Abstract
The emergence of Multidrug Resistance (MDR) strains of bacteria has accelerated the search for new antibacterials. The specific bacterial peptidoglycan biosynthetic pathway represents opportunities for the development of novel antibacterial agents. Among the enzymes involved, Mur ligases, described herein, and especially the amide ligases MurC-F are key targets for the discovery of multi-inhibitors, as they share common active sites and structural features.
1. Introduction
Microorganisms have existed on Earth for centuries, but it was not until the 17th century that Antoni van Leeuwenhoek, a renowned Dutch scientist, made groundbreaking observations identifying various microorganisms, including yeasts and even red blood cells [1]. His work marked an important moment in the history of biology, laying the foundation for microbiology and bacteriology. Following Leeuwenhoek’s discoveries, a series of events triggered a surge of interest in microbiology, leading to significant advancements in the field. In the years that followed, numerous scientists studied microorganisms, particularly those that caused major epidemics. In 1835, Agostino Bassi identified Beauveria bassiana as the microbial origin of the silkworm disease called muscardine [2]. In 1854, Filippo Pacini isolated and identified the Vibrio cholerae as a pathogen [3]; meanwhile, Casimir Davaine discovered the anthrax bacillus [4]. These discoveries propelled microbiology forward, driven by the efforts of figures like Robert Koch [5] and Louis Pasteur [6]. The early 20th century marked a significant period in the history of antibiotics. German scientist Paul Ehrlich created the first effective drugs (Figure 1) against syphilis in 1910 [7], and, later, Alexander Fleming made a groundbreaking discovery in 1928, observing that Penicillium notatum had the ability to inhibit the growth of Staphylococcus aureus (S. aureus). This discovery [8] laid the foundation for the development of penicillin G 3 and other antibiotics such as sulfanilamide 4 [9,10].
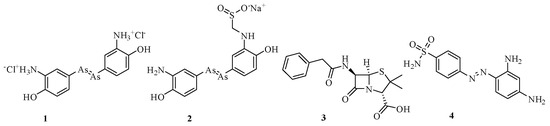
Figure 1.
Structure of Salvarsan (1), Neosalvarsan (2), penicillin G (3), and sulfanilamide (4).
The golden age of antibiotic discovery occurred from the 1940s to the 1960s, with the identification of numerous families of antibiotics 5–15 (Figure 2) [11].
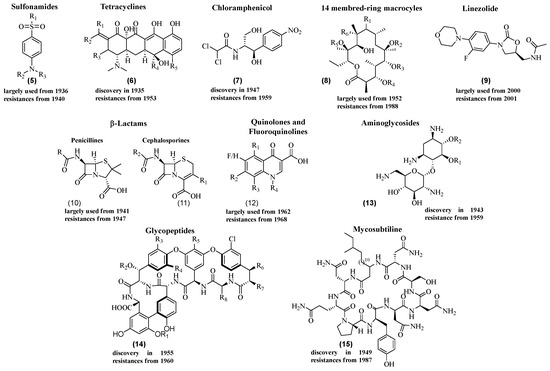
Figure 2.
Some examples of families of antibiotics.
However, bacteria developed resistance mechanisms in response to antibiotic treatments [12,13]. This resistance is a significant global health threat, and the World Health Organization (WHO) considers it one of the most serious challenges to global health. Infections caused by resistant bacteria, coupled with diminishing antibiotic effectiveness, have raised alarms [14,15,16].
Bacterial resistance can occur through different mechanisms [17,18,19,20,21,22,23,24,25,26,27], including extracellular resistance (biofilms), natural or innate resistance (inherent to specific strains), and acquired resistance (mutations or acquisition of resistance genes). Resistance mechanisms involve alterations in genes affecting interactions between bacteria and antibiotics, modifications of antibiotic targets, enzymatic mechanisms to prevent antibiotic binding, the hindrance of antibiotic entry through bacterial membranes, and the efflux of antibiotics from bacterial cells. This modification involves changes in the target’s genetic sequences, resulting in decreased efficacy of the antibiotic. For example, this phenomenon has been observed in a strain of Mycobacterium leprae resistant to rifampicin 16, an antibiotic of the rifamycin family (Figure 3) [27].
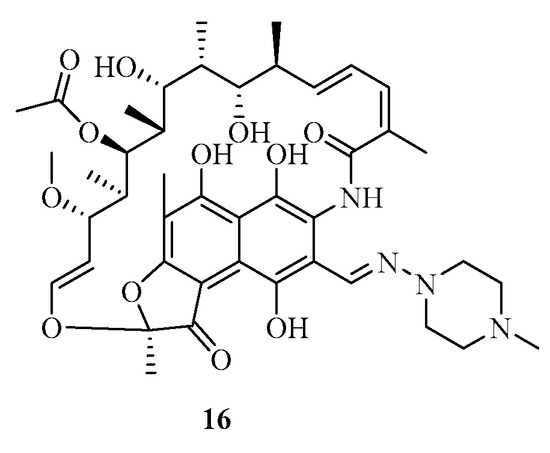
Figure 3.
Structure of rifampicin (16).
Enzymes produced by the bacteria can also inactivate the antibiotic by cleaving it. For example, enzymes known as “β-lactamase” are capable of hydrolyzing β-lactam rings found in antibiotics such as penicillins 17 (Scheme 1) to inactive compounds 18 [28]. β-lactams target bacterial wall biosynthesis by inhibiting the transpeptidase activity of penicillin-binding proteins (PBPs), which are involved in the final steps of peptidoglycan synthesis. The open form of β-lactams results in the loss of their biological activity against PBP transpeptidases.
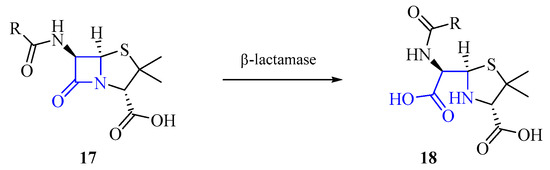
Scheme 1.
Hydrolysis by β-lactamase.
The ability of an antibiotic to reach its target within bacteria can also occur via the bacterial membranes [29,30,31,32,33,34,35,36,37]. Despite the challenges of bacterial resistance, antibiotics remain effective against certain pathogenic bacteria. These antibiotics target specific bacterial processes, such as DNA replication, RNA polymerase activity, protein synthesis, and bacterial wall synthesis. For example, antibiotics can inhibit DNA gyrase and RNA polymerase [38,39,40,41,42], interfere with the metabolism of folic acid [43,44,45,46,47], disrupt protein synthesis [48,49,50,51,52,53], and target the bacterial wall synthesis process [54,55,56,57,58,59]. Efforts to combat resistance involve the development of new antibiotics targeting novel bacterial processes and exploring new therapeutic targets. These ongoing efforts are crucial in the fight against antibiotic-resistant bacteria and the preservation of effective treatment options.
2. The Peptidoglycan Chain
Peptidoglycan, also called murein, is a biopolymer present in all bacteria that provides protection from the external environment, especially osmotic pressure [60]. A notable difference between bacteria is that the cell wall is composed of about 95% peptidoglycan chains for Gram-positive bacteria and about 20% for Gram-negative bacteria, which explains the effectiveness of some antibiotics compared to others. Its complex chemical structure is defined by a cross-linked network in which a repeating unit contains an N-acetylglucosamine (NAG) and an N-acetylmuramic acid (NAM) linked by a β-1 bond → 4 (Figure 4) [61].
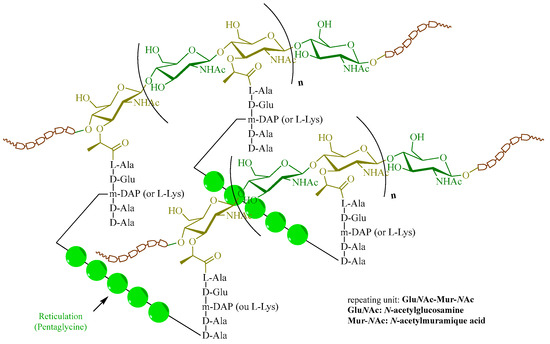
Figure 4.
Peptidoglycan structures.
This peptidoglycan forms a three-dimensional network linked by cross-links simply composed of peptides, a pentaglycine here. This cross-linking is linked on both sides with two small peptides that are grafted by the carboxylic acid function of MurNAc. The chemical nature of these peptides differs according to the bacteria; in Gram-positive bacteria, the unnatural amino acid called meso-diaminopimelic acid (m-DAP) is found, whereas in Gram-negative bacteria, L-Lysine is present. This structure presents an impressive chemical diversity and explains the strength of the peptidoglycan chain in bacteria. Indeed, several elements that compose it testify to this, such as the MurNAc, a saccharide not present in eukaryotic cells, amino acids of the d series, and m-DAP.
The biosynthesis of the peptidoglycan chain (Scheme 2) is a complex process that takes place in every part of the bacterium [62]. It is common to all Gram-positive or Gram-negative bacteria with some variations for Mycobacterium tuberculosis [63]. Each enzyme involved in this biosynthesis is important because the inhibition of one would lead to bacterial lysis and therefore cell death. During this biosynthesis, the formation of several key intermediates is obtained in each site of the bacterium (cytoplasm, membrane, and periplasm). The starting point of this synthesis is the conversion of fructose-6-phosphate (F6P, 19) to uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc, 20). Then, a lactoyl function is introduced on this substrate, followed by a pentapeptide chain to form uridine diphosphate N-acetylmuramoyl-pentapeptide (UDP-MurNAc-pentapeptide, 21). This entity, which is nothing but a part of the peptidoglycan-repeating unit, binds to undecaprenylphosphate (C55P) of the plasma membrane and is then glycosylated to obtain Lipid II (22). An enzyme called flippase will allow Lipid II to pass from the internal face of the membrane to its external face [64]. Finally, this monomer is polymerized in the periplasm by transglycosylation and transpeptidation steps to end up with the mature peptidoglycan (23).
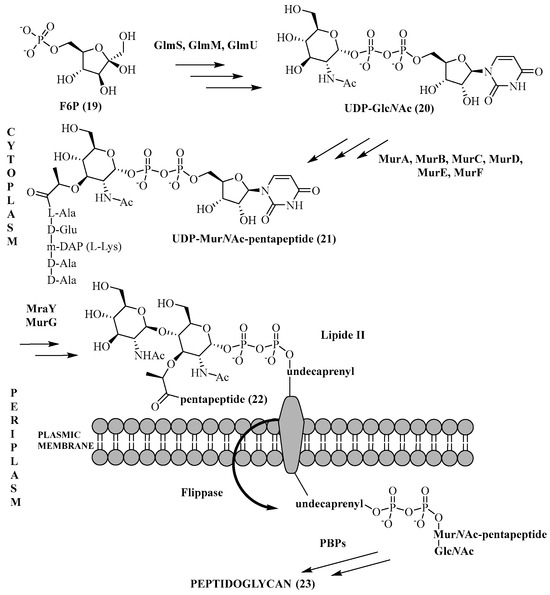
Scheme 2.
Key steps in peptidoglycan biosynthesis.
Mur ligases are involved at several parts of this process; their role is to catalyze the formation of a peptide bond with the corresponding amino acid on the substrate, which is specific to each of these enzymes. These enzymes have all been identified as non-ribosomal ATP-dependent proteins in the cytoplasm, and MurA and MurB allow the biosynthesis of UDP-MurNAc (25) from UDP-GlcNAc (28) (Figure 5).
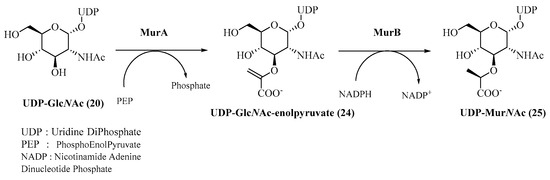
Figure 5.
MurA- and MurB-catalyzed formation of UDP-MurNAc.
