N-Containing α-Mangostin Analogs via Smiles Rearrangement as the Promising Cytotoxic, Antitrypanosomal, and SARS-CoV-2 Main Protease Inhibitory Agents
Abstract
1. Introduction
2. Results and Discussion
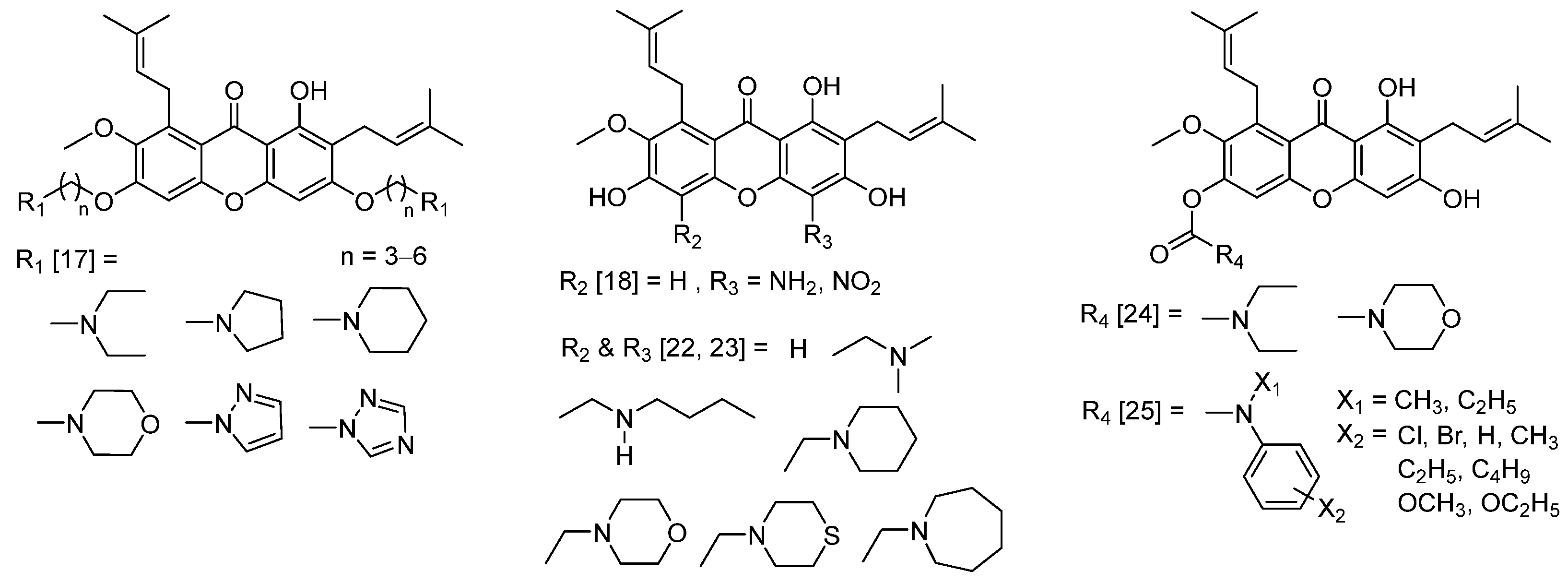
2.1. One-Pot Synthesis of N-Containing α-Mangostin Analogs via Smiles Rearrangement
2.2. Optimization of the Smiles Rearrangement of α-Mangostin (1)
2.3. Evaluation of Cytotoxicity
2.4. Evaluation of Antibacterial Activities
2.5. Evaluation of Antimalarial and Antitrypanosomal Activities
2.6. Evaluation of SARS-CoV-2 Main Protease (3CLpro) Inhibitory Activities
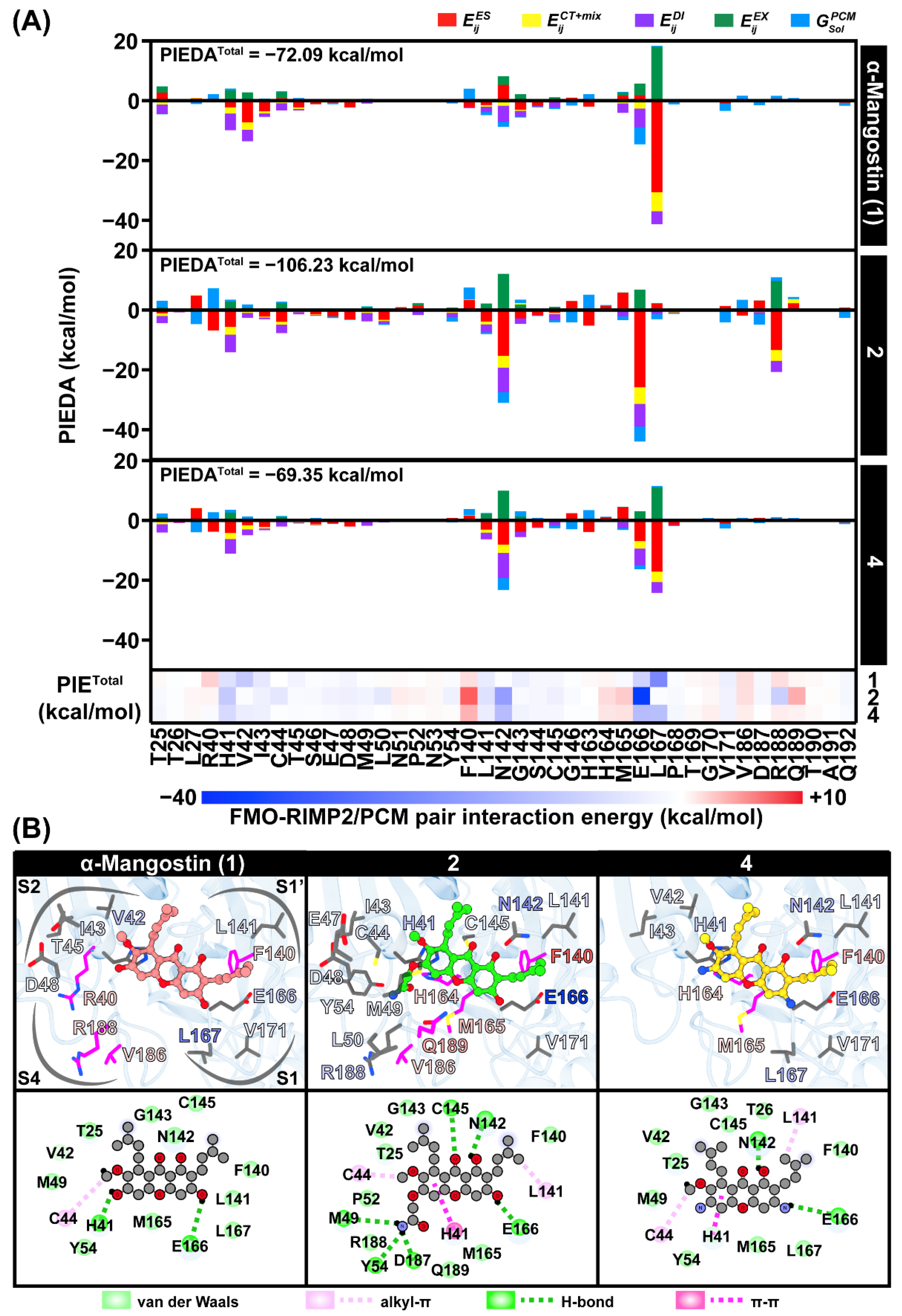
2.7. Interaction Energy Profile of the Potent Compounds toward SARS-CoV-2 3CLpro
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Extraction, Isolation, and Purification of α-Mangostin from Mangosteen Pericarp
3.3. One-Pot Synthesis of N-Containing α-Mangostin Analogs via Smiles Rearrangement
3.4. Cytotoxicity Evaluation against Cancer Cell Lines
3.5. In Vitro Antibacterial Assay
3.6. In Vitro Antimalarial Assay
3.7. In Vitro Antitrypanosomal Assay
3.8. In Vitro SARS-CoV-2 Main Protease (3CLpro) Inhibition Assay
3.9. In Silico Study of Potent Compounds toward SARS-CoV-2 Main Protease (3CLpro)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ibrahim, M.Y.; Hashim, N.M.; Mariod, A.A.; Mohan, S.; Abdulla, M.A.; Abdelwahab, S.I.; Arbab, I.A. α-Mangostin from Garcinia mangostana Linn: An updated review of its pharmacological properties. Arab. J. Chem. 2016, 9, 317–329. [Google Scholar] [CrossRef]
- Failla, M.L.; Gutiérrez-Orozco, F. Mangosteen Xanthones: Bioavailability and bioactivities. Fruit and Vegetable Phytochemicals: Chemistry and Human Health, 2nd ed.; Wiley: New York, NY, USA, 2017; pp. 165–182. [Google Scholar]
- Chen, G.; Li, Y.; Wang, W.; Deng, L. Bioactivity and pharmacological properties of α-mangostin from the mangosteen fruit: A review. Expert Opin. Ther. Pat. 2018, 28, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Narasimhan, S.; Maheshwaran, S.; Abu-Yousef, I.A.; Majdalawieh, A.F.; Rethavathi, J.; Das, P.E.; Poltronieri, P. Anti-bacterial and anti-fungal activity of xanthones obtained via semi-synthetic modification of α-mangostin from Garcinia mangostana. Molecules 2017, 22, 275. [Google Scholar] [CrossRef]
