Identification of a β-Carboline Alkaloid from Chemoselectively Derived Vanilla Bean Extract and Its Prevention of Lipid Droplet Accumulation in Human Hepatocytes (HepG2)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Antioxidant Activity, Total Polyphenol Content (TPC), and Metabolite Profiling and Fingerprinting of Selected Bean Extracts
2.1.1. Antioxidant Activity
2.1.2. Proton Nuclear Magnetic Resonance Profiling and Fingerprinting of Selected Bean Extracts
2.1.3. LC–MS/MS Analysis and Global Natural Product Social-Aided Dereplication of Constituents from Selected Bean Extracts
2.2. Heated Treatment and Acid Modification of Vanilla Bean Extract and Its Dereplication of Metabolites by LC-MS
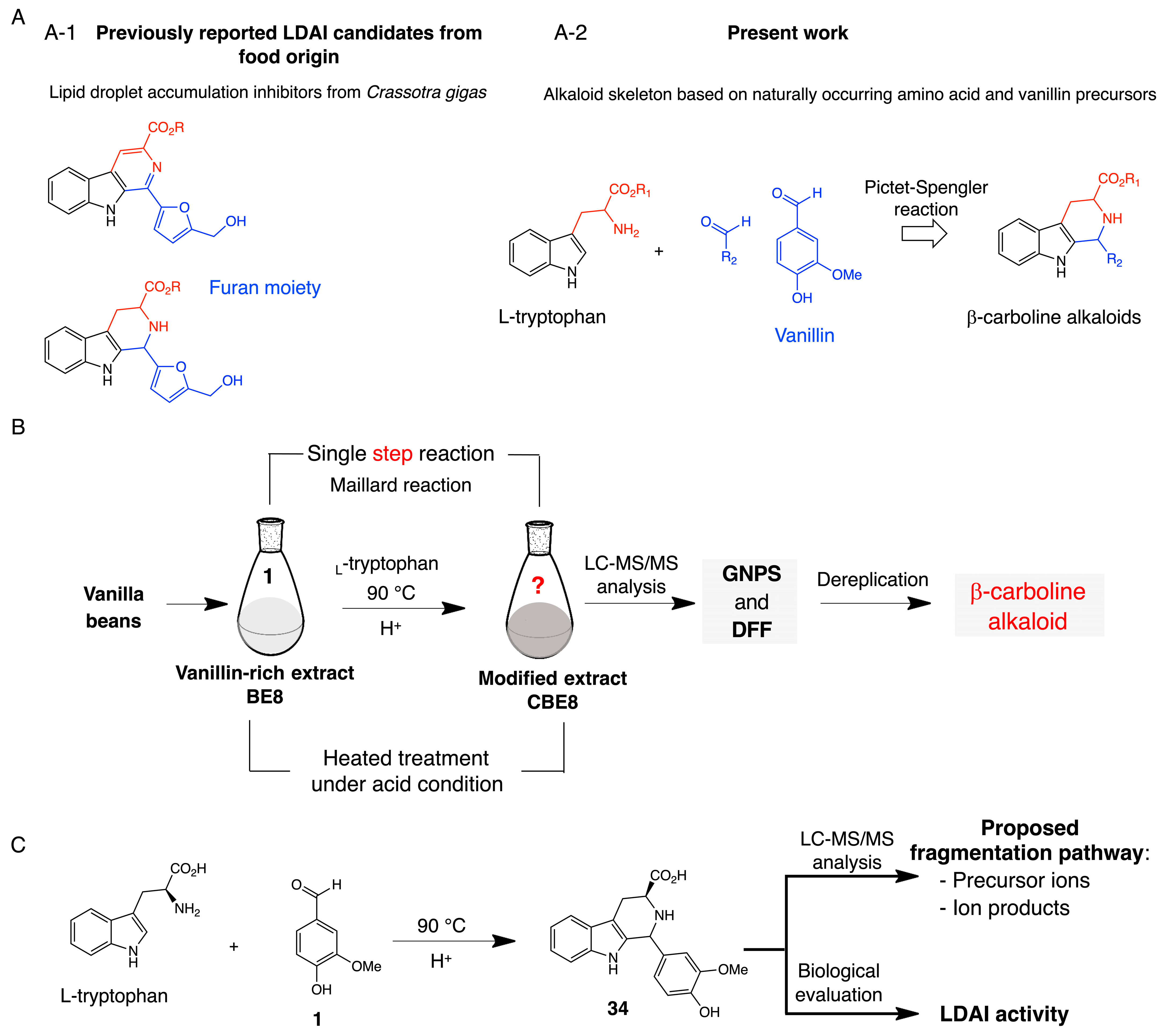
2.2.1. Chemoselective Derivatization of Vanilla Bean Extract
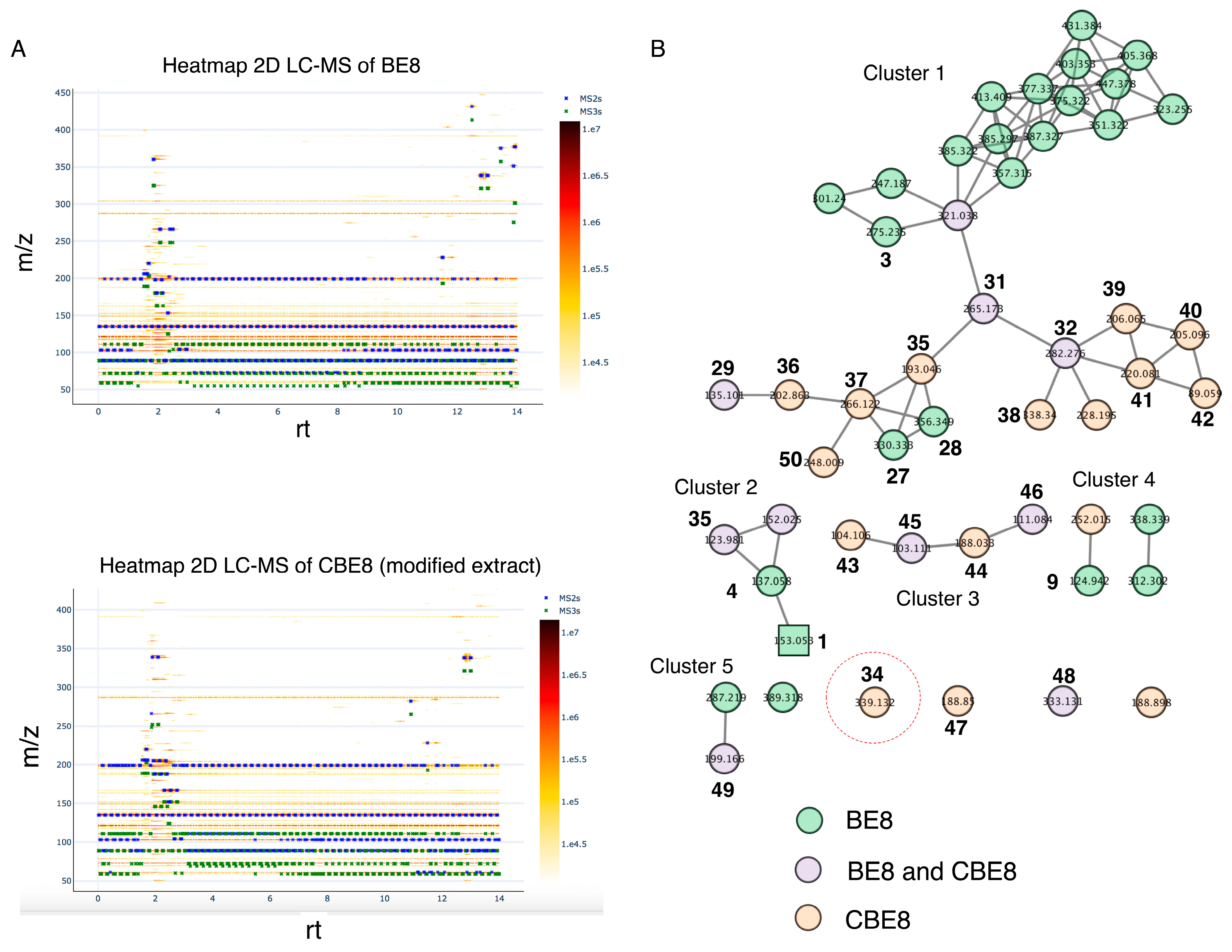
2.2.2. Molecular Networking and Detection of β-Carboline 34 in Modified Bean Extract
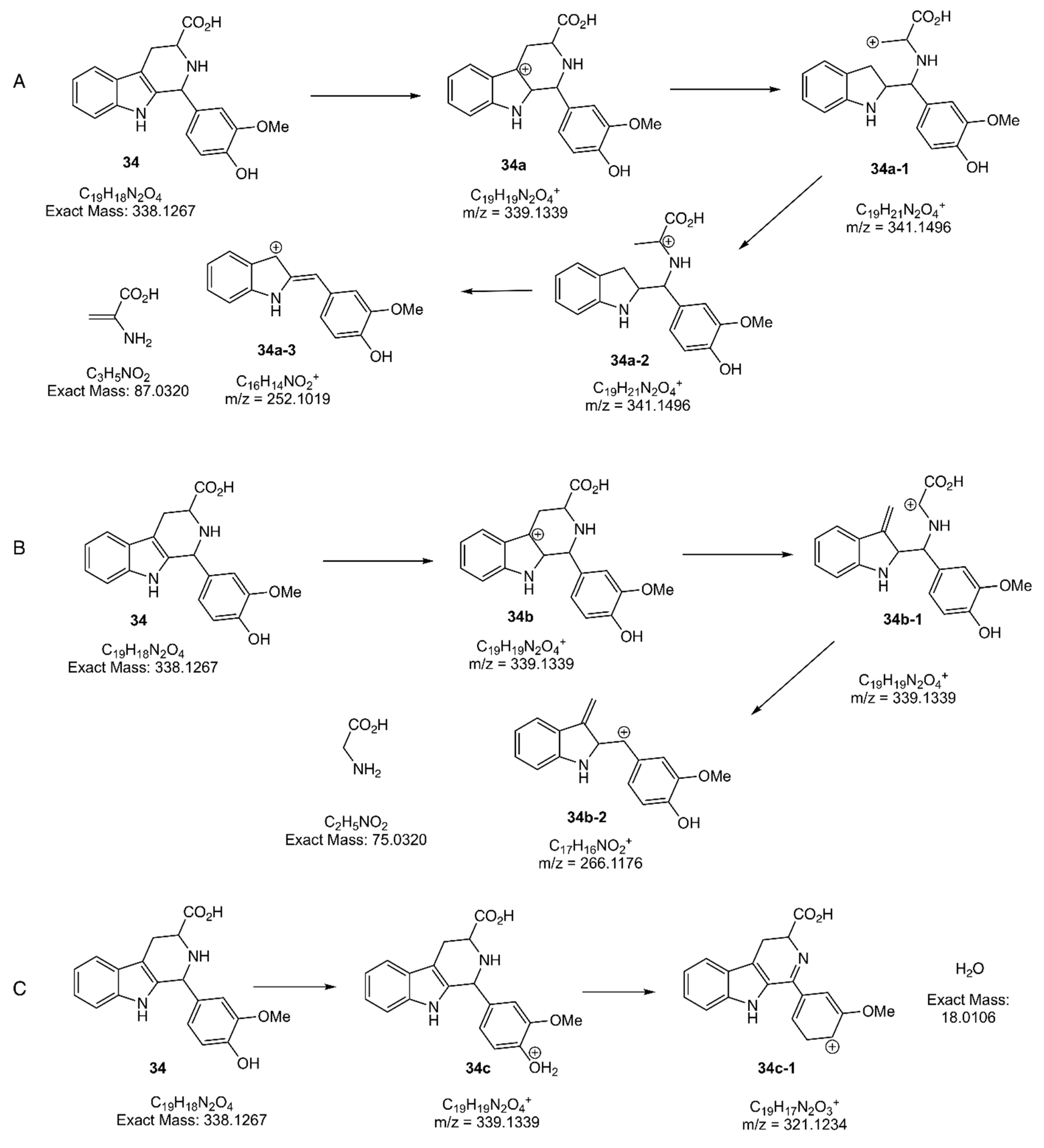