MurA or UDP-GlcNAc enolpyruvyltransferase catalyzes the transfer of an enolpyruvate moiety from phosphoenol pyruvate (PEP) to UDP-GlcNAc (20) with phosphate release. The crystallographic structure [65] of MurA as well as its mechanism [66] have been well identified (Scheme 3). Its mode of action involves an addition–elimination mechanism: the anti-addition of PEP on UDP-GlcNAc (20) is catalyzed by Asp305 (numbering of MurA from Escherichia coli) and Cys115 of MurA to form the corresponding tetrahedral intermediate, and then the syn-elimination allows UDP-GlcNAc enolpyruvate (24) to be obtained.
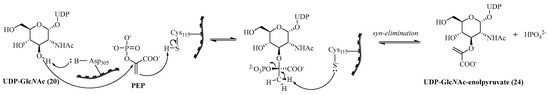
Scheme 3.
Mechanism of MurA−catalyzed UDP-MurNAc enolpyruvate (adapted from Ref. [47]).
Currently, the only antibiotic used that specifically targets MurA is fosfomycin (26) (Figure 6). It is a PEP analog which binds irreversibly to Cys115 (numbering of MurA from Escherichia coli) of the active site of MurA, rendering the enzyme inactive and causing cell death. However, there are many mechanisms of resistance to this compound: mutations of the enzyme, cellular permeability, or an enzymatic action that inactivates the antibiotic. Similarly, innate resistance of certain species such as Mycobacterium tuberculosis or Chlamydia trachomatis exist where an Asp [67] replaces the targeted Cys residue. Numerous covalent and noncovalent inhibitors have been developed [68]. Avenaciolide compounds [69] (isolated from Neosartorya fischeri) are tetrahedral intermediate inhibitors, while quinazolinone analogs [70] are competitive inhibitors. Compound 27 is promising because it shows good biological activities with an MIC of 16 μg/mL−1 and 32 μg/mL−1 on methicillin-resistant Staphylococcus aureus (MRSA) and Bacillus subtilis (B. subtilis), respectively. Furthermore, it was shown to be specific for MurA enzymes with an IC50 of 2.8 μM on MurA from Escherichia coli (E. coli) and 7.9 μM on resistant MurA. Compound 28 is active on Escherichia coli and Staphylococcus aureus, respectively, with an MIC of 1 μg/mL−1 and 8 μg/mL−1 while showing an IC50 on MurA of 47 μM.
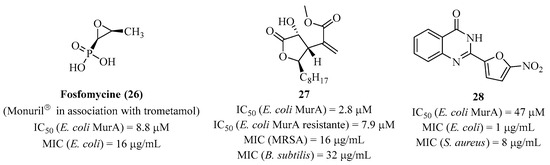
Figure 6.
Structure of fosfomycin (26) and compounds 27 and 28.
MurB or UDP-GlcNAc enolpyruvate reductase catalyzes the stereoselective reduction of UDP-GlcNAc enolpyruvate (24) to UDP-MurNAc (25) with the action of a co-enzyme called Nicotinamide Adenine Dinucleotide Phosphate (NADPH) [71]. This enzyme also contains an active site with the Flavin Adenine Dinucleotide cofactor (FAD). The crystallographic structure [72,73] of MurB and its mechanism [74,75] have been elucidated (Scheme 4). The first step of the mechanism involves the formation of the FAD-MurB complex, which acts as a redox intermediate. Similarly, after the formation of the second NADPH-MurB complex, the reduction of FAD by NADPH leads to the reaction intermediate FADH2 -MurB by transferring the H4 (pro-S) from NADPH to the N of FAD. After the release of NADP+, UDP-GlcNAc enolpyruvate (24) binds to the enzyme. The second step is the reduction of the latter by the FADH2 -MurB thanks to the transfer of hydrogen in C-3 on the enolpyruvate part. After the release of FAD, the enolate intermediate obtained is stabilized by the carboxylic acid of the substrate with the enzyme. Finally, the isomerization of the substrate to the final product UDP-MurNAc (25) requires the presence of water.
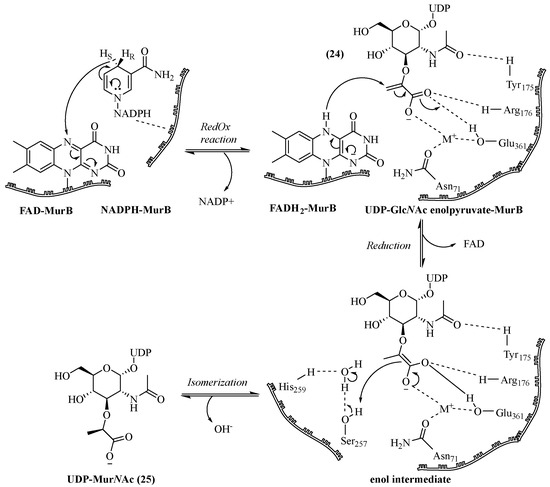
Scheme 4.
MurB-catalyzed mechanism of UDP-MurNAc formation (adapted from ref. [72]).
Currently, there is no antibiotic used for the MurB of bacteria. However, few inhibitors have been developed (Figure 7) such as the imidazolinone 29, one of the first inhibitors targeting the MurB substrate site from Escherichia coli. It shows activity against Staphylococcus aureus strains [76]. Compounds 30 and 31 have been identified as multi-inhibitors of MurA/MurB targeting FAD, with interesting activities on Staphylococcus aureus strains [77,78].
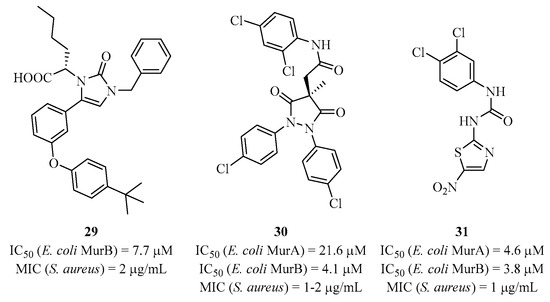
Figure 7.
Structure of some MurB inhibitors.
Finally, the last cytoplasmic steps allowing the formation of UDP-MurNAc pentapeptide from the UDP-MurNAc (25) involve MurC-F, which are grouped into a family of four amino acid ligase enzymes that form a peptide bond in the presence of the corresponding amino acid [79,80]. Other enzymes at the plasma membrane are responsible for the continuation of this synthesis (Scheme 5). Among them, MurG, a glycosyltransferase, catalyzes the glycosylation between Lipid I and a GlcNAc unit from UDP-GlcNAc (20) to form Lipid II. This essential enzyme is highly conserved in all bacterial species. However, glycosyltransferases are present in a large majority of cells in both prokaryotes and eukaryotes, which requires a high specificity of potential MurG inhibitors in drug design [81].
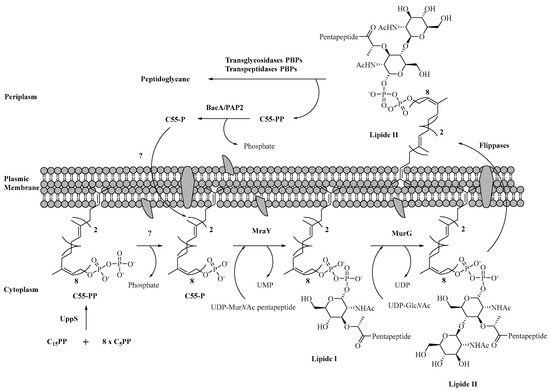
Scheme 5.
Enzymatic synthesis of Lipid II and its translocation.
The crystallographic structure of MurG has been solved, as well as its mechanism, by Walker et al. [82,83,84]. Other teams have shown that the structure of MurG is able to interact with other proteins such as MraY [85], and with the MurE-MurF dimer complex [86], which allows the substrate to evolve with small distances within this protein–protein complex. Several inhibitors targeting MurG have been developed as the pentacyclic compound (32) (Figure 8) [87]. In addition, the use of high-throughput screening has allowed the development of new inhibitors against the MurG, such as the pyrimidinetrione (33) [88,89]; more recently, the diazepanone analog (34) was found to inhibit both MraY and MurG [90].
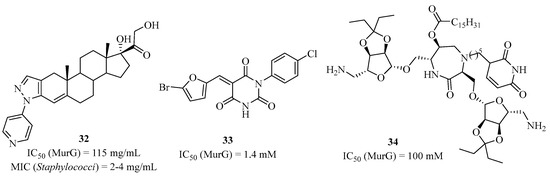
Figure 8.
Selected MurG inhibitors.
3. MurC-F Ligases as New Antibiotic Targets
In the fight against bacterial resistance, another effective way is to go after seldom exploited targets that do not show any resistance mechanism. In this context, enzymes involved in bacterial wall biosynthesis meet this criterion. Among some of these enzymes, resistances have already appeared, such as in the periplasm where penicillin that targets PBPs has become ineffective with the appearance of β-lactamases or with glycopeptides that target Lipid II and has become less active. By contrast, Mur ligases have an activity further upstream in the biosynthesis. These enzymes have many advantages that make them an innovative therapeutic target: they are essential for bacterial survival, ubiquitous within prokaryotes with no equivalent in eukaryotes. To date, they are relevant targets in the development of multi-inhibitors and they do not present any resistance mechanism [91].
MurC-F ligases are responsible for the formation of the Lipid I precursor (UDP-MurNAc-pentapeptide, 38) from UDP-MurNAc (25) by the successive synthesis of tri-, tetra-, and penta-peptides by MurC, MurD, MurE, and MurF (Scheme 6) [92]. The characterization of the protein sequences of MurC-F enzymes has been performed on different bacterial strains, both Gram-positive and Gram-negative [93]. Even if sequential differences between species can be observed, each of them shares the same structural topology, i.e., a protein divided into three different domains with a conserved active site [94]. The main differences in the protein sequences of the Mur ligases are that the amino acid binding site differs depending on which one is used. The MurC enzyme catalyzes the addition of the first amino acid of the peptide chain bound to the UDP-MurNAc (25). In most species, this amino acid is L-Ala, but in very rare cases, other amino acids such as glycine or L-Ser are used. MurD catalyzes the addition of the second amino acid to the substrate. Except for some variations in amino acids due to modifications in biosynthesis, the amino acid is a D-Glu in all species. Studies have shown the importance of having the D-enantiomer of glutamic acid because L-Glu is not a substrate for MurD. For MurE, which catalyzes the third amino acid addition, the amino acid is either a meso-diaminopimelic acid (m-A2pm) in most Gram-negative bacteria and in Bacilli species, while in Gram-positive bacteria, the amino acid is an L-Lys. Studies have shown that MurE contains a very specific active site for its own amino acid; if the wrong amino acid were to be incorporated, it would result in cell lysis. Finally, MurF catalyzes the addition of an unnatural dipeptide D-Ala-D-Ala. In some resistant bacterial species, a modification of this dipeptide by D-Ala-D-Ser or by D-Ala-D-Lac can be observed. To date, each of these enzymes has been purified and studied with the corresponding substrates and co-substrates [95]. This allowed the authors to propose the active sites of the Mur ligases as well as the reaction mechanism where we present these parameters.
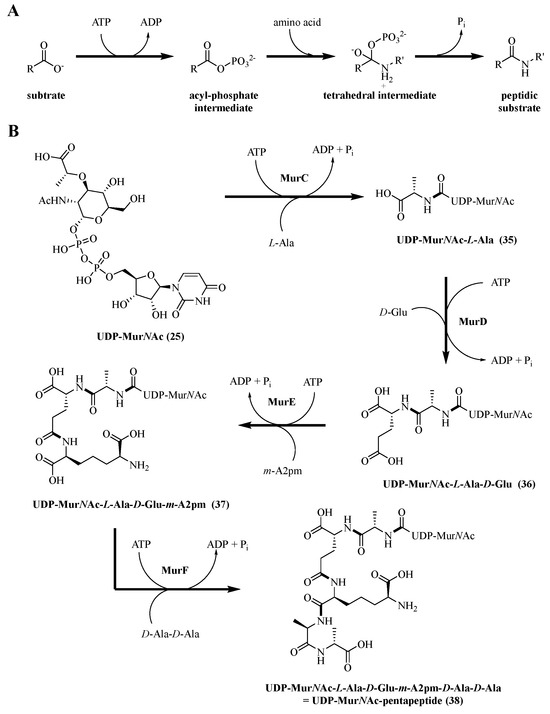
Scheme 6.
