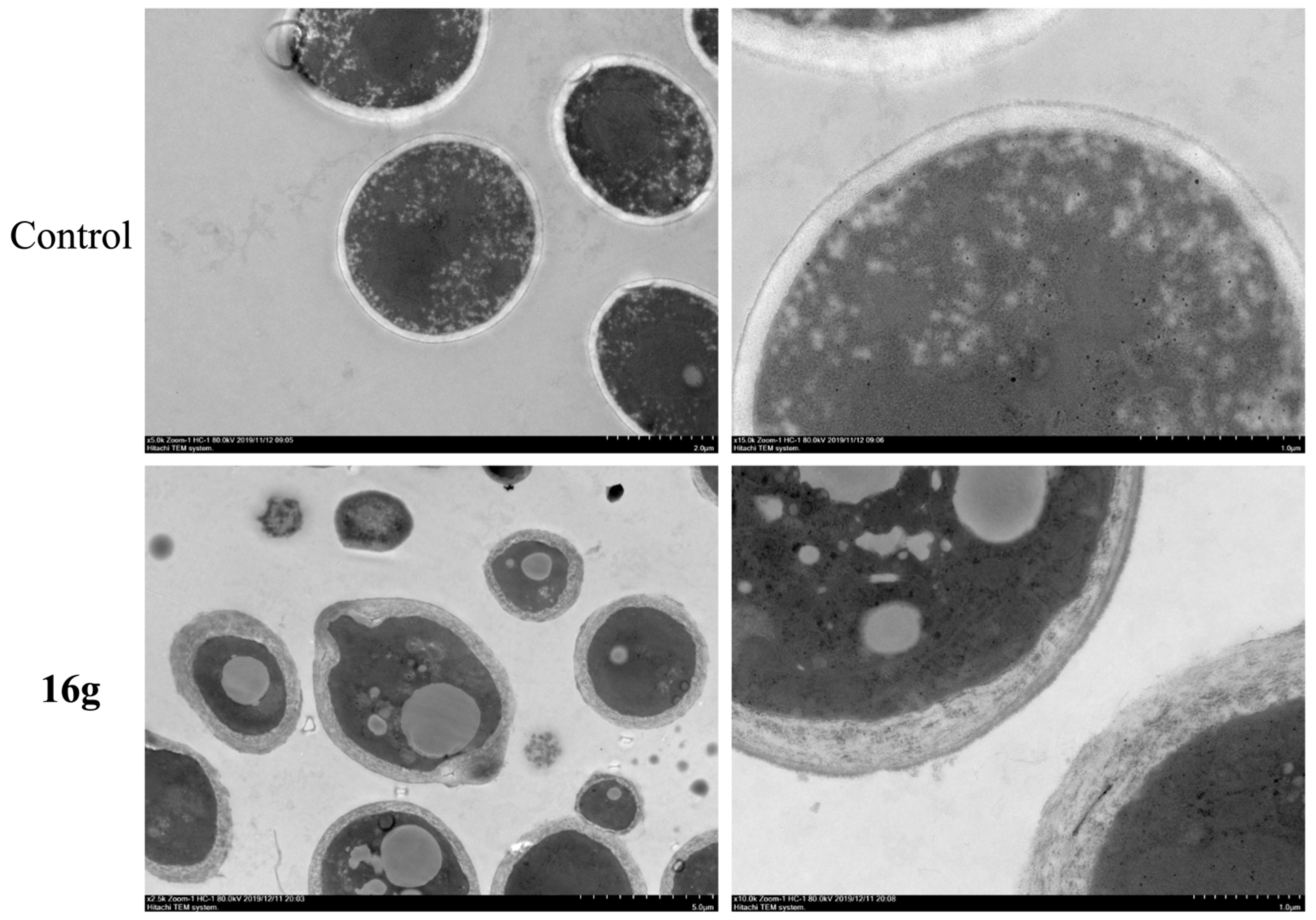
2.9. 16g Treatment Significantly Disrupted the Cell Wall Morphology of C. albicans
2.10. ADMET Prediction
3. Materials and Methods
3.1. General Procedure for the Synthesis of Target Compounds
3.2. Cytotoxicity Tests
3.3. Drug Susceptibility Testing
3.4. Hyphae Formation Assay
3.5. Biofilm Formation Assay
3.6. Time-Kill Curve Studies
3.7. PAINS Screening and ADME/T Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Jermy, A. Stop neglecting fungi. Nat. Microbiol. 2017, 2, 17120. [Google Scholar]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kang, S.; Luo, L.; Shi, Q.; Ma, J.; Yin, J.; Song, B.; Hu, D.; Yang, S. Synthesis and antifungal activities of novel nicotinamide derivatives containing 1,3,4-oxadiazole. Chem. Cent. J. 2013, 7, 64. [Google Scholar] [CrossRef]
- Avenot, H.F.; Michailides, T.J. Resistance to Boscalid Fungicide in Alternaria alternata Isolates from Pistachio in California. Plant Dis. 2007, 91, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Queron, P.Y.; Grosjean-Cournoyer, M.C.; Desbordes, P.; Genix, P.; Hartmann, B.; Mattes, A.; Kunz, K.; Fischer, R.; Gaertzen, O.; Ort, O.; et al. N-[4-(pyridin-2-yl) butyl]-carboxamide derivatives as fungicides, their preparation, fungicidal compositions, and use for the control of phytopathogenic fungi. Chem. Abstr. 2008, 148, 121597. [Google Scholar]
- Nakamoto, K.; Tsukada, I.; Tanaka, K.; Matsukura, M.; Haneda, T.; Inoue, S.; Murai, N.; Abe, S.; Ueda, N.; Miyazaki, M.; et al. Synthesis and evaluation of novel antifungal agents-quinoline and pyridine amide derivatives. Bioorg. Med. Chem. Lett. 2010, 20, 4624–4626. [Google Scholar] [CrossRef]
- Ye, Y.H.; Ma, L.; Dai, Z.C.; Xiao, Y.; Zhang, Y.Y.; Li, D.D.; Wang, J.X.; Zhu, H.L. Synthesis and antifungal activity of nicotinamide derivatives as succinate dehydrogenase inhibitors. J. Agric. Food Chem. 2014, 62, 4063–4071. [Google Scholar] [CrossRef]
- Xing, X.; Liao, Z.; Tan, F.; Zhu, Z.; Jiang, Y.; Cao, Y. Effect of Nicotinamide Against Candida albicans. Front. Microbiol. 2019, 10, 595. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.J.; Lin, G.T.; Wu, J.P.; Xu, G.; Xu, D. Novel N-(1H-Pyrazol-5-yl)nicotinamide Derivatives: Design, Synthesis and Antifungal Activity. Chem. Biodivers. 2022, 19, e202101032. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.Z.; Ni, T.J.; Li, L.P.; Li, T.; Zhang, D.Z.; Jiang, Y.Y. A New Antifungal Agent (4-phenyl-1, 3-thiazol-2-yl) Hydrazine Induces Oxidative Damage in Candida albicans. Front. Cell. Infect. Microbiol. 2020, 10, 578956. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, H.; Zhu, S.; Ji, Z.; Chi, X.; Xie, F.; Hao, Y.; Lu, H.; Yang, F.; Yan, L.; et al. Discovery of novel 7-hydroxy-5-oxo-4,5-dihydrothieno[3,2-b]pyridine-6-carboxamide derivatives with potent and selective antifungal activity against Cryptococcus species. J. Med. Chem. 2022, 65, 11257–11269. [Google Scholar] [CrossRef]
- Bohm, L.; Torsin, S.; Tint, S.H.; Eckstein, M.T.; Ludwig, T.; Perez, J.C. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog. 2017, 13, e1006699. [Google Scholar] [CrossRef]
- Murad, A.M.; Leng, P.; Straffon, M.; Wishart, J.; Macaskill, S.; MacCallum, D.; Schnell, N.; Talibi, D.; Marechal, D.; Tekaia, F.; et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001, 20, 4742–4752. [Google Scholar] [CrossRef]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011, 9, 109–118. [Google Scholar] [CrossRef]
- Han, S.B.; Shin, Y.J.; Hyon, J.Y.; Wee, W.R. Cytotoxicity of voriconazole on cultured human corneal endothelial cells. Antimicrob. Agents Chemother. 2011, 55, 4519–4523. [Google Scholar] [CrossRef]
- Beedie, S.L.; Mahony, C.; Walker, H.M.; Chau, C.H.; Figg, W.D.; Vargesson, N. Shared mechanism of teratogenicity of anti-angiogenic drugs identified in the chicken embryo model. Sci. Rep. 2016, 6, 30038. [Google Scholar]
- Clinical and Laboratory Standards Institute/National Committee for Clinical Laboratory Standards. M27ea3—Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009. [Google Scholar]
- Zhong, H.; Hu, D.D.; Hu, G.H.; Su, J.; Bi, S.; Zhang, Z.E.; Wang, Z.; Zhang, R.L.; Xu, Z.; Jiang, Y.Y.; et al. Activity of Sanguinarine against Candida albicans Biofilms. Antimicrob. Agents Chemother. 2017, 61, e02259-16. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Wickes, B.L.; Lopez-Ribot, J.L. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 2002, 68, 5459–5463. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Zhao, L.X.; Mylonakis, E.; Hu, G.H.; Zou, Y.; Huang, T.K.; Yan, L.; Wang, Y.; Jiang, Y.Y. In vitro and in vivo activities of pterostilbene against Candida albicans biofilms. Antimicrob. Agents Chemother. 2014, 58, 2344–2355. [Google Scholar] [CrossRef] [PubMed]