- Pollitt, E.J.; Szkuta, P.T.; Burns, N.; Foster, S.J. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018, 14, e1007112. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, W.; Hennig, S.; Eckart, M.; Kränzler, H.; Batzilla, C.; Kozitskaya, S. Nosocomial infections by Staphylococcus epidermidis: How a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 2006, 28, 14–20. [Google Scholar] [CrossRef]
- Sivaranjani, M.; Prakash, M.; Gowrishankar, S.; Rathna, J.; Pandian, S.K.; Ravi, A.V. In vitro activity of alpha-mangostin in killing and eradicating Staphylococcus epidermidis RP62A biofilms. Appl. Microbiol. Biotechnol. 2017, 101, 3349–3359. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Ibrahim, S.R.; Shaaban, M.I.; Ross, S.A. Mangostanaxanthones I and II, new xanthones from the pericarp of Garcinia mangostana. Fitoterapia 2014, 98, 215–221. [Google Scholar] [CrossRef]
- Forsyth, V.S.; Armbruster, C.E.; Smith, S.N.; Pirani, A.; Springman, A.C.; Walters, M.S.; Nielubowicz, G.R.; Himpsl, S.D.; Snitkin, E.S.; Mobley, H.L. Rapid growth of uropathogenic Escherichia coli during human urinary tract infection. MBio 2018, 9, e00186-18. [Google Scholar] [CrossRef]
- Upegui, Y.; Robledo, S.M.; Gil Romero, J.F.; Quiñones, W.; Archbold, R.; Torres, F.; Escobar, G.; Nariño, B.; Echeverri, F. In vivo antimalarial activity of α-mangostin and the new xanthone δ-mangostin. Phytother. Res. 2015, 29, 1195–1201. [Google Scholar] [CrossRef]
- Otoguro, K.; Iwatsuki, M.; Ishiyama, A.; Namatame, M.; Nishihara-Tukashima, A.; Kiyohara, H.; Hashimoto, T.; Asakawa, Y.; Ōmura, S.; Yamada, H. In vitro antitrypanosomal activity of plant terpenes against Trypanosoma brucei. Phytochemistry 2011, 72, 2024–2030. [Google Scholar] [CrossRef] [PubMed]
- Al-Massarani, S.M.; El Gamal, A.A.; Al-Musayeib, N.M.; Mothana, R.A.; Basudan, O.A.; Al-Rehaily, A.J.; Farag, M.; Assaf, M.H.; El Tahir, K.H.; Maes, L. Phytochemical, antimicrobial and antiprotozoal evaluation of Garcinia mangostana pericarp and α-mangostin, its major xanthone derivative. Molecules 2013, 18, 10599–10608. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Brunner, I.; Han, A.R.; Hamburger, M.; Kinghorn, A.D.; Frye, R.; Butterweck, V. Pharmacokinetics of α-mangostin in rats after intravenous and oral application. Mol. Nutr. Food Res. 2011, 55 (Suppl. S1), S67–S74. [Google Scholar] [CrossRef]