(A) General mechanism; (B) general scheme of UDP-MurNAc-pentapeptide formation.
The stability parameters of Mur ligases are important to determine the specificity and kinetic parameters. A characterization study of these parameters was performed on MurC-F enzymes from Mycobacterium tuberculosis [96]. The specific activities of MurC-F are calculated by measuring the concentration of the released inorganic phosphate (Scheme 6B). They are estimated to be 1.2, 0.8, 1.3, and 0.9 μmol/min/mg/protein (μmol of phosphate formed per minute per milligram of enzyme) for MurC, MurD, MurE, and MurF, respectively. The stability of each of the proteins showed a great sensitivity according to the temperature and the pH of the medium. Indeed, the enzymatic activity remains stable under 40 °C with an optimal activity between 35 °C and 40 °C, while a higher temperature (between 45 and 70 °C) shows a significant decrease in the activity. Moreover, enzymes are sensitive to the pH of the buffered medium since the optimal pH is estimated between pH 8 and 8.5. Small variations with a more acidic or basic pH are enough to strongly decrease the activity. Mur ligases use the energy of the phosphate bond to catalyze the formation of the amino acid amide bond on the growing peptide chain (Scheme 6). This mechanism, common to all four Mur ligases, starts with the elaboration of a complex with the enzyme and its substrates in the following way: first, the ATP binds to the enzyme, and then the substrate UDP sugar is added, as well as the amino acid at the end. In the active site, the first step is to activate the carboxylic acid of the substrate UDP sugar with ATP (Scheme 6A) to form the acyl phosphate intermediate. Two Mg ions2+ will create a bridge between the negatively charged groups of ATP and the substrate UDP sugar to facilitate the phosphorylation of the carboxylate group of the substrate. Then, the second step involves the amino acid, which displaces the phosphate by a nucleophilic attack with the formation of the tetrahedral intermediate. Finally, the release of the phosphate Pi allows for obtaining the peptide substrate. The catalytic activity of a base is necessary to recover the proton from the amine group. Other studies have shown that the ATP phosphate could replace this base or the enzyme itself since this would better fit the normal energy scale of the reaction [97]. With MurC-F, therefore, L-Ala, D-Glu, m-A2pm, and the dipeptide D-Ala-D-Ala can be introduced successively in the presence of ATP (Scheme 6B).
In addition to the mechanistic data of these enzymes, X-ray structures of MurC-F ligases for several different species have been elucidated [98,99,100,101].
Inhibitors of MurC-F Escherichia coli were developed since they have the advantage of containing multiple active sites per enzyme, allowing researchers to target specific areas of interest to design molecules that may have biological activity benefits. Moreover, the increasing emergence of bacterial resistance challenges the conventional approach to developing antibacterials. Mur ligases are ideal targets for this purpose since they share similar three-dimensional structures, making them a set of targets for a single compound.
3.1. Inhibitors from “Medicinal Chemistry Approach”
This follows a simple mono-therapeutic model aimed at designing a drug for a specific target in a particular region or event. This approach has proven effective in the development of numerous antibiotics. The identification of potential hits (molecules of interest) involves in silico, in vitro, and in vivo studies [102]. The first characterized inhibitors targeted domain 3 of Mur ligases and emerged in the 1990s, along with the initial elucidation of crystallographic structures.
3.1.1. Amino Acid Mimics of Mur Ligases
The exploration of MurC inhibition using analogs of L-Alanine began early, even before the complete structure of the enzyme was known. Takahashi et al. showed that glycine could inhibit the addition of L-Alanine to the UDP-MurNAc substrate as a competitive inhibitor [103]. Several studies a few years later, including those by Parquet [104] et al. and VillaFranca et al. [105], demonstrated that various L-amino acids, without incorporation into the natural substrate, could inhibit MurC. D-configurations of amino acids, however, showed no inhibitory activity. For example, the three following amino acid analogs, 39, 40, and 41 (Figure 9), were the most potent competitive inhibitors, ranging from millimolar to minimum activity for 76.
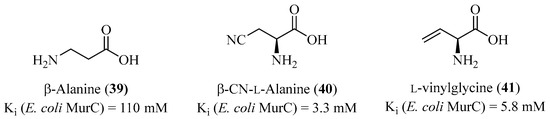
Figure 9.
Structure of L-Alanine analogs.
Inhibitors of MurD using analogs of D- or L-glutamic acid showed similar results as those for MurC. Van Heijenoort et al. demonstrated weakly active analogs, as seen with compounds 42 and 43 with residual activities of 58% and 47%, respectively, at a concentration of 10 mM (Figure 10) [106].
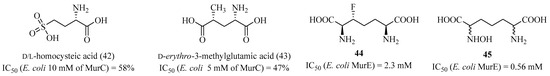
Figure 10.
Structure of D-glutamic acid analogs and of meso-diaminopimelic acid analogs.
This same team conducted similar studies on analogs of meso-diaminopimelic acid on the MurE enzyme [107]. Among synthetized products, compounds 44 and 45 were the best analogs in terms of MurE inhibition, although their activity was weak, with IC50 values of 2.3 mM and 0.56 mM for 44 and 45, respectively. For MurF, the literature does not show results containing derivatives with motifs from the D-Ala-D-Ala dipeptide.
3.1.2. Heterocyclic Inhibitors
In 2001, Gobec et al. based their work on this mechanism with MurD and the tetrahedral transition state to design phosphino-alanine derivatives, but without significant inhibition [108]. In 2007, the same group published small compounds, such as sulfonamides of D-glutamic acid. They co-crystallized inhibitors like compound 46 (Figure 11) with the MurD enzyme to better understand its binding site [109]. This allowed the group to develop other analogs with improved activity and the ability to crystallize the compounds, such as compound 47 with an IC50 of 85 μM [110]. The modification of position 6 of the naphthalene in 46 allowed the crystallographic structure of the inhibitor/MurD complex to show that this position enables interactions between the substituents and the di-phosphate binding site of domain 1. Subsequently, Gobec et al., in collaboration with Mašič’s group, performed ligand-docking studies, leading to the identification of a new interesting structure that could bind to the di-phosphate binding site. These structures 48–50 contained rhodanine-type heterocycles, such as compounds, which was active with an IC50 of 45 μM (for 48).
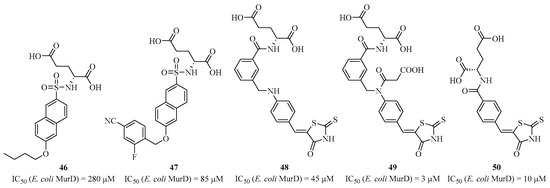
Figure 11.
Structure of sulfonamide and heterocyclic analogs of D-glutamic acid.
Subsequent SAR studies resulted in the discovery of analogs with improved biological activities, such as compounds 49 and 50 with IC50 values in the micromolar range [111,112,113]. These recent studies enabled the synthesis of a large library of hybrid molecules capable of binding both to the active site of the amino acid in domain 3 and to the di-phosphate pocket in domain 1, using in silico analysis and considering that the D-glutamic acid part must remain. However, no molecule with significant in vivo bacterial activity has been identified. Additionally, some synthesized compounds, such as those containing the “ene-rhodanines” heterocycle, may be considered as PAINS (pan-assay interference compounds) that produce false positives in the biological experimentation used to determine IC50 [114]. MurC presents a unique case where Lee et al. synthesized derivatives of benzylidene rhodanines [115]. Gobec et al. attempted to develop sulfonamide inhibitors of D-glutamic acid targeting MurE [116]. Subsequently, they synthesized a second generation of non-D-glutamic acid sulfonamide inhibitors that mimic it with rigid diacids. These inhibitors were found to be active on MurD, not MurE [117]. Compounds 51 and 52 (Figure 12), exhibiting IC50 values of 182 μM and 8.4 μM, respectively, were co-crystallized with the enzyme, and the structures showed that these inhibitors bind at the D-glutamic acid position, where the 2-cyano-4-fluoro-phenyl occupies the uracil pocket.
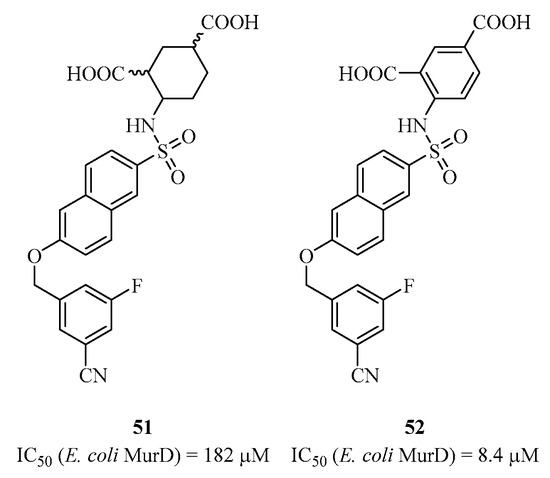
Figure 12.
Structure of analogs 51 and 52 targeting MurD.
3.2. Inhibitors Mimicking the Natural Substrate UDP-MurNAc-(Peptide)
3.2.1. Phosphinic Inhibitors
One of the first families of inhibitors is the “phosphinic” compounds that target the natural substrate-binding site in domain 1 of Mur ligases. The phosphinic acid in these molecules provides a tetrahedral geometry that blocks the enzymatic mechanism, and the uridine part of the compounds allows them to position themselves in the active site [118,119]. For MurC, AstraZeneca published the synthesis of two highly active inhibitors, 53 and 54, with respective IC50 values of 49 nM and 60 μM (Figure 13) [120]. Compound 53 forms a stable enzyme/ATP/inhibitor and enzyme/ADP/inhibitor complex, which stops the enzymatic activity [121]. The first phosphinic inhibitor 55 was synthesized for MurD in 1996 by Blanot et al., which was active with an IC50 in the micromolar range [122]. Similarly, Merck published other inhibitors in 1998 with similar structures. They showed that adding the glucosamine part to compound 56 improved its activity, reaching IC50 values in the nanomolar range [123]. Later, Blanot et al., in collaboration with Gobec’s team, published simplified inhibitors by retaining only the peptide-phosphinic part, based on compound 57, with an IC50 of 20 nM [124]. Even without improving this inhibitory activity, they still published active compounds like compound 58, which was the most active with an IC50 of 78 μM.
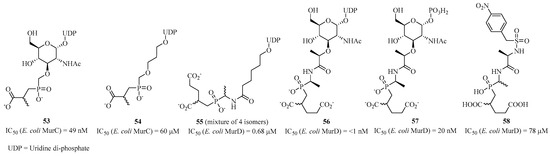
Figure 13.
Structure of phosphinic analogs of MurC and MurD.
For MurE, Merck’s group synthesized compounds 59 and 60 with respective IC50 values of 1.1 μM and 700 μM (Figure 14), similar to compound 56 for MurC [125]. Similarly, Gobec et al. used 60 as a starting point to add various substituents, but without significant inhibition, with compound 61 showing the best residual activity at 71% at 1 mM [126]. Docking studies of an analog of 61 showed that this family of molecules is not transition-state inhibitors but rather competitive inhibitors of the substrate.
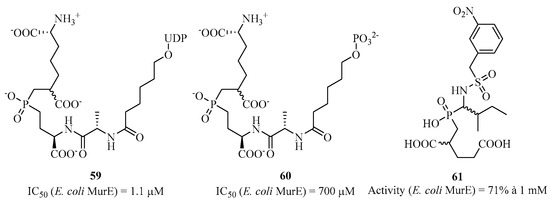
Figure 14.
Structure of phosphinic analogs of MurE.