2.2.3. Structure Characterization and Proposed Fragmentation Pathway of Detected β-Carboline 34
2.2.4. Identification of 34 in BE8 Using the Specific Tandem Mass Spectrometry Fragmentation
2.3. Cell Viability, Lipid Droplet Accumulation Inhibition Activity, and the Effect of β-Carboline Alkaloid 34 on Lipid Metabolism
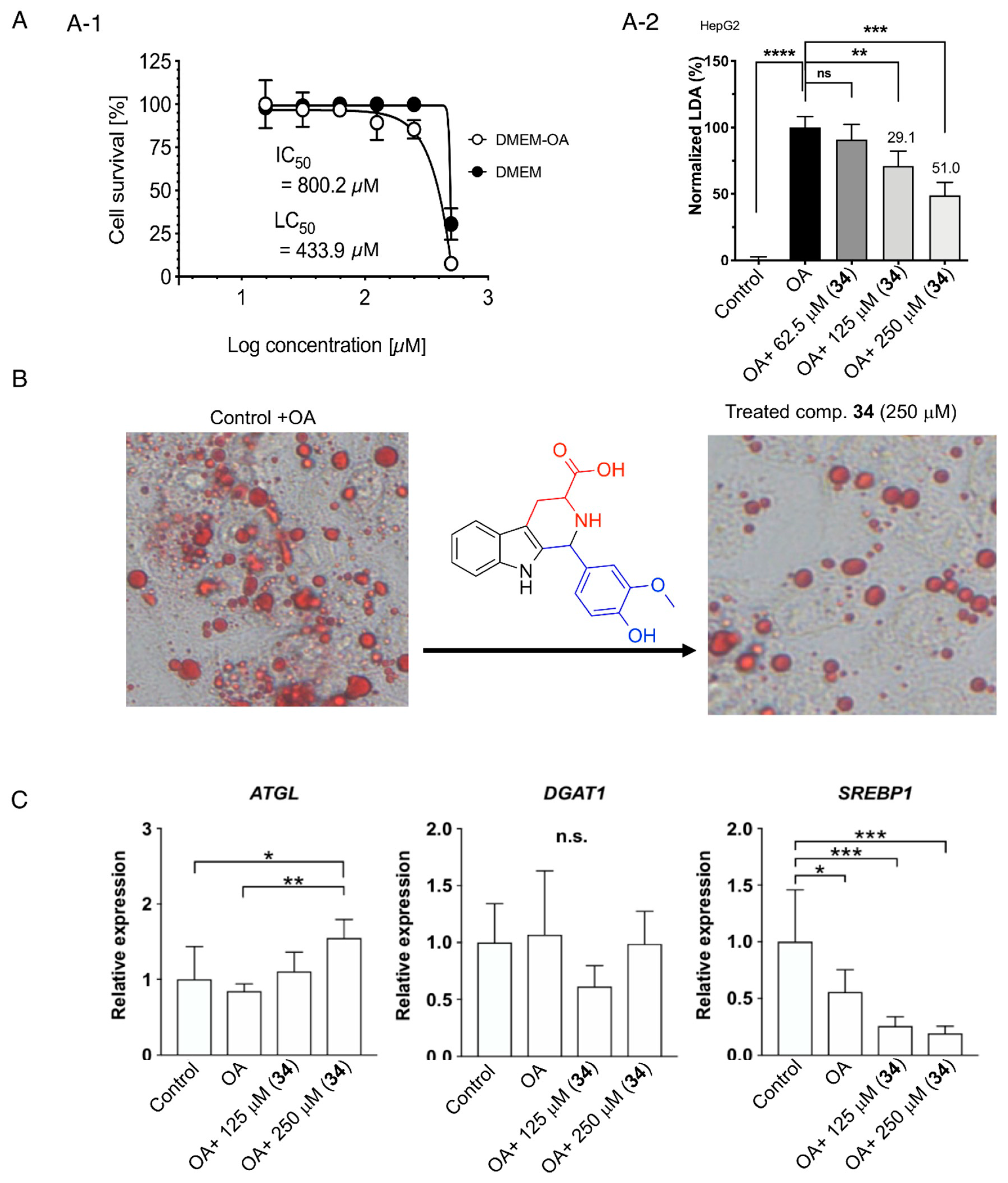
2.3.1. Cell Viability: Evaluation of Cytotoxicity and Lipocytotoxicity of β-Carboline Alkaloid 34 in Oleic Acid-Loaded HepG2 Cells
2.3.2. Lipid Droplet Accumulation Inhibition Activity and the Effect of β-Carboline Alkaloid 34 on Lipolysis and Lipogenesis in Oleic Acid-Loaded HepG2 Cells
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Determination of Antioxidant Activity and TPC
3.3. Metabolite Profiling of Vanilla Bean Extracts
3.3.1. Nuclear Magnetic Resonance Profiling of Vanilla Bean Extract
3.3.2. Liquid Chromatography/Mass Spectrometry Profiling of Vanilla Bean Extract
- A.