- Petiwala, S.M.; Li, G.; Ramaiya, A.; Kumar, A.; Gill, R.K.; Saksena, S.; Johnson, J.J. Pharmacokinetic characterization of mangosteen (Garcinia mangostana) fruit extract standardized to α-mangostin in C57BL/6 mice. Nutr. Res. 2014, 34, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Gu, X.; Song, Q.; Wang, X.; Huang, M.; Hu, M.; Hou, L.; Kang, T.; Chen, J.; Chen, H. Nanoformulated alpha-mangostin ameliorates Alzheimer’s disease neuropathology by elevating LDLR expression and accelerating amyloid-beta clearance. J. Control. Release 2016, 226, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Koh, J.-J.; Li, J.; Qiu, S.; Aung, T.T.; Lin, H.; Lakshminarayanan, R.; Dai, X.; Tang, C.; Lim, F.H. Design and synthesis of amphiphilic xanthone-based, membrane-targeting antimicrobials with improved membrane selectivity. J. Med. Chem. 2013, 56, 2359–2373. [Google Scholar] [CrossRef]
- Fei, X.; Jo, M.; Lee, B.; Han, S.-B.; Lee, K.; Jung, J.-K.; Seo, S.-Y.; Kwak, Y.-S. Synthesis of xanthone derivatives based on α-mangostin and their biological evaluation for anti-cancer agents. Bioorganic Med. Chem. Lett. 2014, 24, 2062–2065. [Google Scholar] [CrossRef]
- Morelli, C.F.; Biagiotti, M.; Pappalardo, V.M.; Rabuffetti, M.; Speranza, G. Chemistry of α-mangostin. Studies on the semisynthesis of minor xanthones from Garcinia mangostana. Nat. Prod. Res. 2015, 29, 750–755. [Google Scholar] [CrossRef]
- Pennington, L.D.; Moustakas, D.T. The necessary nitrogen atom: A versatile high-impact design element for multiparameter optimization. J. Med. Chem. 2017, 60, 3552–3579. [Google Scholar] [CrossRef]
- Boger, D.L. The difference a single atom can make: Synthesis and design at the chemistry–biology interface. J. Org. Chem. 2017, 82, 11961–11980. [Google Scholar] [CrossRef]
- Buravlev, E.V.; Shevchenko, O.G.; Kutchin, A.V. Synthesis and membrane-protective activity of novel derivatives of α-mangostin at the C-4 position. Bioorganic Med. Chem. Lett. 2015, 25, 826–829. [Google Scholar] [CrossRef]