Besides the good enzymatic activities of these phosphinic analogs, none have shown antibacterial activity, which could be explained by their low cell wall penetration. However, an important point to consider in the design of these derivatives is that the uridine part is beneficial for retaining good IC50 values for each of the Mur ligases. Moreover, the total synthesis of phosphinic compounds proves to be challenging in terms of the number of steps. Several research groups have explored the laboratory synthesis of various natural substrates of Mur ligases, which has served as a synthetic tool, knowing that the nucleosidic synthesis of di-phosphate derivatives is a true challenge [127,128,129,130,131].
3.2.2. Peptidic Inhibitors
In 2003, Fishwick et al. published small peptides as analogs of the UDP sugar substrate targeting MurD [132]. This peptidic macrocycle 62 was designed based on the enzyme’s crystallographic structure and the docking of this potential inhibitor in the active site. Several analogs were synthesized, with compound 63 showing the best activity with an IC50 of 0.7 μM (5.1 μM for 60) (Figure 15). There are also a few examples where authors developed peptidic derivatives targeting MurE [133,134].
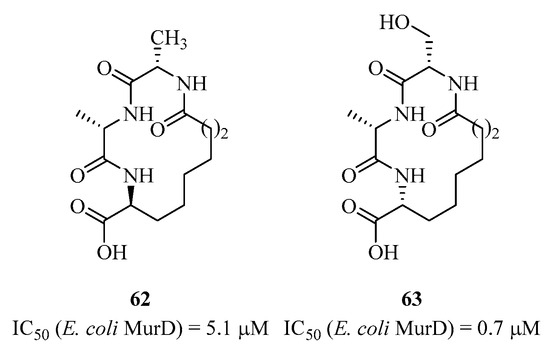
Figure 15.
Structure of peptides 62 and 63 targeting MurD.
On the other hand, several groups of biologists have been interested in peptide synthesis using the phage display technique with different peptide libraries. Several studies have identified peptides targeting MurC [135], MurD [136], MurE [137,138], and MurF [139], inhibiting with weak IC50 values in the mM range.
3.2.3. Heterocyclic Inhibitors
A very rapid technique to obtain inhibitors is to screen molecules through biological assays. AstraZeneca did this by screening its compound library targeting MurC [140]. Compound 64 was identified as an inhibitor with an IC50 of 2.3 μM (Figure 16). Its action seems to target the di-phosphate binding site, but the mode of inhibition has not been clearly elucidated. Moreover, it shows binding affinities to other proteins, such as bovine serum albumin. For MurD, Obreza’s team synthesized sulfono-hydrazine derivatives that also mimic the UDP sugar substrate’s di-phosphates [141]. Compound 65 showed the best IC50 of 30 μM but did not exhibit antibacterial activity. Additionally, the same team developed other similar analogs, like compound 66, but without improving its activity [142]. For the same enzyme, Gobec’s team performed virtual screening of a molecule library, which led to the identification of inhibitors [143].
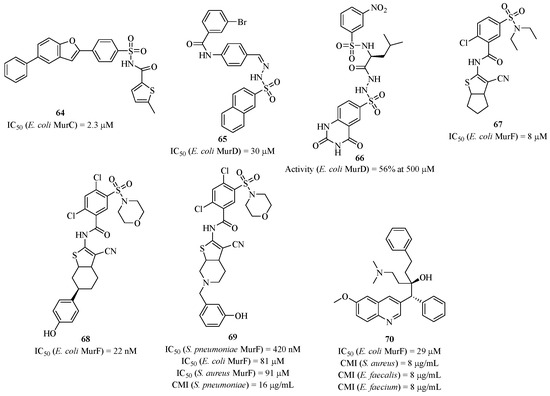
Figure 16.
Structure of compound 64 targeting MurC, compounds 65 and 66 targeting MurD, and compounds 67–70 targeting MurF.
A research group from Abbott Laboratories conducted a high-throughput affinity screening of a molecule library targeting MurF, which highlighted molecule 67 as an attractive candidate [144,145]. Enzymatic studies revealed an IC50 of 8 μM, and through SAR studies, this result was optimized with compound 68, which had an IC50 of 22 nM. Compound 67 was co-crystallized with MurF and bound to the active site in place of the natural UDP sugar substrate in a conformation called “closed” of the enzyme [146,147]. However, these compounds did not exhibit antibacterial activity, even in the presence of permeable strains, which may indicate non-specific interactions with other proteins. Based on these results, Gobec’s team decided to take up the structural motif of compound 68 to improve its antibacterial activities [148]. This led to compound 69, which was active against Staphylococcus pneumoniae strains with an MIC of 16 μg/mL. However, 69 and its analogs did not exhibit activity against other bacterial strains. The authors published a second generation of these compounds active against several strains [149], where their antibacterial action resulted from membrane degradation. Other heterocyclic analogs were synthesized by Gobec’s laboratory with a few antibacterial activities [150,151]. In 2006, a research group from Johnson & Johnson developed pyrimidine derivatives that inhibited MurF but showed no in vivo activity [152]. Only from 2007 did the same group identify, through molecule screening, hydroxyquinoline-type derivatives of interest, such as compound 70 with an IC50 of 29 μM. Although this compound exhibited activity against both Gram-positive and Gram-negative bacterial strains, it is possible that compound 70 interacted with other proteins other than MurF [153,154].
3.3. Inhibitors Mimicking the Co-Substrate ATP
There are very few inhibitors of this type in the literature targeting Mur ligases. This can be explained by the fact that there are many ATP-dependent enzymes, including kinases, resulting in a very severe lack of selectivity. Therefore, the few synthesized inhibitors were obtained through screening techniques of molecule libraries. In 2008, Dougherty’s team from the pharmaceutical group Pfizer carried out a screening of its molecule library and managed to demonstrate that compound 71 is a competitive inhibitor of ATP, selectively targeting MurC from only a few enterobacterial strains closely related to Escherichia coli (Figure 17). However, this compound did not show antibacterial activity [155].
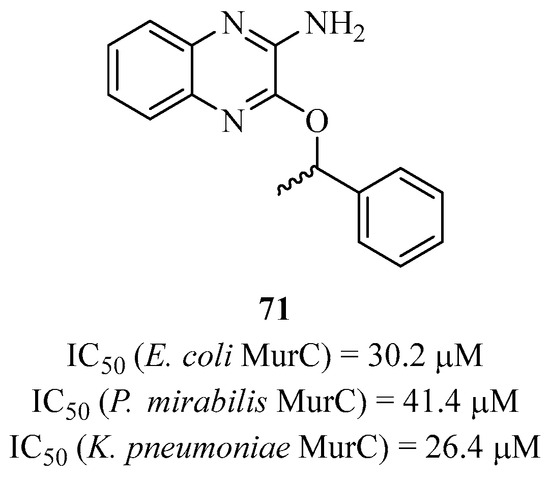
Figure 17.
Structure of compound 71 targeting MurC.
3.4. Natural Inhibitors
Using the same molecule screening technique (in this case, from plants), a research group identified natural compounds like 72 and 73 that were found to be active against MurE (Figure 18) [156,157,158]. Compound 72 showed an IC50 of 67 μM against MurE from M. tuberculosis and weakly inhibited the growth of a mutant strain of Mycobacterium tuberculosis. As for compound 73, it had an IC50 of 75 μM against MurE and was active against a panel of Gram-positive and Gram-negative strains. It also acted on efflux by inhibiting a protein responsible for NorA.
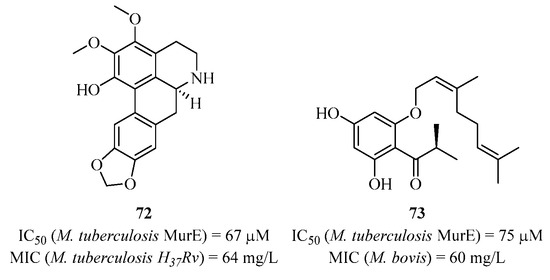
Figure 18.
Structure of natural compounds 72 and 73 targeting MurE.
3.5. Multi-Target Synthetic Approaches
The synthesis of multi-inhibitor compounds involves a “multi-therapeutic” model, where a molecule is produced to inhibit multiple biological processes by targeting several enzymes simultaneously. Generally, the choice is made on similar enzymes with close activities, as demonstrated with the Mur ligases. The reason behind this concept is to combat bacterial resistance to the purported new antibacterial compound built on these criteria. The advantage of multi-inhibition is that it leaves no chance for bacteria to react or adapt when exposed to this toxic agent. In the literature, there are a few relatively recent examples of multi-targeted Mur ligases that we present. This principle poses a challenge in developing such compounds while retaining attractive biological properties.
3.5.1. Mimicking the Amino Acid of Mur Ligases
Solmajer et al. and Gobec et al. developed new structures derived from D-glutamic acid using several in silico approaches, as they had done for classical inhibitors. Among these previous studies, they also identified multi-active compounds, and SAR studies were carried out based on these results [159]. The 1,3-phenyl-dicarboxylic acid, which is the cyclic mimic of D-glutamic acid, seems to play an important role in the binding to each of the Mur ligases. Initially, the team developed analogs of this type targeting MurD and MurE, as shown by compound 74 with an IC50 of 270 μM on MurD and an IC50 of 32 μM on MurE [160]. Subsequently, the group developed other analogs by modifying the heterocyclic part that fits into the di-phosphate pocket while retaining the 1,3-phenyl-dicarboxylic acid [161]. They synthesized compound 75, which is the best in the series and capable of inhibiting all four Mur ligases with IC50 values of 41 μM, 60 μM, 93 μM, and 89 μM for MurC, MurD, MurE, and MurF, respectively (Figure 19). Although these molecules are multi-active, with compound 75 being one of the best multi-inhibitors synthesized to date, there is no revealed antibacterial activity. The group attempted to measure activities on different strains with analogs close to 75 (without the di-carboxylic acid groups), but the results only showed weak activities.
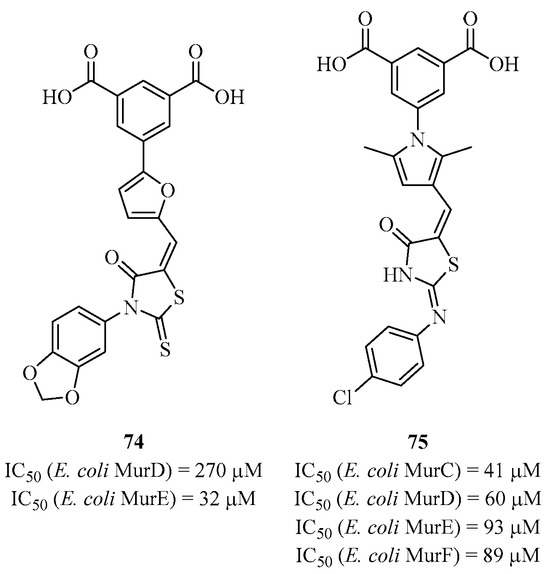
Figure 19.
Structure of multi-inhibitors 74 and 75 targeting MurC-F.
3.5.2. Mimicking the Natural Substrate UDP-MurNAc(-Peptide)
Phosphonic Acid Inhibitors
Gobec et al. developed a series of phosphoric acid analogs of hydroxyethylamines as a bioisostere of the tetrahedral intermediate of the UDP sugar substrate [162]. Compound 76 is the best analog in this series, active against all four enzymes with IC50 values of 26 μM, 530 μM, 160 μM, and 150 μM for MurC, MurD, MurE, and MurF, respectively (Figure 20). However, these compounds do not show antibacterial activities. These analogs, including 76, are the only phosphorylated derivatives that are multi-active against Mur ligases.
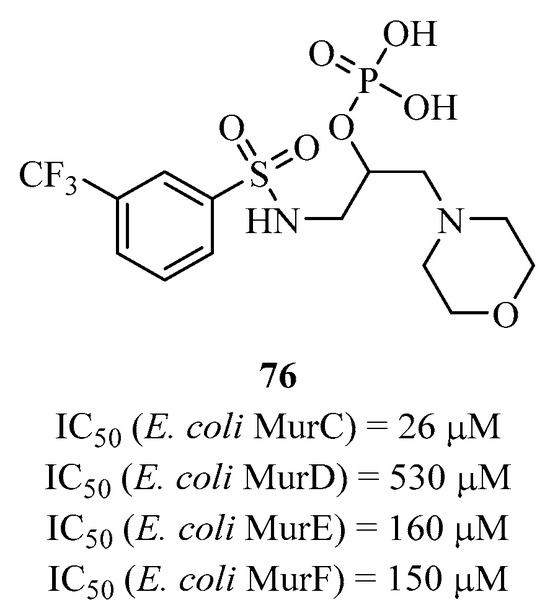
Figure 20.
