Exploring Cinnamoyl-Substituted Mannopyranosides: Synthesis, Evaluation of Antimicrobial Properties, and Molecular Docking Studies Targeting H5N1 Influenza A Virus
Abstract
1. Introduction
2. Results
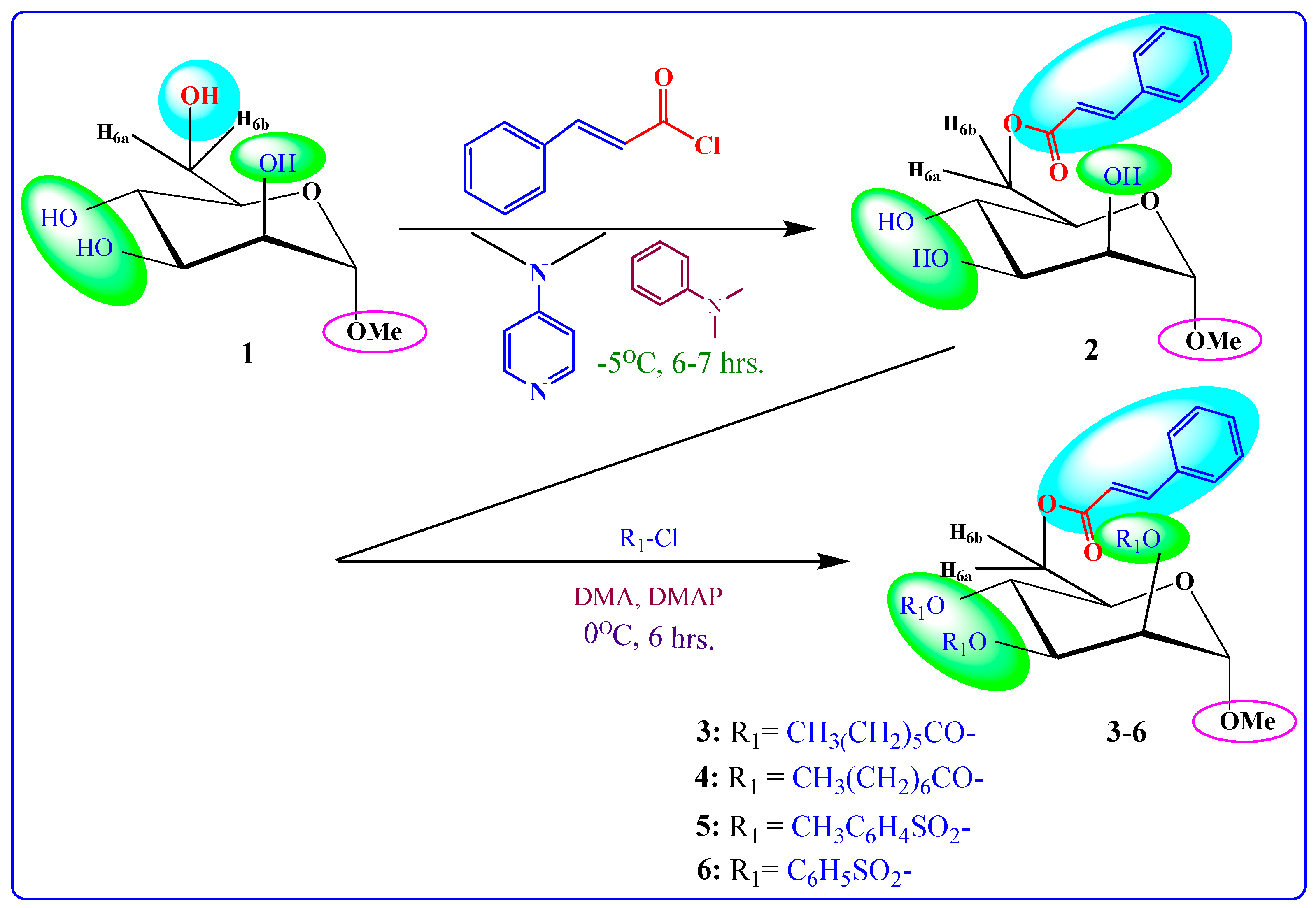
2.1. Chemistry