- Canton, E.; Peman, J.; Hervas, D.; Espinel-Ingroff, A. Examination of the in vitro fungicidal activity of echinocandins against Candida lusitaniae by time-killing methods. J. Antimicrob. Chemother. 2013, 68, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Gil-Alonso, S.; Jauregizar, N.; Canton, E.; Eraso, E.; Quindos, G. Comparison of the in vitro activity of echinocandins against Candida albicans, Candida dubliniensis, and Candida africana by time-kill curves. Diagn. Microbiol. Infect. Dis. 2015, 82, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]


| Compd | Structure | MIC (μg/mL) | Compd | Structure | MIC (μg/mL) |
|---|---|---|---|---|---|
| C. alb SC5314 | C. alb SC5314 | ||||
| 7 |  | 16 | 16f |  | >16 |
| 16a | 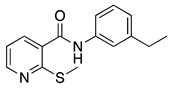 | >16 | 16g | 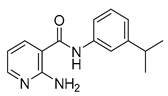 | 0.25 |
| 16b | 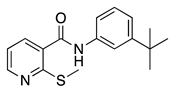 | >16 | 16h | 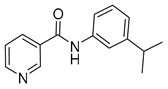 | >16 |
| 16c | 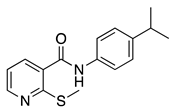 | >16 | FLC | - | 0.25 |
| 16d | 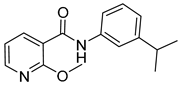 | >16 | 1 | - | >16 |
| 16e | 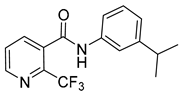 | >16 | 3 | - | >16 |
| Compd. | Structure | MIC (μg/mL) | Compd. | Structure | MIC (μg/mL) |
|---|---|---|---|---|---|
| C. alb SC5314 | C. alb SC5314 | ||||
| 16g | 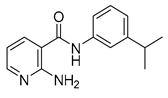 | 0.25 | 16w | 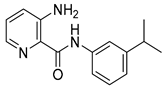 | 64 |
| 16i |  | >64 | 16x | 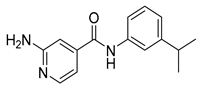 | >64 |
| 16j |  | 0.5 | 16y |  | >64 |
| 16k | 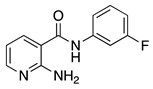 | >64 | 16z | 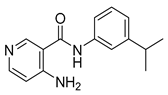 | 16 |
| 16l | 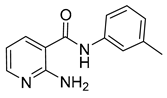 | 16 | 17 |  | 64 |
| 16m | 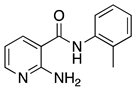 | >64 | 18a |  | 16 |
| 16n | 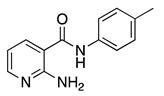 | 64 | 18b | 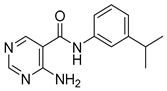 | 64 |
| 16o | 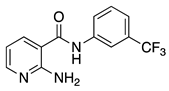 | >64 | 19 |  | 4 |
| 16p | 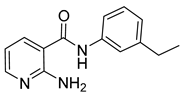 | 4 | 20 | 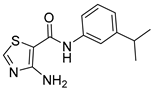 | >64 |
| 16q | 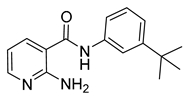 | >64 | 21 | 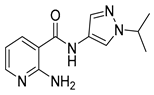 | >64 |
| 16r | 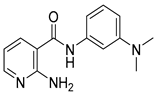 | 16 | 22a | 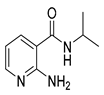 | >64 |
| 16s |  | 8 | 22b | 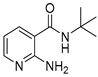 | >64 |