Structure of multi-inhibitor 76 targeting MurC-F.
Heterocyclic Inhibitors
After observing that the N-acyl hydrazone structure has potential in terms of biological activity in medicinal chemistry, Gobec’s group designed analogs around this structure and showed that some are active, like compound 77 with IC50 values of 123 μM and 230 μM for MurC and MurD, respectively (Figure 21) [163]. Moreover, this compound showed weak antibacterial activities against Escherichia coli and Staphylococcus aureus. The group also designed other heterocyclic structures to adopt a more closed conformation than with the natural substrates of enzymes MurD and MurF but still show activities on different strains.
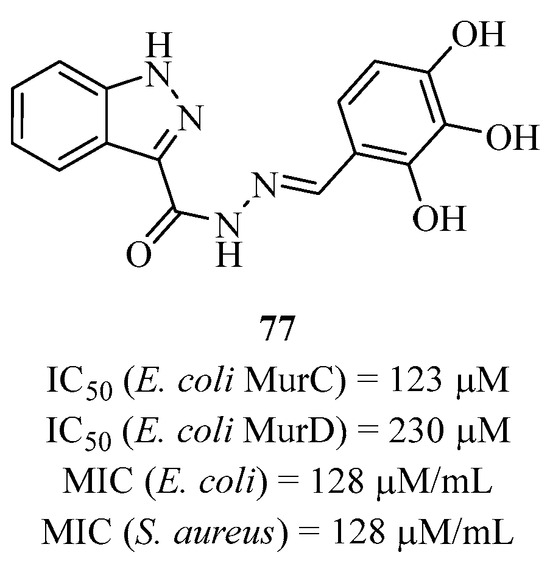
Figure 21.
Structure of multi-inhibitor 77 targeting MurC and D.
Gobec et al., who synthesized a good number of benzylidenesulfonyl hydrazine analogs, showed that some of them were multi-inhibitors [164]. They performed SAR studies to optimize enzymatic activities but without antibacterial activities. Singh’s laboratory identified several multi-active structures of Mur ligases through different screenings, including pulvinones [165] and phenyl dihydrothienopyrazolol [166] with activities against multiple strains. Recently, they identified a naphthyl-type tetronic acid structure capable of inhibiting not only all four Mur ligases but also MurA and MurB [167]. Compound 78 showed the best activities in the series with an IC50 in the range of 20 μM for each enzyme, and it inhibited the bacterial growth of Escherichia coli and Staphylococcus aureus strains (Figure 22).
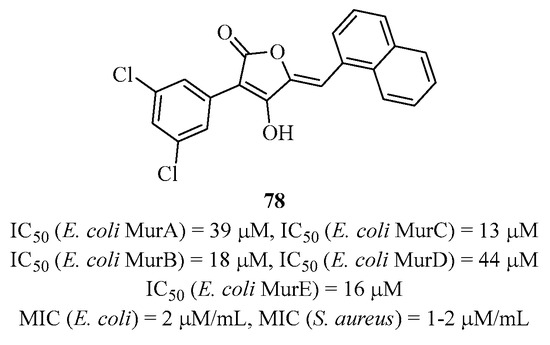
Figure 22.
Structure of multi-inhibitor 78 targeting MurA-F.
3.5.3. Mimicking the Co-Substrate ATP
Very recently, Zega et al. identified several compounds with different structures that exhibit multi-inhibitory activities against MurC to MurF. These compounds were identified through screening a collection of molecules originally designed to inhibit kinases by the competitive inhibition of ATP [168]. Compounds 79 and 80 are multi-inhibitors of MurC-F with IC50 values ranging from 10 to 368 μM (Figure 23). Because these compounds mimic ATP, the authors conducted kinetic and NMR studies on compound 79 and showed that it actually binds to the amino acid binding site. They also performed additional biological analyses with eukaryotic kinases and demonstrated that these molecules are specific and promising for the development of new antibacterials.
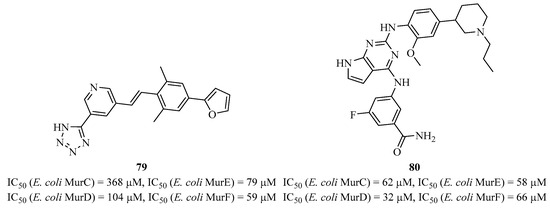
Figure 23.
Structure of multi-inhibitor compounds 79 and 80 targeting MurC-F.
3.5.4. Natural Analogs
Süssmuth et al. identified feglymycin (81), isolated from Streptomyces strains, as an active inhibitor of both MurA and MurC [169]. Feglymycin, a 13-mer peptide, exhibited activity with an IC50 in the range of μM for MurA and in the range of mM for MurC (Figure 24).
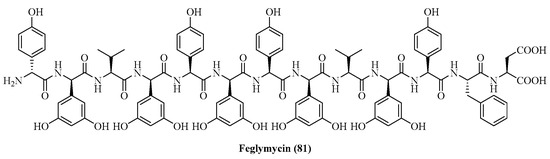
Figure 24.
Structure of feglymycin (81) targeting MurA and C.
4. Conclusions
Over the last few decades, the peptidoglycan biosynthetic pathway has emerged as a promising and attractive target for antibacterial drug discovery. Among the various enzymes involved, the Mur ligase family has drawn attention due to its exclusive presence in bacteria (not found in human cells) and as they are essential for bacterial cell wall biosynthesis. An in-depth understanding of their structures has led to the development of powerful new antibacterial agents. While each Mur ligase can be considered a unique antibacterial target, MurC-F ligases have highly conserved amino acid regions in their active sites. This characteristic can be used in the design of promising Mur ligase multi-inhibitors. A number of inhibitors from different chemical families were developed against Mur ligases, with significant inhibitory activity. Modern techniques remain highly utilized in the design of potentially active molecules, particularly with virtual screening methods [170,171]. However, none has yet been shown to have antibacterial activity. One reason could be the difficulty for these compounds to cross the bacterial membrane and reach the cytoplasm where Mur ligases activities are located. Another possibility explaining the lack of antibacterial activity may be the complexity of the Mur ligase pathway, which is high, making it difficult for the inhibitor to reach the various sites.
In the future, a better understanding of the protein–protein interactions of the Mur ligase pathway, combined with the consideration of factors enabling better penetration of the bacterial wall, will enable the design of Mur ligase inhibitors with proven antibacterial activity.
Funding
This research was funded by EUROFERI program (Region Centre-Val de Loire/FEDER EX010351). The general functioning of ICOA comes from CHemBio (FEDER-FSE 2014-2020-EX003677), Techsab (FEDER-FSE 2014-2020-EX011313), RTR Motivhealth (2019-00131403), and Labex programs SYNORG (ANR-11-LABX-0029) and IRON (ANR-11-LABX-0018-01).
Acknowledgments
Luigi A Agrofoglio is grateful to the members of the consortium of the EUROFERI program for sharing points of view concerning “one-health” and antibiotics. V.H. thanks SYNORG for financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Porter, J.R. Antony Van Leeuwenhoek: Tercentenary of his discovery of bacteria. Microbiol. Rev. 1976, 40, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Mazzarello, P. Bassi, Agostino; John Wiley & Sons, Ltd.: Chischester, UK, 2013. [Google Scholar]
- Pacini, F. Osservazioni microscopiche e deduzioni patologiche sul cholera asiatico. In Memoria del Dott. Filippo Pacini: Lettra alla Societa Medico-Fisica di Firenze Nella Seduta Tip; Tipografia di Federigo Bencini; Medical-Physical Society of Florence: Florence, Italy, 1854; pp. 397–405. [Google Scholar]
- Jean, T. Casimir Davaine (1812–1882) and the therapeutics of anthrax and livestock septicemia. Rev. D’histoire Pharm. 1973, 61, 334–339. [Google Scholar]
- Koch, R. Berliner Klinische Wochen-Schrift. 1882, pp. 428–445. Available online: https://babel.hathitrust.org/cgi/pt?id=mdp.39015020075001&seq=7 (accessed on 8 November 2023).