- LC-MS instrument conditions and processing
- B.
- Molecular network of BE (BE1–BE8) samples
- C.
- Compounds dereplication using LC-MS
3.3.3. Chemo-Selective Derivatization of Vanilla Bean Extracts under Acidic Conditions and LC-MS/MS Analysis of the Modified Vanilla Bean Extract
3.3.4. Preparation Procedure of Identified β-Carboline Alkaloid 34 Using Acidic Conversion
3.4. Evaluation of Cell Viability
3.5. Lipid Metabolism-Related Gene Expression
3.6. Lipid Droplet Accumulation Inhibition Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teodoro, A.J. Bioactive compounds of food: Their role in prevention and treatment of diseases. Oxid. Med. Cell. Longev. 2019, 2019, 3765986. [Google Scholar] [CrossRef]
- Amirkia, V.; Heinrich, M. Alkaloids as drug leads—A preventive structural and biodiversity-based analysis. Phytochem. Lett. 2014, 10, xlviii–liii. [Google Scholar] [CrossRef]
- Sorrenti, V.; Burò, I.; Consoli, V.; Vanella, L. Recent Advances in Health Benefits of Bioactive Compounds from Food Wastes and By-Products: Biochemical Aspects. Int. J. Mol. Sci. 2023, 24, 2019. [Google Scholar] [CrossRef] [PubMed]
- Samtiya, M.; Aluko, R.E.; Dhewa, T.; Moreno-Rojas, J.M. Potential Health Benefits of Plant Food-Derived Bioactive Components: An Overview. Foods 2021, 10, 839. [Google Scholar] [CrossRef] [PubMed]
- Ortega, C.A.; Favier, L.S.; Cifuente, D.A. Chemical derivatization of natural extracts obtained from Larrea divaricate Cav. Increase in the antioxidant activity and protein precipitating capacity. Acad. J. Sci. Res. 2017, 5, 197–202. [Google Scholar]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Socaciu, C. From phytochemistry to metabolomics: Eigth decades of research in plant and food science. Studia UBB Chemia, LXIV 2019, 4, 205–224. [Google Scholar] [CrossRef]
- Ramalo, I.A.; Salazar, M.O.; Mendez, L.; Furlan, R.L.E. Chemically Engineered Extracts: Source of Bioactive Compounds. Acc. Chem. Res. 2011, 44, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.O.; Ramallo, I.A.; Micheloni, O.; Gonzalez Sierra, M.; Furlan, R.L.E. Chemically engineered extracts: Bioactivity alteration through sulfonylation. Bioorg. Med. Chem. Lett. 2009, 19, 5067–5507. [Google Scholar] [CrossRef]
- Del Vitto, L.A.; Petenatti, E.M.; Petenatti, M.E. Recursos Herbolarios de San Luis (República Argentina) Primera Parte: Plantas Nativas. Multequina 1997, 6, 49–66. [Google Scholar]
- Amaya, D.R.; Farfan, J.F. (Eds.) Chemical Changes during Processing and Storage of Foods. Implications for Food Quality and Human Health, 1st ed.; Academic Press: Cambridge, MA, USA, 2020; Print Book & E-Book; ISBN 9780128173800/9780128173817. Available online: https://www.elsevier.com/books/chemical-changes-during-processing-and-storage-of-foods/rodriguez-amaya/978-0-12-817380-0 (accessed on 21 September 2022).