- Buravlev, E.V.; Shevchenko, O.G.; Anisimov, A.A.; Suponitsky, K.Y. Novel Mannich bases of α- and γ-mangostins: Synthesis and evaluation of antioxidant and membrane-protective activity. Eur. J. Med. Chem. 2018, 152, 10–20. [Google Scholar] [CrossRef]
- Kim, H.-J.; Fei, X.; Cho, S.-C.; Choi, B.Y.; Ahn, H.-C.; Lee, K.; Seo, S.-Y.; Keum, Y.-S. Discovery of α-mangostin as a novel competitive inhibitor against mutant isocitrate dehydrogenase-1. Bioorganic Med. Chem. Lett. 2015, 25, 5625–5631. [Google Scholar] [CrossRef]
- Chen, Y.; Bian, Y.; Wang, J.-W.; Gong, T.-T.; Ying, Y.-M.; Ma, L.-F.; Shan, W.-G.; Xie, X.-Q.; Zhan, Z.-J. Effects of α-Mangostin Derivatives on the Alzheimer’s Disease Model of Rats and Their Mechanism: A Combination of Experimental Study and Computational Systems Pharmacology Analysis. ACS Omega 2020, 5, 9846–9863. [Google Scholar] [CrossRef] [PubMed]
- Truce, W.E.; Kreider, E.M.; Brand, W.W. The Smiles and related rearrangements of aromatic systems. Org. React. 2004, 18, 99–215. [Google Scholar]
- Holden, C.M.; Greaney, M.F. Modern aspects of the Smiles rearrangement. Chem. Eur. J. 2017, 23, 8992–9008. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.-S.; Vijaykumar, B.; Jang, K.; Shin, H.-H.; Zuo, H.; Shin, D.-S. One-pot conversion of phenols to anilines via Smiles rearrangement. Tetrahedron Lett. 2013, 54, 5151–5154. [Google Scholar] [CrossRef]
- Mayer, S.; Keglevich, P.; Ábrányi-Balogh, P.; Szigetvári, Á.; Dékány, M.; Szántay, C., Jr.; Hazai, L. Synthesis and in vitro anticancer evaluation of novel chrysin and 7-aminochrysin derivatives. Molecules 2020, 25, 888. [Google Scholar] [CrossRef] [PubMed]
- Suksamrarn, S.; Suwannapoch, N.; Ratananukul, P.; Aroonlerk, N.; Suksamrarn, A. Xanthones from the Green Fruit Hulls of Garcinia m angostana. J. Nat. Prod. 2002, 65, 761–763. [Google Scholar] [CrossRef] [PubMed]
- Suksamrarn, S.; Suwannapoch, N.; Phakhodee, W.; Thanuhiranlert, J.; Ratananukul, P.; Chimnoi, N.; Suksamrarn, A. Antimycobacterial activity of prenylated xanthones from the fruits of Garcinia mangostana. Chem. Pharm. Bull. 2003, 51, 857–859. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.-J.; Qiu, S.; Zou, H.; Lakshminarayanan, R.; Li, J.; Zhou, X.; Tang, C.; Saraswathi, P.; Verma, C.; Tan, D.T. Rapid bactericidal action of alpha-mangostin against MRSA as an outcome of membrane targeting. Biochim. Biophys. Acta 2013, 1828, 834–844. [Google Scholar] [CrossRef]