- Pasteur, L. Oeuvres de Pasteur; Masson: Paris, France, 1922; Volume 2. [Google Scholar]
- Lloyd, N.C.; Morgan, H.W.; Nicholson, B.K.; Rominus, R.S. The composition of Ehrlich’s Salvarsan: Resolution of a century-old debate. Angew. Chem. Int. Ed. 2005, 44, 941–944. [Google Scholar] [CrossRef]
- Ligon, B.L. Penicillin: Its discovery and early development. Sem. Pediatr. Infect. Dis. 2004, 15, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, P. L’aventure des Sulfamides: Daniel Bovet, une chimie qui guérit. History of the discovery of sulfonamides. Rev. D’histoire Pharm. 1990, 78, 227–228. [Google Scholar]
- McDermott, W.; Rogers, D.E. Social ramifications of control of microbial disease. Johns Hopkins Med. J. 1982, 151, 302–312. [Google Scholar] [PubMed]
- Walsh, C.T.; Wright, G. Introduction: Antibiotic Resistance. Chem. Rev. 2005, 105, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic Resistance Crisis Part 1: Causes and threats. Pharm. Therapeut. 2015, 40, 277–283. [Google Scholar]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance; Global Report on Surveillance; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- WHO. Ranking of medically important antimicrobials of risks management of antimicrobial resistance due to non-human use. In Critically Important Antimicrobials for Human Medicine 5th Revision; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- FAO. The FAO Action Plan on Antimicrobial Resistance 2021–2025; FAO: Rome, Italy, 2021. [Google Scholar]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hasen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A review on antibiotic resistance: Alarm Bells are ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Shi, Q.-S.; Huang, X.-M.; Xie, X.-B. The three bacterial lines of defense against antimicrobial agents. Int. J. Mol. Sci. 2015, 16, 21711–21733. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Dis. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef]
- Obst, U.; Schwartz, T.; Volkmann, H. Antibiotic resistant pathogenic Bacteria and their resistance genes in bacterialbiofilms. Int. J. Artif. Org. 2006, 29, 387–394. [Google Scholar] [CrossRef]
- Vedithi, S.C.; Malhotra, S.; Das, M.; Daniel, S.; Kishore, N.; George, A.; Arumugam, S.; Rajan, L.; Ebenezer, M.; Asher, D.B.; et al. Structural Implications of Mutations Conferring Rifampin Resistance in Mycobacterium leprae. Sci. Rep. 2018, 8, 5016. [Google Scholar] [CrossRef]
- Frère, J.-M. Beta-lactamases and bacterial resistance to antibiotics. Mol. Microbiol. 1995, 16, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J.M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Nikaido, H. Prevention of drugs access to bacterial targets: Permeability barriers and active efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, D.; Payne, D.J.; Holmes, D.J.; Rosenberg, M. Novel targets for the future development of antibacterial agents. J. Appl. Microbiol. Symp.Supp. 2002, 92, 28S–34S. [Google Scholar] [CrossRef]
- Hards, K.; Cook, G.M. Targeting bacterial energetics to produce new antimicrobials. Drug Resist. Updates 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Kingwell, K. New antibiotic hits Gram-negative bacteria. Nat. Rev. Drug Discov. 2018, 17, 785. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanisms of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Lewis, R.J.; Tsai, F.T.F.; Wigley, D.B. Molecular mechanisms of drug inhibition of DNA gyrase. Bioessays 1996, 18, 661–671. [Google Scholar] [CrossRef]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef]
- Andersson, M.I.; MacGowan, A.P. Development of the quinolones. J. Antimicrob. Chemother. 2003, 51, 1–11. [Google Scholar] [CrossRef]
- Chopra, L. Bacterial RNA polymerase: A promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs 2007, 8, 600–607. [Google Scholar]
- Bertacine Dias, M.V.; Santos, J.C.; Libreros-Zúñiga, G.A.; Ribeiro, J.A.; Chavez-Pacheco, S.M. Folate biosynthesis pathway: Mechanisms and insights into drug design for infectious diseases. Future Med. Chem. 2018, 10, 935–959. [Google Scholar] [CrossRef]
- Bourne, C.R. Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics 2014, 3, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Berminghan, A.; Derrick, J.P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 2002, 24, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Achari, A.A.; Somers, D.O.; Champness, J.N.; Bryant, P.K.; Rosemond, J.; Stammers, D.K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struc. Biol. 1997, 4, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, F. Synthesis of Nucleotide Analogues Targeting the Inhibition of Flavin-Dependent Thymidylate Synthase. Ph.D. Thesis, University of Orleans, Orleans, France, 2018. [Google Scholar]
- Dunkle, J.A.; Xiong, L.; Mankin, A.S.; Cate, J.H.D. Structure of Escherichia coli ribosome with antibiotics bond near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA 2010, 107, 17152–17157. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistanc. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-targeting antibiotics: Modes of action, mechanism of resistance, and implications for drug design. Ann. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 2017, 117, 12415–12474. [Google Scholar] [CrossRef]
- Seiple, I.B.; Zhang, Z.; Jakubec, P.; Langlois-Mercier, A.; Wright, P.M.; Hog, D.T.; Yabu, K.; Rao Allu, S.; Fukuzaki, T.; Carlsen, P.N.; et al. A platform for the discovery of new macrolide antibiotics. Nature 2016, 533, 338–345. [Google Scholar] [CrossRef]
- Veve, M.P.; Wagner, J.L. Lefamulin: Review of a promising novel pleuromutilin antibiotic. Pharmocotherapy 2018, 38, 935–946. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- McKenna, M. Antibiotic resistance: The last resort. Nature 2013, 499, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.F.; Meroueh, S.O.; Mobashery, S. Bacterial resistance to β-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005, 105, 395–424. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.M.; Martin, N.I. The calcium-dependent lipopeptide antibiotics: Structure, mechanism, & medicinal chemistry. Med. Chem. Commun. 2019, 10, 634–646. [Google Scholar]
- Baltz, R.H.; Miao, V.; Wrigley, S.K. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005, 22, 717–741. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.; Hansford, K.A.; Butler, M.S.; Jia, Z.J.; Mark, A.E.; Cooper, M.A. Developments in glycopeptide antibiotics. ACS Infect. Dis. 2018, 4, 715–735. [Google Scholar] [CrossRef]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2011, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Van Heijenoort, J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 2001, 18, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Maitra, A.; Munshi, T.; Healy, J.; Martin, L.T.; Vollmer, W.; Keep, N.H.; Bhakta, S. Cell wall peptidoglycan in Mycobacterium tuberculosis: An Achilles’ heel for the TB-causing pathogen. FEMS Microbiol. Rev. 2019, 43, 548–575. [Google Scholar] [CrossRef]
- Ruiz, N. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 2015, 8, 21–31. [Google Scholar] [CrossRef]
- Skarzynski, T.; Mistry, A.; Wonacott, A.; Hutchinson, S.E.; Kelly, V.A.; Duncan, K. Structure of UDP-N-AcGlc enolpyruvyltransferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-AcGlc and the drug fosfomycin. Structure 1996, 4, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Skarzynski, T.; Kim, D.H.; Lees, W.J.; Walsh, C.T.; Duncan, K. Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analog of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA. Biochemistry 1998, 37, 2572–2577. [Google Scholar] [CrossRef]
- Castañeda-Garcia, A.; Blázquez, J.; Rodríguez-Rojas, A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2013, 2, 217–236. [Google Scholar] [CrossRef]
- Hrast, M.; Sosič, I.; Šink, R.; Gobec, S. Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg. Chem. 2014, 55, 2–15. [Google Scholar] [CrossRef]
- Chang, C.-M.; Chern, J.; Chen, M.-Y.; Huang, K.-F.; Chen, C.-H.; Yang, Y.-L.; Wu, S.-H. Avenaciolides: Potencial MurA targeted inhibitors against peptidoglycan biosynthesis in methicillin-resistant Staphylococcus aureus (MRSA). J. Am. Chem. Soc. 2015, 137, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Hrast, M.; Rožman, K.; Jukič, M.; Patin, D.; Gobec, S.; Sova, M. Synthesis and structure-activity relationship study of novel quinazolinone-based inhibitors of MurA. Bioorg. Med. Chem. Lett. 2017, 27, 3529–3533. [Google Scholar] [CrossRef]
- Farmer, B.T., II; Constantine, K.L.; Goldfarb, V.; Friedrichs, M.S.; Wittekind, M.; Yanchunas, J., Jr.; Robertson, J.G.; Mueller, L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struc. Biol. 1996, 3, 995–997. [Google Scholar] [CrossRef] [PubMed]
- Benson, T.E.; Marquardt, J.L.; Marquardt, A.C.; Etzkorn, F.A.; Walsh, C.T. Overexpression, purification, and mechanism study of UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry 1993, 32, 2024–2030. [Google Scholar] [CrossRef]
- Benson, T.E.; Walsh, C.T.; Hogle, J.M. The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls. Structure 1996, 4, 47–54. [Google Scholar] [CrossRef]
- Dhalla, A.M.; Yanchunas, J., Jr.; Ho, H.T.; Falk, P.J.; Villafranca, J.J.; Robertson, J.G. Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry 1995, 34, 5390–5402. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Kurokawa, K.; Matsuo, M.; Sakamoto, K.; Ueno, K.; Kita, K.; Sekimizu, K. Identification and characterization of amino acid residues essential for the active site of UDP-N-acetyl-enolpyruvylglucosamine reductase (MurB) from S. aureus. J. Biol. Chem. 2006, 281, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Bronson, J.J.; DenBleyker, K.L.; Falk, P.J.; Mate, R.A.; Ho, H.-T.; Pucci, M.J.; Snyder, L.B. Discovery of the first antibacterial small molecule inhibitors of MurB. Bioorg. Med. Chem. Lett. 2003, 13, 873–875. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, A.M.; Failli, A.; Shumsky, J.; Yang, Y.; Severin, A.; Singh, G.; Hu, W.; Keeney, D.; Petersen, P.J.; Katz, A.H. Pyrazolidine-3,5-dione and 5-hydroxy-1H-pyrazol-3-(2H)-ones, inhibitors of UDP-N-acetylenolpyruvyl glucosamine reductase. J. Med. Chem. 2006, 49, 6027–6036. [Google Scholar] [CrossRef]
- Francisco, G.D.; Li, Z.; Donald Albright, J.; Eudy, N.H.; Katz, A.H.; Peterson, P.J.; Labthavikul, P.; Singh, G.; Yang, Y.; Rasmussen, B.A.; et al. Phenyl thiazolyl urea and carbamate derivatives as new inhibitors of bacterial cell-wall biosynthesis. Bioorg. Med. Chem. Lett. 2004, 14, 235–238. [Google Scholar] [CrossRef]
- El Zoeiby, A.; Sanschagrin, F.; Levesque, R.C. Structure and function of the Mur enzymes: Development of novel inhibitors. Mol. Microbiol. 2003, 47, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kouidmi, I.; Levesque, R.C.; Paradis-Bleau, C. The biology of Mur ligases as an antibacterial target. Mol. Microbiol. 2014, 94, 242–253. [Google Scholar] [CrossRef]
- Ha, S.; Gross, B.; Walker, S.E. coli MurG: A paradigm for a superfamily of glycosyltransferase. Curr. Drug Targ. Infect. Disord. 2001, 1, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Chang, E.; Lo, M.-C.; Men, H.; Park, P.; Ge, M.; Walker, S. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J. Am. Chem. Soc. 1999, 121, 8415–8426. [Google Scholar] [CrossRef]