2.2. Characterization
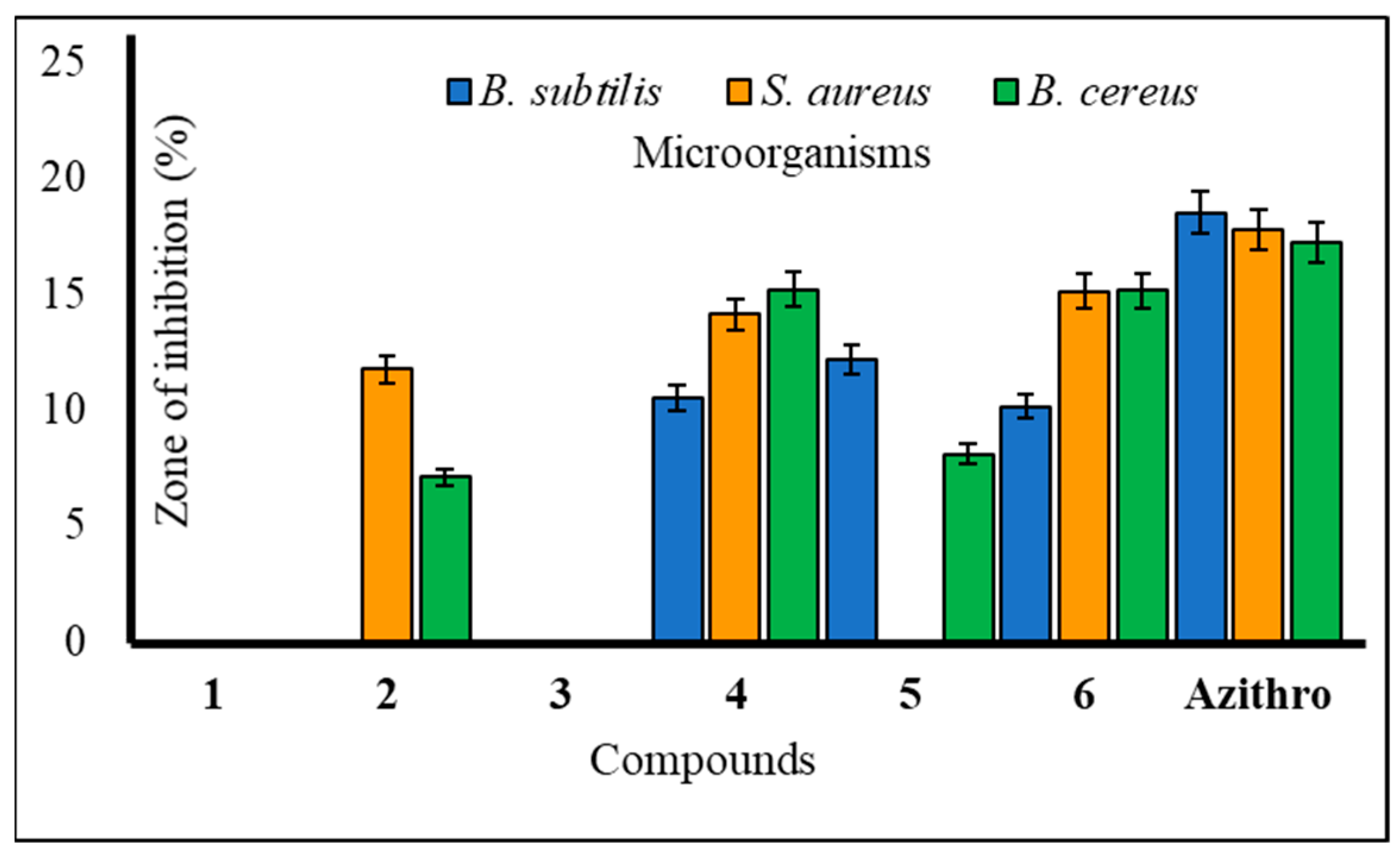
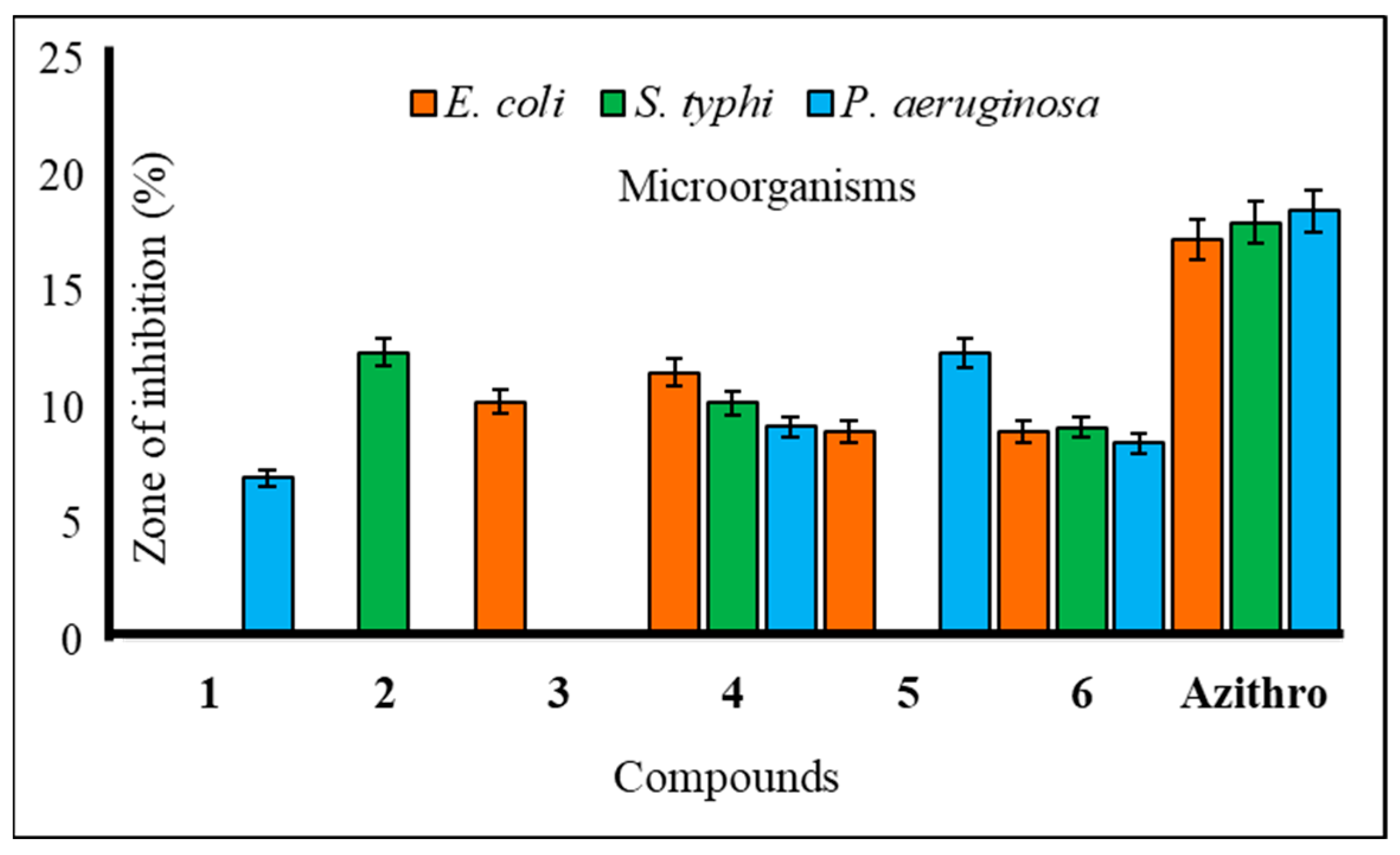
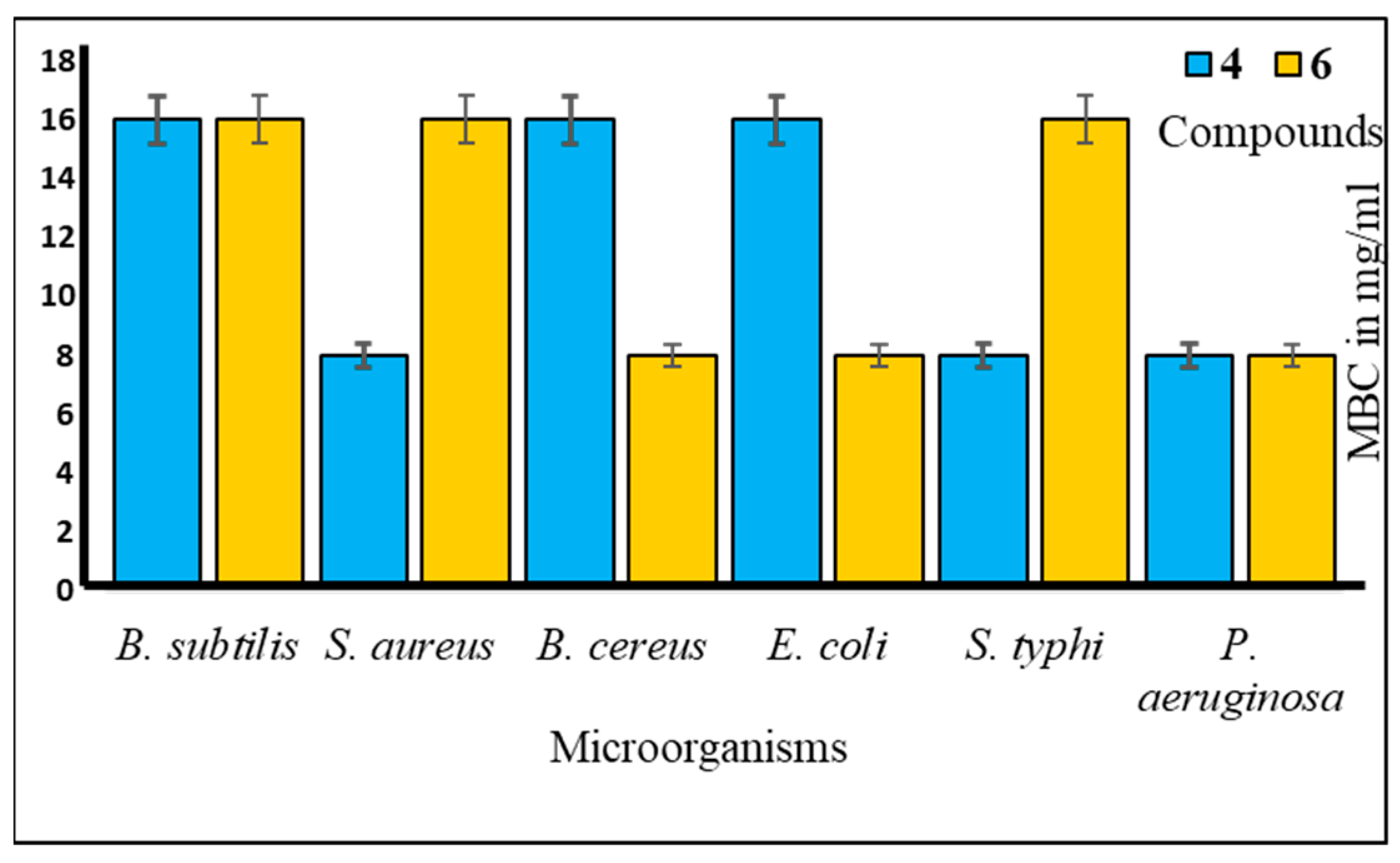