- Sakagami, Y.; Iinuma, M.; Piyasena, K.; Dharmaratne, H. Antibacterial activity of α-mangostin against vancomycin resistant Enterococci (VRE) and synergism with antibiotics. Phytomedicine 2005, 12, 203–208. [Google Scholar] [CrossRef]
- Deetanya, P.; Hengphasatporn, K.; Wilasluck, P.; Shigeta, Y.; Rungrotmongkol, T.; Wangkanont, K. Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification. Comput. Struct. Biotechnol. J. 2021, 19, 3364–3371. [Google Scholar] [CrossRef]
- Wansri, R.; Lin, A.C.K.; Pengon, J.; Kamchonwongpaisan, S.; Srimongkolpithak, N.; Rattanajak, R.; Wilasluck, P.; Deetanya, P.; Wangkanont, K.; Hengphasatporn, K. Semi-Synthesis of N-Aryl Amide Analogs of Piperine from Piper nigrum and Evaluation of Their Antitrypanosomal, Antimalarial, and Anti-SARS-CoV-2 Main Protease Activities. Molecules 2022, 27, 2841. [Google Scholar] [CrossRef]
- Hengphasatporn, K.; Harada, R.; Wilasluck, P.; Deetanya, P.; Sukandar, E.R.; Chavasiri, W.; Suroengrit, A.; Boonyasuppayakorn, S.; Rungrotmongkol, T.; Wangkanont, K.; et al. Promising SARS-CoV-2 main protease inhibitor ligand-binding modes evaluated using LB-PaCS-MD/FMO. Sci. Rep. 2022, 12, 17984. [Google Scholar] [CrossRef]
- Sanachai, K.; Somboon, T.; Wilasluck, P.; Deetanya, P.; Wolschann, P.; Langer, T.; Lee, V.S.; Wangkanont, K.; Rungrotmongkol, T.; Hannongbua, S. Identification of repurposing therapeutics toward SARS-CoV-2 main protease by virtual screening. PLoS ONE 2022, 17, e0269563. [Google Scholar] [CrossRef]
- Biovia, D. Discovery Studio Visualizer; Dassault Systems: San Diego, CA, USA, 2021. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Smilkstein, M.; Sriwilaijaroen, N.; Kelly, J.X.; Wilairat, P.; Riscoe, M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004, 48, 1803–1806. [Google Scholar] [CrossRef]