- Ha, S.; Walker, D.; Shi, Y.; Walker, S. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Prot. Sci. 2000, 9, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, L.; Ha, S.; Gross, B.; Falcone, B.; Walker, D.; Mokhtarzadeh, M.; Walker, S. Crystal structure of the MurG: UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc. Natl. Acad. Sci. USA 2003, 100, 845–849. [Google Scholar] [CrossRef]
- Mohammadi, T.; Karczmarek, A.; Crouvoisier, M.; Bouhss, A.; Mengin-Lecreulx, D.; Den Blaauwen, T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved the lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 2007, 65, 1106–1121. [Google Scholar] [CrossRef]
- Laddomada, F.; Miyachiro, M.M.; Jessop, M.; Patin, D.; Job, V.; Megin-Lecreulx, D.; Le Roy, A.; Ebel, C.; Breyton, C.; Gutsche, I.; et al. The MurG glycosyltransferase provides an oligomeric scaffold for the cytoplasmic steps of peptidoglycan biosynthesis in the human pathogen Bordetella pertussis. Sci. Rep. 2019, 9, 4656. [Google Scholar] [CrossRef]
- Mann, P.A.; Müller, A.; Xiao, L.; Pereira, P.M.; Yang, C.; Ho Lee, S.; Wang, H.; Trzeciak, J.; Schneeweis, J.; Moreira dos Santos, M.; et al. Murgocil is a highly bioactive Staphylococcal-specific inhibitor of the peptidoglycan glycosyltransferase enzyme MurG. ACS Chem. Biol. 2013, 8, 2442–2451. [Google Scholar] [CrossRef]
- Helm, J.S.; Hu, Y.; Chen, L.; Gross, B.; Walker, S. Identification of active-site inhibitors of MurG using a generalizable, high-throughput glycosyltransferase screen. J. Am. Chem. Soc. 2003, 125, 11168–11169. [Google Scholar] [CrossRef]
- Hu, Y.; Helm, J.S.; Chen, L.; Ginsberg, C.; Gross, B.; Kraybill, B.; Tiyanont, K.; Fang, X.; Wu, T.; Walker, S. Identification of selective inhibitors for the glycosyltransferase MurG via highthroughput screening. Chem. Biol. 2004, 11, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Mravljak, J.; Monasson, O.; Al-Dabbagh, B.; Crouvoisier, M.; Bouhss, A.; Gravier-Pelletier, C.; Le Merrer, Y. Synthesis and biological evaluation of a diazepanone-based library of liposidomycin analogs as MraY inhibitors. Eur. J. Med. Chem. 2011, 46, 1582–1592. [Google Scholar] [CrossRef]
- Belete, T.M. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microbiome J. 2019, 11, 100052. [Google Scholar] [CrossRef]
- Das, D.; Hervé, M.; Feuerhelm, J.; Farr, C.L.; Chiu, H.-J.; Elsliger, M.-A.; Knuth, M.W.; Klock, H.E.; Miller, M.D.; Godzik, A.; et al. Structure and function of the first full-length murein peptide ligase (Mpl) cell wall recycling protein. PLoS ONE 2011, 6, e17624. [Google Scholar] [CrossRef]
- Smith, C.A. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J. Mol. Biol. 2006, 362, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.A.; Auger, G.; Fanchon, E.; Martin, L.; Blanot, D.; Van Heijenoort, J.; Dideberg, O. Crystal structure of UDP-N-acetylmuramoyl-L-alanine: D-glutamate ligase from Escherichia coli. EMBO J. 1997, 16, 3416–3425. [Google Scholar] [CrossRef] [PubMed]
- Patin, D.; Boniface, A.; Kovač, A.; Hervé, M.; Dementin, S.; Barreteau, H.; Mengin-Lecreulx, D.; Blanot, D. Purification and biochemical characterization of Mur ligases from Staphylococcus aureus. Biochemistry 2010, 92, 1793–1800. [Google Scholar] [CrossRef]
- Munshi, T.; Gupta, A.; Evangelopoulos, D.; David Guzman, J.; Gibbons, S.; Keep, N.H.; Bhakta, S. Characterization of ATP-dependent Mur ligases involved in the biogenesis of cell wall peptidoglycan in Mycobacterium tuberculosis. PLoS ONE 2013, 8, e60143. [Google Scholar] [CrossRef] [PubMed]
- Perdih, A.; Hodoscek, M.; Solmajer, T. MurD ligase from E. coli: Tetrahedral intermediate formation study by hybrid quantum mechanical/molecular mechanical replica path method. Proteins 2009, 74, 744–759. [Google Scholar] [CrossRef] [PubMed]
- Deva, T.; Baker, E.N.; Squire, C.J.; Smith, C.A. Structure of Escherichia coli UDP-N-acetylmuramoyl:L-alanine (MurC). Acta Cryst. D 2006, 62, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.A.; Auger, G.; Martin, L.; Fanchon, E.; Blanot, D.; Le Beller, D.; Van Heijenoort, J.; Dideberg, O. Determination of the MurD mechanism through crystallographic analysis of enzyme complexes. J. Mol. Biol. 1999, 289, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.; Flouret, B.; Chantalat, L.; Van Heijenoort, J.; Mengin-Lecreulx, D.; Dideberg, O. Crystal structure of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: Meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 2001, 276, 10999–11006. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Munshi, S.; Leiting, B.; Anderson, M.S.; Chrzas, J.; Chen, Z. Crystal structure of Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme (MurF) at 2.3 A resolution. J. Mol. Biol. 2000, 304, 435–445. [Google Scholar] [CrossRef]
- Wang, C.; Xu, P.; Zhang, L.; Huang, J.; Zhu, K.; Luo, C. Current strategies and applications for precision drug design. Front. Pharmacol. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Hishinuma, F.; Izaki, K.; Takahashi, H. Inhibition of l-alanine adding enzyme by glycine. Agric. Biol. Chem. 1971, 35, 2050–2058. [Google Scholar] [CrossRef]
- Liger, D.; Masson, A.; Blanot, D.; Van Heijenoort, J.; Parquet, C. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Eur. J. Biochem. 1995, 230, 80–87. [Google Scholar]
- Emanuele, J.J.; Jin, H.; Jacobson, B.L.; Chang, C.Y.; Einspahr, H.M.; VillaFranca, J.J. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Prot. Sci. 1996, 5, 2566–2574. [Google Scholar] [CrossRef]
- Pratviel-Sosa, F.; Acher, F.; Trigalo, F.; Blanot, D.; Azerad, R.; Van Heijenoort, J. Effect of various analogues of d-glutamic acid on the d-glutamate-adding enzyme from Escherichia coli. FEMS Microbiol. Lett. 1994, 115, 223–228. [Google Scholar]
- Auger, G.; Van Heijenoort, J.; Vederas, J.C.; Blanot, D. Effect of analogues of diaminopimelic acid on the meso-diaminopimelate-adding enzyme from Escherichia coli. FEBS Lett. 1996, 391, 171–174. [Google Scholar] [CrossRef]
- Gobec, S.; Urleb, U.; Auger, G.; Blanot, D. Synthesis and biochemical evaluation of some novel N-acyl phosphono and phosphinoalanine derivatives as potential inhibitors of the d-glutamic acid-adding enzyme. Pharmazie 2001, 56, 295–297. [Google Scholar] [CrossRef]
- Kotnik, M.; Humljan, J.; Contreras-Martel, C.; Oblak, M.; Kristan, K.; Hervé, M.; Blanot, D.; Urleb, U.; Gobec, S.; Dessen, A.; et al. Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase. J. Mol. Biol. 2007, 370, 107–115. [Google Scholar] [CrossRef]
- Humljan, J.; Kotnik, M.; Contreras-Martel, C.; Blanot, D.; Urleb, U.; Dessen, A.; Solmajer, T.; Gobec, S. Novel naphthalene-N-sulfonyl-d-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme. J. Med. Chem. 2008, 51, 7486–7494. [Google Scholar] [CrossRef] [PubMed]
- Zidar, N.; Tomašić, T.; Šink, R.; Rupnik, V.; Kovač, A.; Turk, S.; Patin, D.; Blanot, D.; Contreras-Martel, C.; Dessen, A.; et al. Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase. J. Med. Chem. 2010, 53, 6584–6594. [Google Scholar] [CrossRef]
- Tomašić, T.; Zidar, N.; Šink, R.; Kovač, A.; Blanot, D.; Contreras-Martel, C.; Dessen, A.; Müller Premru, M.; Zega, A.; Gobec, S.; et al. Structure-based design of a new series of d-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase (MurD). J. Med. Chem. 2011, 54, 4600–4610. [Google Scholar] [CrossRef] [PubMed]
- Tomašić, T.; Kovač, A.; Simčič, M.; Blanot, D.; Golič Grdadolnik, S.; Gobec, S.; Kikelj, D.; Pertelin Mašić, L. Novel 2-thioxothiazolidin-4-one inhibitors of bacterial MurD ligase targeting d-glu and diphosphate-binding sites. Eur. J. Med. Chem. 2011, 46, 3964–3975. [Google Scholar] [CrossRef]
- Baell, J.; Walters, M.A. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Sim, M.M.; Ng, S.B.; Buss, A.D.; Crasta, S.C.; Goh, K.L.; Lee, S.K. Benzylidene rhodanines as novel inhibitors of UDP-N-acetylmuramate/l-Alanine ligase. Bioorg. Med. Chem. Lett. 2002, 12, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Humljan, J.; Kotnik, M.; Boniface, A.; Šolmajer, T.; Urleb, U.; Blanot, D.; Gobec, S. A new approach towardspeptidosulfonamides: Synthesis of potential inhibitors of bacterial peptidoglycan biosynthesis enzymes MurD and MurE. Tetrahedron 2006, 62, 10980–10988. [Google Scholar] [CrossRef]
- Sosič, I.; Barreteau, H.; Simčič, M.; Šink, R.; Cesar, J.; Zega, A.; Golič Grdadolnik, S.; Contreras-Martel, C.; Dessen, A.; Amoroso, A.; et al. Second-generation sulfonamide inhibitors of d-glutamic acid-adding enzyme: Activity optimization with conformationnally rigid analogues of d-glutamic acid. Eur. J. Med. Chem. 2011, 46, 2880–2894. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Wu, Z. Phosphinate analogs of d-, d-dipeptides: Slow-binding inhibition and proteolysis protection of VanX, a d-, d-dipeptidase required for vancomycin resistance in Enterococcus faecium. Proc. Natl. Acad. Sci. USA 1995, 92, 11603–11607. [Google Scholar]
- Ellsworth, B.A.; Tom, N.J.; Bartlett, P.A. Synthesis and evaluation of inhibition of bacterial d-alanine:d-alanine ligases. Chem. Biol. 1996, 3, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Reck, F.; Marmor, S.; Fisher, S.; Wuonola, M.A. Inhibitors of the cell wall biosynthesis enzyme MurC. Bioorg. Med. Chem. Lett. 2001, 11, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Marmor, S.; Peterson, C.P.; Reck, F.; Yang, W.; Gao, N.; Fisher, S.L. Biochemical characterization of a phosphinate inhibitor of Escherichia coli MurC. Biochemistry 2001, 40, 12207–12214. [Google Scholar] [CrossRef] [PubMed]
- Tanner, M.E.; Vaganay, S.; Van Heijenoort, J.; Blanot, D. Phosphinate inhibitors of the d-glutamic acid-adding enzyme of peptidoglycan biosynthesis. J. Org. Chem. 1996, 61, 1756–1760. [Google Scholar] [CrossRef] [PubMed]
- Gegnas, L.D.; Waddell, S.T.; Chabin, R.M.; Reddy, S.; Wong, K.K. Inhibitors of the bacterial cell wall biosynthesis enzyme Mur D. Bioorg. Med. Chem. Lett. 1998, 8, 1643–1648. [Google Scholar] [CrossRef]
- Štrancar, K.; Blanot, D.; Gobec, S. Design, synthesis and structure-activity relationship of new phosphinate inhibitors of MurD. Bioorg. Med. Chem. Lett. 2006, 16, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Wong, K.K.; Pompliano, D.L.; Reddy, S.; Tanner, M.E. A phophinate inhibitor of the meso-diaminopimelic acid-adding enzyme (MurE) of peptidoglycan biosynthesis. J. Org. Chem. 1998, 63, 10081–10086. [Google Scholar] [CrossRef]
- Štrancar, K.; Boniface, A.; Blanot, D.; Gobec, S. Phosphinate inhibitors of UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: L-lysine ligase (MurE). Arch. Pharm. Int. J. Pharm. Med. Chem. 2007, 340, 127–134. [Google Scholar] [CrossRef]
- Blanot, D.; Auger, G.; Liger, D.; Van Heijenoort, J. Synthesis of α and β anomers of UDP-N-acetylmuramic acid. Carb. Res. 1994, 252, 107–115. [Google Scholar] [CrossRef]
- Hitchcock, S.A.; Eid, C.N.; Aikins, J.A.; Zia-Ebrahimi, M.; Blaszczak, L.C. The first total synthesis of bacterial cell wall precursor UDP-N-acetylmuramyl-pentapeptide (Park nucleotide). J. Am. Chem. Soc. 1998, 120, 1916–1917. [Google Scholar] [CrossRef]
- Liu, H.; Sadamoto, R.; Sears, P.S.; Wong, C.-H. An efficient chemoenzymatic strategy for the synthesis of wild-type and vancomycin-resitant bacterial cell-wall precursors: UDP-N-acetylmuramyl-peptides. J. Am. Chem. Soc. 2001, 123, 9916–9917. [Google Scholar] [CrossRef] [PubMed]
- Lioux, T.; Busson, R.; Rozenski, J.; Nguyen-Distèche, M.; Frère, J.-M.; Herdewijn, P. Synthesis of peptidoglycan units with UDP at the anomeric position. Collect. Czech. Chem. Commun. 2005, 70, 1615–1641. [Google Scholar] [CrossRef]