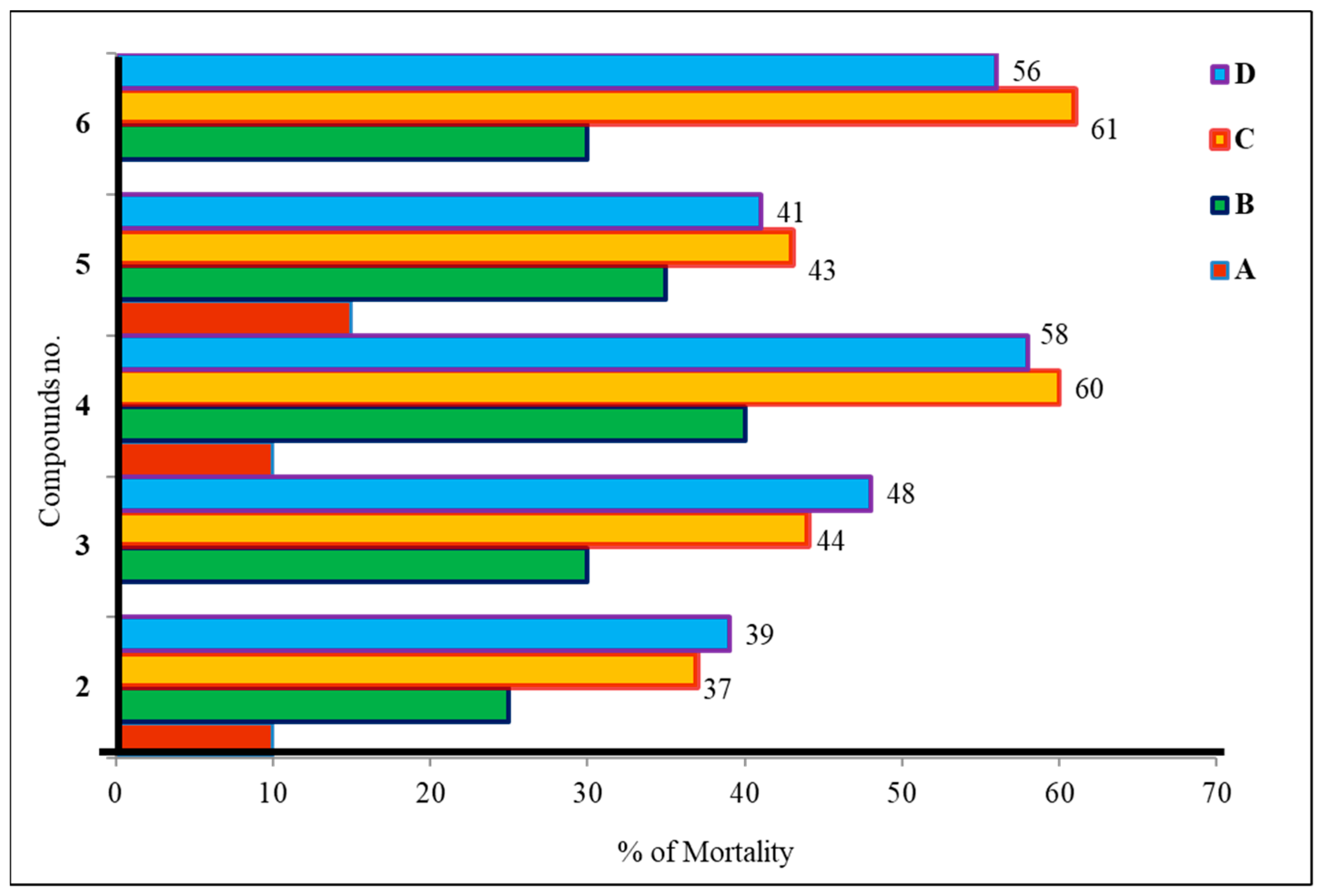
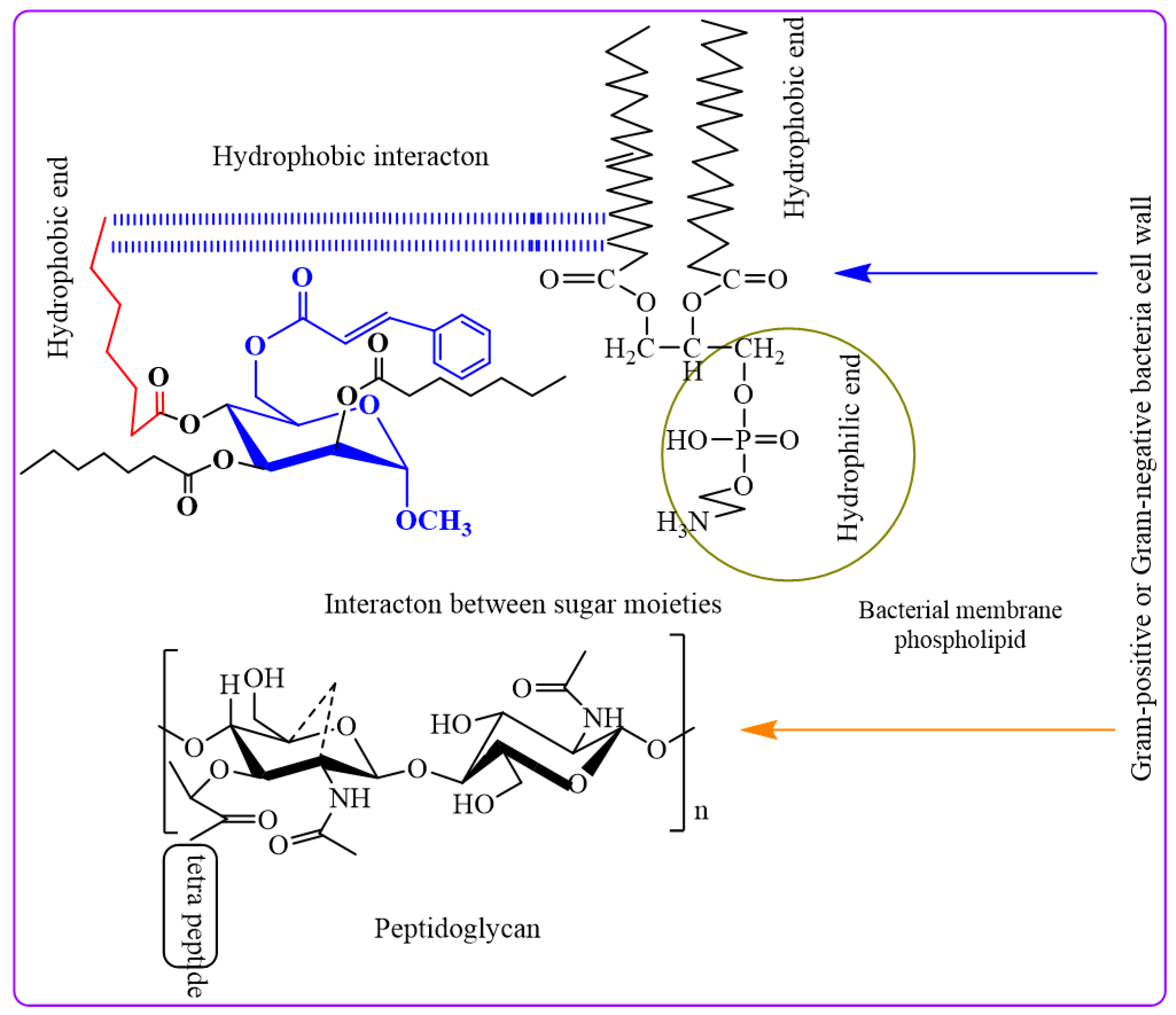
2.3. Antibacterial Potentiality