| 16t | 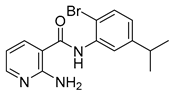 | 4 | 22c | 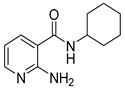 | >64 |
| 16u | 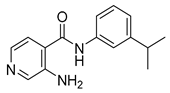 | 32 | 22d | 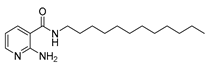 | >64 |
| 16v | 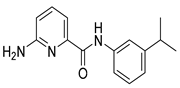 | >64 | FLC | - | 0.25 |
| C. albicans | Isolate | MIC (μg/mL) | |
|---|---|---|---|
| 16g | FLC | ||
| fluconazole-sensitive (2) | Y0109 | 0.125 | 0.125 |
| 465 | 0.5 | 0.125 | |
| fluconazole-resistant (6) | 862 | 0.5 | >64 |
| 786 | 0.5 | >64 | |
| 100 | 1 | >64 | |
| 385 | 0.125 | >64 | |
| 898 | 0.5 | >64 | |
| 504 | 0.25 | >64 | |
| Species | Isolate | MIC (μg/mL) | |
|---|---|---|---|
| 16g | FLC | ||
| C. parapsilosis | 22019 | 4 | 1 |
| 660 | 2 | 0.25 | |
| C. krusei | 463 | 8 | 4 |
| 629 | 16 | 2 | |
| C. glabrata | 537 | 8 | 0.5 |
| C. tropicalis | 752 | 0.5 | 0.5 |
| 112936 | 2 | 2 | |
| C. neoformans | 32609 | 32 | 1 |
| 34877 | 16 | 2 | |
| 56992 | 8 | 1 | |
| A. fumigatus | 7544 | >32 | >64 |
| 023-2 | >32 | >64 | |
| T.mentagrophyton | T5A | 16 | 16 |
| T5B | 32 | 16 | |
| T5E | 32 | 16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, T.; Xie, F.; Li, L.; Hao, Y.; Chi, X.; Yan, L.; Zhang, D.; Jiang, Y.; Lv, Q. Design, Synthesis and Structure-Activity Relationship Studies of Nicotinamide Derivatives as Potent Antifungal Agents by Disrupting Cell Wall. Molecules 2023, 28, 1135. https://doi.org/10.3390/molecules28031135
Ni T, Xie F, Li L, Hao Y, Chi X, Yan L, Zhang D, Jiang Y, Lv Q. Design, Synthesis and Structure-Activity Relationship Studies of Nicotinamide Derivatives as Potent Antifungal Agents by Disrupting Cell Wall. Molecules. 2023; 28(3):1135. https://doi.org/10.3390/molecules28031135
Chicago/Turabian StyleNi, Tingjunhong, Fei Xie, Liping Li, Yumeng Hao, Xiaochen Chi, Lan Yan, Dazhi Zhang, Yuanying Jiang, and Quanzhen Lv. 2023. "Design, Synthesis and Structure-Activity Relationship Studies of Nicotinamide Derivatives as Potent Antifungal Agents by Disrupting Cell Wall" Molecules 28, no. 3: 1135. https://doi.org/10.3390/molecules28031135
APA StyleNi, T., Xie, F., Li, L., Hao, Y., Chi, X., Yan, L., Zhang, D., Jiang, Y., & Lv, Q. (2023). Design, Synthesis and Structure-Activity Relationship Studies of Nicotinamide Derivatives as Potent Antifungal Agents by Disrupting Cell Wall. Molecules, 28(3), 1135. https://doi.org/10.3390/molecules28031135