- Herraiz, T.; Galisteo, J.; Chamorro, C. L-tryptophan Reacts with Naturally Occurring and Food-Occurring Phenolic Aldehydes to Give Phenolic Tetrahydro-β-Carboline Alkaloids: Activity as Antioxidants and Free Radical Scavengers. J. Agric. Food Chem. 2003, 51, 2168–2173. [Google Scholar] [CrossRef]
- Herraiz, T.; Ough, C.S. Chemical and technological factors determining tetrahydro-b-carboline-3-carboxylic acid content in fermented alcoholic beverages. J. Agric. Food Chem. 1993, 41, 959–964. [Google Scholar] [CrossRef]
- Herraiz, T.; Peña, A.; Mateo, H.; Herraiz, M.; Salgado, A. Formation, Characterization, and Occurrence of β-Carboline Alkaloids Derived from α-Dicarbonyl Compounds and l-Tryptophan. J. Agric. Food Chem. 2022, 70, 9143–9153. [Google Scholar] [CrossRef]
- Dibwe, D.F.; Oba, S.; Takeishi, N.; Sakurai, T.; Tsukui, T.; Chiba, H.; Hui, S.P. Food-Derived β-Carboline Alkaloids Ameliorate Lipid Droplet Accumulation in Human Hepatocytes. Pharmaceuticals. 2022, 15, 578. [Google Scholar] [CrossRef] [PubMed]
- Fuda, H.; Watanabe, M.; Hui, S.P.; Joko, S.; Okabe, H.; Jin, S.; Takeda, S.; Miki, E.; Watanabe, T.; Chiba, H. Anti-apoptotic Effects of Novel Phenolic Antioxidant Isolated from the Pacific Oyster (Crassostrea gigas) on Cultured Human Hepatocytes Under Oxidative Stress. Food Chem. 2015, 176, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; González-Ruiz, V.; Rudaz, S. Implementation of Liquid Chromatography-High Resolution Mass Spec-Trometry Methods for Untargeted Metabolomic Analyses of Biological Samples: A Tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef]
- Gao, Y.; Hou, L.; Gao, J.; Li, D.; Tian, Z.; Fan, B.; Wang, F.; Li, S. Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review. Foods 2021, 10, 2294. [Google Scholar] [CrossRef] [PubMed]
- González-Peña, D.; Brennan, L. Recent Advances in the Application of Metabolomics for Nutrition and Health. Annu. Rev. Food Sci. Technol. 2019, 10, 479–519. [Google Scholar] [CrossRef]
- Selamat, J.; Rozani, N.A.A.; Murugesu, S. Application of the Metabolomics Approach in Food Authentication. Molecules 2021, 26, 7565. [Google Scholar] [CrossRef]
- Gauglitz, J.M.; Aceves, C.M.; Aksenov, A.A.; Aleti, G.; Almaliti, J.; Bouslimani, A.; Brown, E.A.; Campeau, A.; Caraballo-Rodríguez, A.M.; Chaar, R.; et al. Untargeted mass spectrometry-based metabolomics approach unveils molecular changes in raw and processed foods and beverages. Food Chem. 2020, 302, 125290. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef]
- Caldas, L.A.; Zied, D.C.; Sartorelli, P. Dereplication of Extracts from Nutraceutical Mushrooms Pleurotus Using Molecular Network Approach. Food Chem. 2022, 370, 131019. [Google Scholar] [CrossRef] [PubMed]
- Saia, S.; Fragasso, M.; De Vita, P.; Beleggia, R. Metabolomics Provides Valuable Insight for the Study of Durum Wheat: A Review. J. Agric. Food Chem. 2019, 67, 3069–3085. [Google Scholar] [CrossRef]
- Kaur, C.; Kapoor, H.C. Antioxidant Activity and Total Phenolic Content of Some Asian Vegetables. Int. J. Food Sci. Technol. 2002, 37, 153–161. [Google Scholar] [CrossRef]
- Villaño, D.; Fernández-Pachón, M.S.; Moyá, M.L.; Troncoso, A.M.; Garcıa-Parrilla, M.C. Radical Scavenging Ability of Polyphenolic Compounds Towards DPPH Free Radical. Talanta. 2007, 71, 230–235. [Google Scholar] [CrossRef]
- Cardador-Martínez, A.C.; Loarca-Piña, G.; Oomah, B.D. Antioxidant Activity in Common Beans (Phaseolus vulgaris L.). J. Agric. Food Chem. 2002, 50, 6975–6980. [Google Scholar] [CrossRef]
- Gowd, V.; Karim, N.; Shishir, M.R.I.; Xie, L.; Chen, W. Dietary Polyphenols to Combat the Metabolic Diseases via Altering Gut Microbiota. Trends Food Sci. Technol. 2019, 93, 81–93. Available online: https://www.sciencedirect.com/science/article/pii/S0308814622017307#bi005 (accessed on 2 March 2023). [CrossRef]