- Wangchuk, P.; Keller, P.A.; Pyne, S.G.; Taweechotipatr, M.; Tonsomboon, A.; Rattanajak, R.; Kamchonwongpaisan, S. Evaluation of an ethnopharmacologically selected Bhutanese medicinal plants for their major classes of phytochemicals and biological activities. J. Ethnopharmacol. 2011, 137, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yang, H.; Shen, W.; Zhao, Q.; Li, J.; Yang, K.; Chen, C.; Jin, Y.; Bartlam, M.; Rao, Z. Production of authentic SARS-CoV Mpro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007, 366, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Patigo, A.; Hengphasatporn, K.; Cao, V.; Paunrat, W.; Vijara, N.; Chokmahasarn, T.; Maitarad, P.; Rungrotmongkol, T.; Shigeta, Y.; Boonyasuppayakorn, S.; et al. Design, synthesis, in vitro, in silico, and SAR studies of flavone analogs towards anti-dengue activity. Sci. Rep. 2022, 12, 21646. [Google Scholar] [CrossRef]
- Hengphasatporn, K.; Wilasluck, P.; Deetanya, P.; Wangkanont, K.; Chavasiri, W.; Visitchanakun, P.; Leelahavanichkul, A.; Paunrat, W.; Boonyasuppayakorn, S.; Rungrotmongkol, T.; et al. Halogenated Baicalein as a Promising Antiviral Agent toward SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 2022, 62, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G.; Nakatsuji, H. Gaussian 16; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Hengphasatporn, K.; Garon, A.; Wolschann, P.; Langer, T.; Yasuteru, S.; Huynh, T.N.; Chavasiri, W.; Saelee, T.; Boonyasuppayakorn, S.; Rungrotmongkol, T. Multiple virtual screening strategies for the discovery of novel compounds active against dengue virus: A hit identification study. Sci. Pharm. 2020, 88, 2. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]

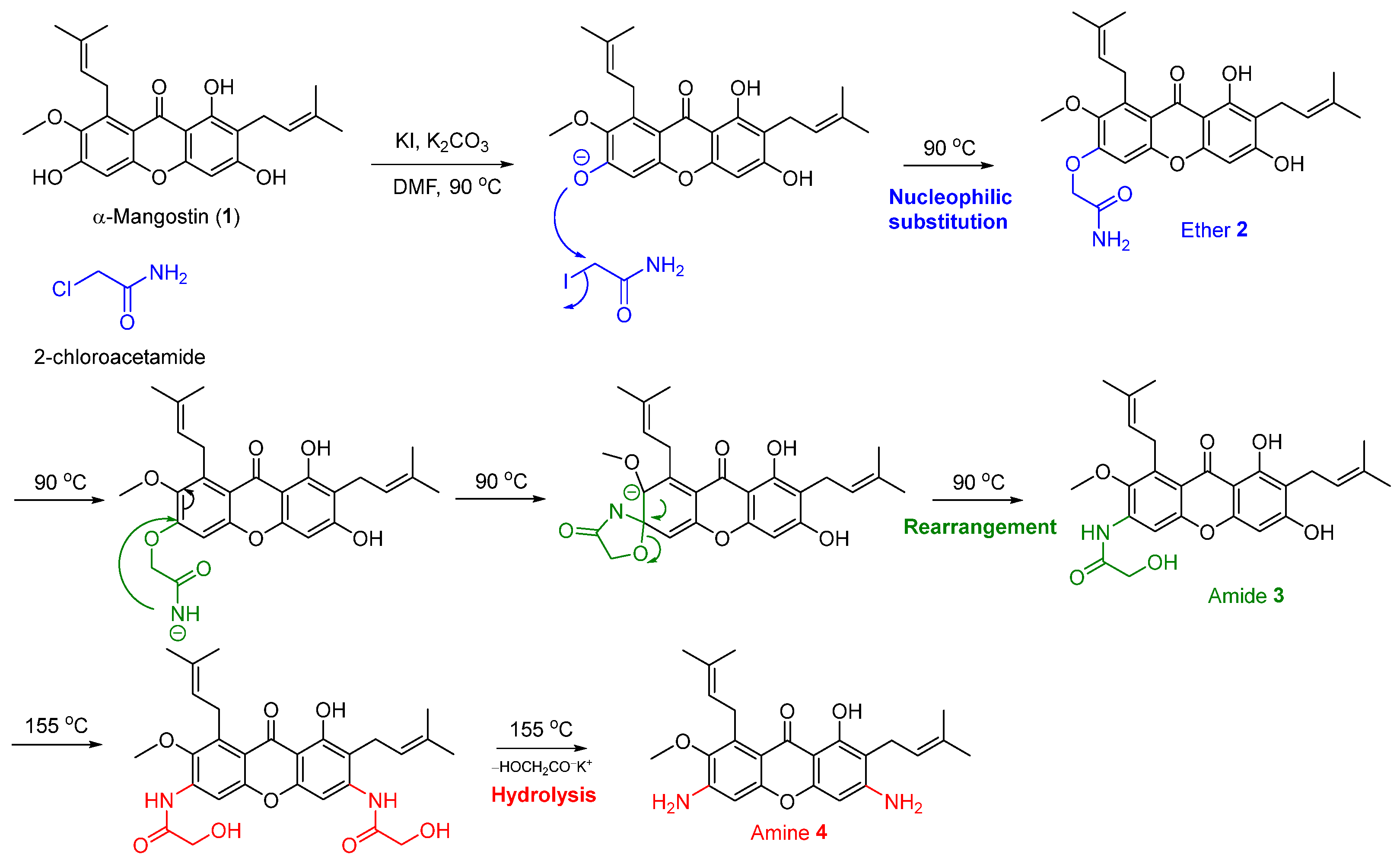


| Entry | 2-Chloro Acetamide (Equiv) | Base a | KI (Equiv) | Heating (°C) | Time (h) | % Yield of Analogs b | Ratio of 2:3:4 | Overall Yield (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | ||||||||
| 1 | 1.2 | K2CO3 | 0.2 | Reflux | 4 | 20 | 17 | 9 | 2:2: 1 | 46 |
| 2 | 2.5 | K2CO3 | 0.2 | Reflux | 4 | 8 | 25 | 20 | 1:3:3 | 53 |
| 3 | 2.5 | K2CO3 | 0.5 | Reflux | 4 | 27 | 26 | 12 | 2:2:1 | 65 |
| 4 | 2.5 | K2CO3 | 1 | Reflux | 4 | 25 | 21 | 17 | 2:1:1 | 63 |
| 5 | 2.5 | K2CO3 | 1 | Reflux | 10 | 20 | 18 | 16 | 1:1:1 | 54 |
| 6 | 2.5 | K2CO3 | 0.2 | Microwave | 0.2 c | 4 | 3 | 17 | 1:1:6 | 24 |
| 7 | 1.2 | Cs2CO3 | 0.2 | Reflux | 4 | 14 | 36 | 35 | 1:3:3 | 85 |
| 8 | 2.5 | Cs2CO3 | 0.2 | Reflux | 4 | 13 | 12 | 27 | 1:1:3 | 50 |
| 9 | 1.2 | Cs2CO3 | 1 | Reflux | 4 | 20 | 33 | 4 | 5:8:1 | 57 |