- Humljan, J.; Starčević, Š.; Car, V.; Anderluth, Š.; Kocjan, D.; Jenko, B.; Urleb, U. Optimization of UDP-N-acetylmuramic acid synthesis. Pharmazie 2008, 2, 102–106. [Google Scholar]
- Horton, J.R.; Bostock, J.M.; Chopra, I.; Hesse, L.; Phillips, S.E.V.; Adams, D.J.; Peter Johnson, A.; Fishwick, C.W.G. Macrocyclic inhibitors of the bacterial cell wall biosynthesis enzyme MurD. Bioorg. Med. Chem. Lett. 2003, 13, 1557–1560. [Google Scholar] [CrossRef] [PubMed]
- Frlan, R.; Perdith, F.; Cirkvenčič, N.; Pečar, S.; Obreza, A. Design an d synthesis of novel UDP-Mur-NAc, UDP-Mur-NAc-l-Ala and UDP-Mur-NAc-l-Ala-d-Glu mimetics. Acta Chim. Slov. 2009, 56, 580–590. [Google Scholar]
- Zivec, M.; Turk, S.; Blanot, D.; Gobec, S. Design and synthesis of new peptidomimetics as potencial inhibitors of MurE. Acta Chim. Slov. 2011, 58, 95–109. [Google Scholar]
- El Zoeiby, A.; Sanschagrin, F.; Darveau, A.; Brisson, J.-R.; Levesque, R.C. Identification of novel inhibitors of Pseudomonas aeruginosa MurC enzyme derived from phage-displayed peptide librairies. J. Antimicrob. Chemother. 2003, 51, 531–543. [Google Scholar] [CrossRef][Green Version]
- Paradis-Bleau, C.; Beaumont, M.; Boudreault, L.; Lloyd, A.; Sanschagrin, F.; Bugg, T.D.H.; Levesque, R.C. Selection of peptide inhibitors against the Pseudomonas aeruginosa MurC cell wall enzyme. Peptides 2006, 27, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Paradis-Bleau, C.; Lloyd, A.; Sanschagrin, F.; Maaroufi, H.; Clarke, T.; Blewett, A.; Dowson, C.; Roper, D.I.; Bug, T.D.; Levesque, R.C. Pseudomonas aeruginosa MurE amide ligase: Enzyme kinetics and peptide inhibitor. Biochem. J. 2009, 421, 263–272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bratkovič, T.; Lunder, M.; Urleb, U.; Štrukelj, B. Peptide inhibitors of MurD and MurE, essential enzymes of bacterial cell wall biosynthesis. J. Bas. Microbiol. 2008, 48, 202–206. [Google Scholar] [CrossRef]
- Paradis-Bleau, C.; Lloyd, A.; Sanschagrin, F.; Clarke, T.; Blewett, A.; Bugg, T.D.; Levesque, R.C. Phage display-derived inhibitor of the essential cell wall biosynthesis enzyme MurF. BMC Biochem. 2008, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, D.E.; Demeritt, J.E.; Hull, K.G.; Fisher, S.L. Biochemical characterization of an inhibitor of Escherichia coli UDP-N-acetylmuramyl-l-alanine ligase. Biochim. Biophy. Acta 2004, 1698, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Frlan, R.; Kovač, A.; Blanot, D.; Gobec, S.; Pečar, S.; Obreza, A. Design and synthesis of novel N-benzylidenesulfonohydrazide inhibitors of MurC and MurD as potential antibacterial agents. Molecules 2008, 13, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Frlan, R.; Kovač, A.; Blanot, D.; Gobec, S.; Pečar, S.; Obreza, A. Design, synthesis and in vitro biochemical activity of novel amino acid sulfonohydrazideinhibitors of MurC. Acta Chim. Slov. 2011, 58, 295–310. [Google Scholar]
- Turk, S.; Kovač, A.; Boniface, A.; Bostock, J.M.; Chopra, I.; Blanot, D.; Gobec, S. Discovery of new inhibitors of the bacterial peptidoglycan biosynthesis enzymes MurD and MurF by structure-based virtual screening. Bioorg. Med. Chem. 2009, 17, 1884–1889. [Google Scholar] [CrossRef]
- Gui Gu, Y.; Florjancic, A.S.; Clark, R.F.; Zhang, T.; Cooper, C.S.; Anderson, D.D.; Lerner, C.G.; Owen McCall, J.; Cai, Y.; Black-Schaefer, C.L.; et al. Structure-activity relationships of novel potent MurF inhibitors. Bioorg. Med. Chem. Lett. 2004, 14, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Comess, K.M.; Schurdak, M.E.; Voorbach, M.J.; Coen, M.; Trumbull, J.D.; Yang, H.; Gao, L.; Tang, H.; Cheng, X.; Lerner, C.G.; et al. An ultraefficient affinity-based high-throughout screening process: Application to bacterial cell wall biosynthesis enzyme MurF. J. Biomol. Screen. 2006, 11, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Longenecker, K.L.; Stamper, G.F.; Hajduk, P.J.; Fry, E.H.; Jakob, C.G.; Harlan, J.E.; Edalji, R.; Bartley, D.M.; Walter, K.A.; Solomon, L.R.; et al. Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor interdomain closure. Prot. Sci. 2005, 14, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Stamper, G.F.; Longenecker, K.L.; Fry, E.H.; Jacok, C.G.; Florjancic, A.S.; Gui Gu, Y.; Anderson, D.D.; Cooper, C.S.; Zhang, T.; Clark, R.F.; et al. Structure-based optimization of MuF inhibitors. Chem. Biol. Drug Des. 2005, 67, 58–65. [Google Scholar] [CrossRef]
- Hrast, M.; Turk, S.; Sosič, I.; Knez, D.; Randall, C.P.; Barreteau, H.; Contreras-Martel, C.; Dessen, A.; O’Neil, A.J.; Mengin-Lecreulx, D.; et al. Structure-activity relationships of new cyanothiophene inhibitors of the essential peptidoglycan biosynthesis enzyme MurF. Eur. J. Med. Chem. 2013, 66, 32–45. [Google Scholar] [CrossRef]
- Hrast, M.; Anderluth, M.; Knez, D.; Randall, C.P.; Barreteau, H.; O’Neil, A.J.; Blanot, D.; Gobec, S. Design, synthesis and evaluation of second generation MurF inhibitors based on cyanothiophene scaffold. Eur. J. Med. Chem. 2014, 73, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Turk, S.; Hrast, M.; Sosič, I.; Barreteau, H.; Mengin-Lecreulx, D.; Blanot, D.; Gobec, S. Biochemical characterization of MurF from Streptococcus pneumoniae and the identification of a new MurF inhibitor through ligand-based virtual screening. Acta Chim. Slov. 2013, 60, 294–299. [Google Scholar] [PubMed]
- Sosič, I.; Štefane, B.; Kovač, A.; Turk, S.; Blanot, D.; Gobec, S. The synthesis of novel 2,4,6-trisubstituted 1,3,5-triazines: A search for potential MurF enzyme inhibitors. Heterocycles 2010, 81, 91–115. [Google Scholar]
- Baum, E.Z.; Crespo-Carbone, S.M.; Abbanat, D.; Foleno, B.; Maden, A.; Goldschmidt, R.; Bush, K. Utility of muropeptide ligase for identification of inhibitors of the cell wall biosynthesis enzyme MurF. Antimicrob. Agents Chemother. 2006, 50, 230–236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baum, E.Z.; Crespo-Carbone, S.M.; Klinger, A.; Foleno, B.D.; Turchi, I.; Macielag, M.; Bush, K. A MurF inhibitor that disrupts cell wall biosynthesis in Escherichia coli. Antimicrob. Agents Chemother. 2007, 51, 4420–4426. [Google Scholar] [PubMed]
- Baum, E.Z.; Crespo-Carbone, S.M.; Foleno, B.D.; Simon, L.D.; Guillemont, J.; Macielag, M.; Bush, K. MurF inhibitors with antibacterial activity: Effect on muropeptide levels. Antimicrob. Agents Chemother. 2009, 53, 3240–3247. [Google Scholar] [CrossRef]
- Zawadzke, L.E.; Norcia, M.; Desbonnet, C.R.; Wang, H.; Freeman-Cook, K.; Dougherty, T.J. Identification of an inhibitor of the MurC enzyme, which catalyzes an essential step in the peptidoglycan precursor synthesis pathway. Assay Drug. Dev. Technol. 2008, 6, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Guzman, J.D.; Gupta, A.; Evangelopoulos, D.; Basavannacharya, C.; Pabon, L.C.; Plazas, E.A.; Muñoz, D.R.; Delgado, W.A.; Cuca, L.E.; Ribon, W.; et al. Anti-tubercular screening of natural products from Colombian plants: 3-methoxynordomesticine, an inhibitor of MurE ligase of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2010, 65, 2101–2107. [Google Scholar] [CrossRef]
- Shiu, W.K.P.; Malkinson, J.P.; Mukhlesur Rahman, M.; Curry, J.; Stapleton, P.; Gunaratnam, M.; Neidle, S.; Mushtaq, S.; Warner, M.; Livermore, D.M.; et al. A new plant-derived antibacterial is an inhibitor of efflux pumps in Staphylococcus aureus. Int. J. Antimicrob. Agents 2013, 42, 513–518. [Google Scholar] [CrossRef]
- Guzman, J.D.; Pesnot, T.; Barrera, D.A.; Davies, H.M.; MacMahon, E.; Evangelopoulos, D.; Mortazavi, P.N.; Munshi, T.; Maitra, A.; Lamming, E.D.; et al. Tetrahydroisoquinolines affect the whole-cell phenotype of Mycobacterium tuberculosis by inhibiting the ATP-dependent MurE ligase. J. Antimicrob. Chemother. 2015, 70, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Tomašić, T.; Šink, R.; Zidar, N.; Fic, A.; Contreras-Martel, C.; Dessen, A.; Patin, D.; Blanot, D.; Müller-Premru, M.; Gobec, S.; et al. Dual inhibitor of MurD and MurE ligases from Escherichia coli and Straphylococcus aureus. ACS Med. Chem. Lett. 2012, 3, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Perdih, A.; Kovač, A.; Wolber, G.; Blanot, D.; Gobec, S.; Solmajer, T. Discovery of novel benzene 1,3-dicarboxylic acid inhibitors of bacterial MurD and MurE ligases by structure-based virtual screening approach. Bioorg. Med. Chem. Lett. 2009, 19, 2668–2673. [Google Scholar] [CrossRef]
- Perdih, A.; Hrast, M.; Barreteau, H.; Gobec, S.; Wolber, G.; Solmajer, T. Benzene 1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorg. Med. Chem. 2014, 22, 4124–4134. [Google Scholar] [CrossRef]
- Sova, M.; Kovač, A.; Turk, S.; Hrast, M.; Blanot, D.; Gobec, S. Phosphorylated hydroxyethylamines as novel inhibitors of the bacterial cell wall biosynthesis enzymes MurC to MurF. Bioorg. Chem. 2009, 37, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Šink, R.; Kovač, A.; Tomašić, T.; Rupnik, V.; Boniface, A.; Bostock, J.; Chopra, I.; Blanot, D.; Peterlin Mašič, L.; Gobec, S.; et al. Synthesis and biological evaluation of N-acylhydrazones as inhibitors of MurC and MurD ligases. ChemMedChem 2008, 3, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Frlan, R.; Vobovnik, N.; Kovač, A.; Blanot, D.; Pečar, S.; Gobec, S. New Arylsulfonohydrazide Inhibitors of Enzymes MurC and MurD. Patent EP1845083 A2 2006-04-13 11 April 2017. [Google Scholar]
- Antane, S.; Caufield, C.E.; Hu, W.; Keeney, D.; Labthavikul, P.; Morris, K.; Naughton, S.M.; Peterson, P.J.; Rasmussen, B.A.; Singh, G.; et al. Pulvinones as bacterial cell wall biosynthesis inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Francisco, G.D.; Hu, W.; Labthavikul, P.; Peterson, P.J.; Severin, A.; Singh, G.; Yang, Y.; Rasmussen, B.A.; Lin, Y.-I.; et al. 2-phenyl-5,6-dihydro-2H-thieno [3,2-c]pyrazol-3-ol derivatives as new inhibitors of bacterial cell wall biosynthesis. Bioorg. Med. Chem. Lett. 2003, 13, 2591–2594. [Google Scholar] [CrossRef] [PubMed]
- Mansour, T.S.; Caufield, C.E.; Rasmussen, B.A.; Chopra, R.; Krishnamurthy, G.; Morris, K.M.; Svenson, K.; Bard, J.; Smeltzer, C.; Naughton, S.; et al. Naphthyl tetronic acid as multi-target inhibitors of bacterial peptidoglycan biosynthesis. ChemMedChem 2007, 2, 1414–1417. [Google Scholar] [CrossRef]
- Hrast, M.; Rožman, K.; Ogris, I.; Škedelj, V.; Patin, D.; Sova, M.; Barreteau, H.; Gobec, S.; Golič Grdadolnik, S.; Zega, A. Evaluation of the published kinase inhibitor set to identify multiple inhibitors of bacterial ATP-dependent mur ligases. J. Enzym. Inhib. Med. Chem. 2019, 34, 1010–1017. [Google Scholar] [CrossRef]
- Raush, S.; Hänchen, A.; Denisiuk, A.; Löhken, M.; Schneider, T.; Süssmuth, R.D. Feglymycin is an inhibitor of the enzyme MurA and MurC of the peptidoglycan biosynthesis pathway. ChemBioChem 2011, 12, 1171–1173. [Google Scholar] [CrossRef]
- Tomašič, T.; Kovač, A.; Klebe, G.; Blanot, D.; Gobec, S.; Kikelj, D.; Peterlin Mašič, L. Virtual screening for potential inhibitors of bacterial MurC and MurD ligases. J. Mol. Model. 2012, 18, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Chakkyarath, V.; Natarajan, J. Identification of ideal multi-targeting bioactive compounds against Mur ligases of Enterobacter aerogenes and its bonding mechanism in comparison with chemical inhibitors. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 135–144. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).