- Lin, D.; Xiao, M.; Zhao, J.; Li, Z.; Xing, B.; Li, X.; Kong, M.; Li, L.; Zhang, Q.; Liu, Y.; et al. An Overview of Plant Phenolic Compounds and Their Importance in Human Nutrition and Management of Type 2 Diabetes. Molecules 2016, 21, 1374. [Google Scholar] [CrossRef]
- Palama, T.L.; Khatib, A.; Choi, Y.; Côme, B.; Fock, I.; Verpoorte, R.; Kodja, H. Metabolic characterization of green pods from Vanilla planifolia accessions grown in La Réunion. Environ. Exp. Bot. 2011, 72, 258–265. [Google Scholar] [CrossRef]
- Palama, T.L.; Khatib, A.; Choi, Y.; Côme, B.; Fock, I.; Verpoorte, R.; Kodja, H. Metabolic Changes in Different Developmental Stages of Vanilla planifolia Pods. J. Agric. Food Chem. 2009, 57, 7651–7658. [Google Scholar] [CrossRef]
- Caldas, L.A.; Soares, D.M.M.; Menolli, N.; Stevani, C.V.; Sartorelli, P. Metabolomics of the Wild Mushroom Gymnopilus imperialis (Agaricomycetes, Basidiomycota) by UHPLC-HRMS/MS Analysis and Molecular Network. Fungal Biol. 2022, 126, 132–138. [Google Scholar] [CrossRef]
- Walsh, J.P.; Renaud, J.B.; Hoogstra, S.; McMullin, D.R.; Ibrahim, A.I.; Visagie, C.M.; Tanney, J.B.; Yeung, K.K.; Sumarah, M.W. Diagnostic Fragmentation Filtering for the Discovery of New Chaetoglobosins and Cytochalasins. Rapid Commun. Mass Spectrom. 2019, 33, 133–139. [Google Scholar] [CrossRef]
- Goh, T.B.; Mordi, M.M.; Mansor, S.M. Mass Spectrometry (LC-MS-MS) as a Tool in the Maillard Reaction Optimisation and Characterisation of New 6-Methoxy-Tetrahydro-β-Carboline Derivatives. Sains Malays. 2015, 44, 127–137. [Google Scholar] [CrossRef]
- Miller Crotti, A.E.; Gates, P.J.; Lopes, J.L.; Lopes, N.P. Electrospray MS-Based Characterization of β-Carbolines- Mutagenic Constituents of Thermally Processes Meat. Mol. Nutr. Food Res. 2010, 54, 433–439. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial Dysfunction and Oxidative Stress in Metabolic Disorders—A Step Towards Mitochondria Based Therapeutic Strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Gluchowski, N.L.; Becuwe, M.; Walther, T.C.; Farese, R.V. Lipid Droplets and Liver Disease: From Basic Biology to Clinical Implications. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 343–355. [Google Scholar] [CrossRef]
- Krahmer, N.; Farese, R.V.; Walther, T.C. Balancing the Fat: Lipid Droplets and8 Human Disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef]
- Sumida, Y.; Yoneda, M. Current and Future Pharmacological Therapies for NAFLD/NASH. J. Gastroenterol. 2018, 53, 362–376. [Google Scholar] [CrossRef]
- Younossi, Z.M. Non-alcoholic Fatty Liver Disease—A Global Public Health Perspective. J. Hepatol. 2019, 70, 531–544. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, K.I. An Overview of NAFLD/NASH in Japan. Yakugaku Zasshi 2016, 136, 565–572. [Google Scholar] [CrossRef]
- Santos, L.F.; Hernández, G.; Puerta, A.V.; Beltrán, Ó.; Botero, R.C.; Mejía, G. Non-Alcoholic Fatty Liver Disease. The New Millennium Pandemia. Rev. Colomb. Gastroenterol. 2010, 25, 373–391. [Google Scholar]
- Torres, D.M.; Williams, C.D.; Harrison, S.A. Features, Diagnosis, and Treatment of Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2012, 10, 837–858. [Google Scholar] [CrossRef]
- Basaranoglu, M.; Neuschwander-Tetri, B.A. Nonalcoholic Fatty Liver Disease: Clinical Features and Pathogenesis. Gastroenterol. Hepatol. 2006, 2, 282–291. [Google Scholar]
- El-Garawani, I.; Hassab El-Nabi, S.; El Kattan, A.; Sallam, A.; Elballat, S.; Abou-Ghanima, S.; El Azab, I.H.; R El-Seedi, H.; A M Khalifa, S.; El-Shamy, S. The Ameliorative Role of Acacia Senegal Gum Against the Oxidative Stress and Genotoxicity Induced by the Radiographic Contrast Medium (Ioxitalamate) in Albino Rats. Antioxidants 2021, 10, 221. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Available online: http://gnps.ucsd.edu (accessed on 2 March 2023).