| 10 | 1.2 | Cs2CO3 | 0.2 | Microwave | 0.2 c | 7 | 8 | 17 | 1:1:2 | 32 |
| 11 | 1.2 | KOH | 0.2 | Reflux | 4 | 3 | 13 | 23 | 1:4:7 | 39 |
| 12 | 2.5 | KOH | 0.2 | Reflux | 4 | 8 | 30 | 4 | 2:7:1 | 42 |
| 13 | 2.5 | KOH | 1 | Reflux | 4 | 17 | 21 | 16 | 1:1:1 | 54 |
| 14 | 2.5 | KOH | 0.2 | Microwave | 0.2 c | 5 | 11 | 18 | 1:2:4 | 34 |
| Cell Lines a | IC50 ± S.D. (µM) | ||||
|---|---|---|---|---|---|
| α-Mangostin (1) | 2 | 3 | 4 | Cisplatin | |
| H292 | 26.16 ± 0.74 | >500 | 265.48 ± 25.22 | 26.50 ± 12.42 | 55.71 ± 0.26 |
| H460 | 25.07 ± 2.15 | 297.66 ± 37.34 | 280.48 ± 17.59 | 13.89 ± 2.55 | 48.81 ± 0.93 |
| SW-1088 | 29.99 ± 3.12 | 78.45 ± 4.70 | 40.39 ± 2.17 | 117.05 ± 15.15 | 275.58 ± 18.33 |
| U87-MG | 30.71 ± 1.53 | 92.40 ± 2.30 | 45.81 ± 2.55 | 97.68 ± 2.00 | 362.02 ± 14.26 |
| MDA-MB-231 | 31.07 ± 1.56 | 77.85 ± 5.59 | 40.51 ± 2.74 | 94.49 ± 2.36 | 230.08 ± 14.26 |
| MCF-7 | 31.58 ± 0.40 | 93.17 ± 2.68 | 43.61 ± 0.20 | 85.48 ± 4.63 | 88.12 ± 1.67 |
| HuH-7 | 24.90 ± 3.33 | 72.55 ± 1.04 | 36.35 ± 0.58 | 64.62 ± 2.99 | 47.99 ± 5.3 |
| HepG2 | 33.11 ± 0.93 | 173.88 ± 14.59 | 42.44 ± 1.51 | 91.05 ± 1.88 | 49.50 ± 6.54 |
| HT-29 | 12.28 ± 1.32 | 72.11 ± 7.80 | 31.43 ± 0.79 | 58.79 ± 0.31 | 132.42 ± 17.29 |
| HCT-116 | 30.94 ± 2.12 | 89.85 ± 1.09 | 37.76 ± 2.22 | 88.82 ± 1.11 | 112.85 ± 12.17 |
| Compounds | Minimal Inhibitory Concentration (µM) | |||||
|---|---|---|---|---|---|---|
| Staphylococcus aureus ATCC 25923 | Staphylococcus epidermidis ATCC 12228 | Kocuria rhizophila ATCC 9341 | Bacillus subtilis ATCC 6633 | Klebsiella pneumoniae ATCC 13883 | Escherichia coli ATCC 25922 | |
| α-Mangostin (1) | 1.22 | 0.61 | 0.61 | 1.22 | >2.50 mM | >2.50 mM |
| 2 | >2.50 mM | 78.13 | 9.76 | 312.5 | >2.50 mM | >2.50 mM |
| 3 | >2.50 mM | >2.50 mM | 78.13 | >2.50 mM | >2.50 mM | >2.50 mM |
| 4 | >2.50 mM | 19.53 | 4.88 | 39.06 | >2.50 mM | >2.50 mM |
| Vancomycin | 0.61 | 1.22 | 0.61 | 1.22 | n.d. | n.d. |
| Erythromycin | n.d. a | n.d. | n.d. | n.d. | 1.22 | 78.13 |
| Gentamicin | n.d. | n.d. | n.d. | n.d. | 0.15 | 1.22 |
| Compounds | IC50 ± S.E.M. (µM) | |
|---|---|---|
| Antimalarial against 3D7 P. falciparum | Antitrypanosomal against T. brucei rhodesiense | |
| α-Mangostin (1) | 5.45 ± 0.88 | 6.74 ± 0.46 |
| 2 | 7.79 ± 1.64 | 7.04 ± 0.57 |
| 3 | 4.25 ± 0.60 | 2.68 ± 0.57 |
| 4 | 13.00 ± 0.98 | 2.32 ± 0.28 |
| Pentamidine a | n.d. | 0.016 ± 0.002 |
| Pyrimethamine b | 0.117 ± 0.017 | n.d. c |
| Compounds | Relative Protease Activity at 100 μM. (%RFU/s a ± S.D.) | IC50 ± S.E. (µM) |
|---|---|---|
| α-Mangostin (1) | 32.4 ± 0.8 | 61.6 ± 1.1 |
| 2 | 25.1 ± 1.9 | 24.6 ± 1.1 |
| 3 | 55.2 ± 0.5 | 325.1 ± 1.3 |
| 4 | 21.2 ± 1.3 | 63.8 ± 1.2 |
| Rutin b | 60.3 ± 2.4 | 103.7 ± 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyae, N.Y.L.; Maiuthed, A.; Phongsopitanun, W.; Ouengwanarat, B.; Sukma, W.; Srimongkolpithak, N.; Pengon, J.; Rattanajak, R.; Kamchonwongpaisan, S.; Ei, Z.Z.; et al. N-Containing α-Mangostin Analogs via Smiles Rearrangement as the Promising Cytotoxic, Antitrypanosomal, and SARS-CoV-2 Main Protease Inhibitory Agents. Molecules 2023, 28, 1104. https://doi.org/10.3390/molecules28031104
Pyae NYL, Maiuthed A, Phongsopitanun W, Ouengwanarat B, Sukma W, Srimongkolpithak N, Pengon J, Rattanajak R, Kamchonwongpaisan S, Ei ZZ, et al. N-Containing α-Mangostin Analogs via Smiles Rearrangement as the Promising Cytotoxic, Antitrypanosomal, and SARS-CoV-2 Main Protease Inhibitory Agents. Molecules. 2023; 28(3):1104. https://doi.org/10.3390/molecules28031104
Chicago/Turabian StylePyae, Nan Yadanar Lin, Arnatchai Maiuthed, Wongsakorn Phongsopitanun, Bongkot Ouengwanarat, Warongrit Sukma, Nitipol Srimongkolpithak, Jutharat Pengon, Roonglawan Rattanajak, Sumalee Kamchonwongpaisan, Zin Zin Ei, and et al. 2023. "N-Containing α-Mangostin Analogs via Smiles Rearrangement as the Promising Cytotoxic, Antitrypanosomal, and SARS-CoV-2 Main Protease Inhibitory Agents" Molecules 28, no. 3: 1104. https://doi.org/10.3390/molecules28031104
APA StylePyae, N. Y. L., Maiuthed, A., Phongsopitanun, W., Ouengwanarat, B., Sukma, W., Srimongkolpithak, N., Pengon, J., Rattanajak, R., Kamchonwongpaisan, S., Ei, Z. Z., Chunhacha, P., Wilasluck, P., Deetanya, P., Wangkanont, K., Hengphasatporn, K., Shigeta, Y., Rungrotmongkol, T., & Chamni, S. (2023). N-Containing α-Mangostin Analogs via Smiles Rearrangement as the Promising Cytotoxic, Antitrypanosomal, and SARS-CoV-2 Main Protease Inhibitory Agents. Molecules, 28(3), 1104. https://doi.org/10.3390/molecules28031104