- Chen, H.; Gao, P.; Zhang, M.; Liao, W.; Zhang, J. Synthesis and Biological Evaluation of a Novel Class of β-Carboline Derivatives. New J. Chem. 2014, 38, 4155–4166. [Google Scholar] [CrossRef]
- Sakurai, T.; Chen, Z.; Yamahata, A.; Hayasaka, T.; Satoh, H.; Sekiguchi, H.; Chiba, H.; Hui, S.P. A Mouse Model of Short-Term, Diet-Indced Fatty Liver with Abnormal Cardiolipin Remodeling via Downregulated Tafazzin Gene Expression. J. Sci. Food Agric. 2021, 101, 4995–5001. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sakurai, T.; Chen, Z.; Inoue, N.; Chiba, H.; Hui, S.P. Lysophosphatidylethanolamine Affects Lipid Accumulation and Metabolism in a Human Liver-Derived Cell Line. Nutrients 2022, 14, 579. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Shang, N.; Kerek, E.; Wu, K.; Wu, J. Mitofusion is required for MOTS-c induced GLUT4 translocation. Sci. Rep. 2021, 11, 14291. [Google Scholar] [CrossRef]

| Bean Extracts (BEs) | Total Polyphenol Content (μg/mg) | nmolTE/mg Bean Extracts | Bean Extracts (BEs) | Total Polyphenol Content (μg/mg) | nmolTE/mg Bean Extracts |
|---|---|---|---|---|---|
| BE1 | 7.80 | 4.26 | BE5 | 59.7 | 0.85 |
| BE2 | 11.8 | 3.40 | BE6 | 5.40 | 5.00 |
| BE3 | 41.5 | 5.96 | BE7 | 12.7 | 7.38 |
| BE4 | 40.3 | 3.69 | BE8 | 60.8 | 7.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dibwe, D.F.; Takeishi, N.; Oba, S.; Sakurai, A.; Sakurai, T.; Tsukui, T.; Chiba, H.; Hui, S.-P. Identification of a β-Carboline Alkaloid from Chemoselectively Derived Vanilla Bean Extract and Its Prevention of Lipid Droplet Accumulation in Human Hepatocytes (HepG2). Molecules 2023, 28, 8024. https://doi.org/10.3390/molecules28248024
Dibwe DF, Takeishi N, Oba S, Sakurai A, Sakurai T, Tsukui T, Chiba H, Hui S-P. Identification of a β-Carboline Alkaloid from Chemoselectively Derived Vanilla Bean Extract and Its Prevention of Lipid Droplet Accumulation in Human Hepatocytes (HepG2). Molecules. 2023; 28(24):8024. https://doi.org/10.3390/molecules28248024
Chicago/Turabian StyleDibwe, Dya Fita, Nire Takeishi, Saki Oba, Akiko Sakurai, Toshihiro Sakurai, Takayuki Tsukui, Hitoshi Chiba, and Shu-Ping Hui. 2023. "Identification of a β-Carboline Alkaloid from Chemoselectively Derived Vanilla Bean Extract and Its Prevention of Lipid Droplet Accumulation in Human Hepatocytes (HepG2)" Molecules 28, no. 24: 8024. https://doi.org/10.3390/molecules28248024
APA StyleDibwe, D. F., Takeishi, N., Oba, S., Sakurai, A., Sakurai, T., Tsukui, T., Chiba, H., & Hui, S.-P. (2023). Identification of a β-Carboline Alkaloid from Chemoselectively Derived Vanilla Bean Extract and Its Prevention of Lipid Droplet Accumulation in Human Hepatocytes (HepG2). Molecules, 28(24), 8024. https://doi.org/10.3390/molecules28248024