2.4. Antifungal Susceptibility
2.5. Cytotoxic Activity of Mannopyranoside Compounds
2.6. Structure–Activity Relationship Study
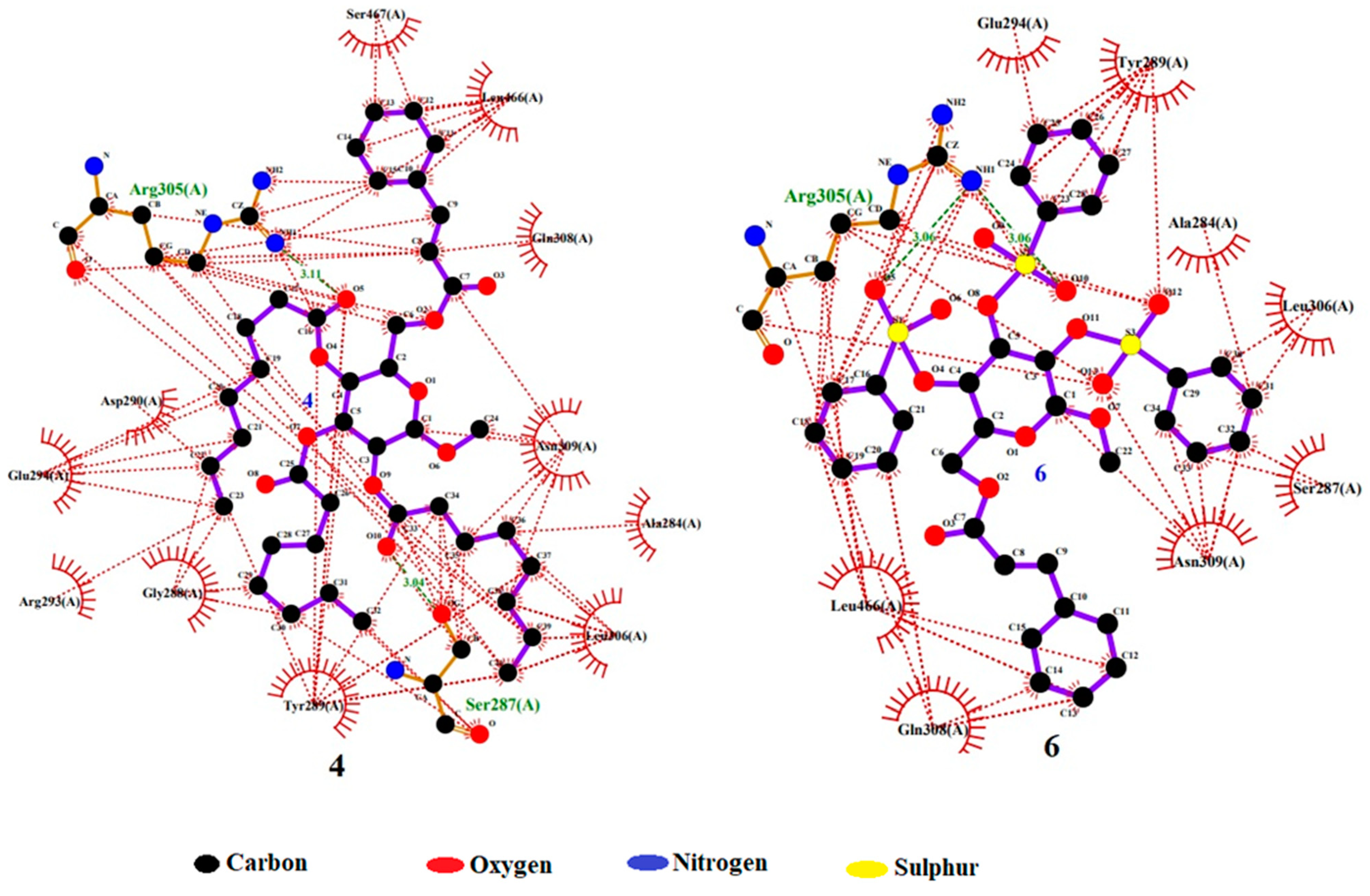
2.7. Molecular Docking Simulation
2.8. Molecular Dynamics
2.9. MM/PBSA Analysis
2.10. Frontier Molecular Orbital (FMO) Analysis
2.11. Molecular Electrostatic Potential (MEP) Investigation
2.12. Pharmacokinetics Properties
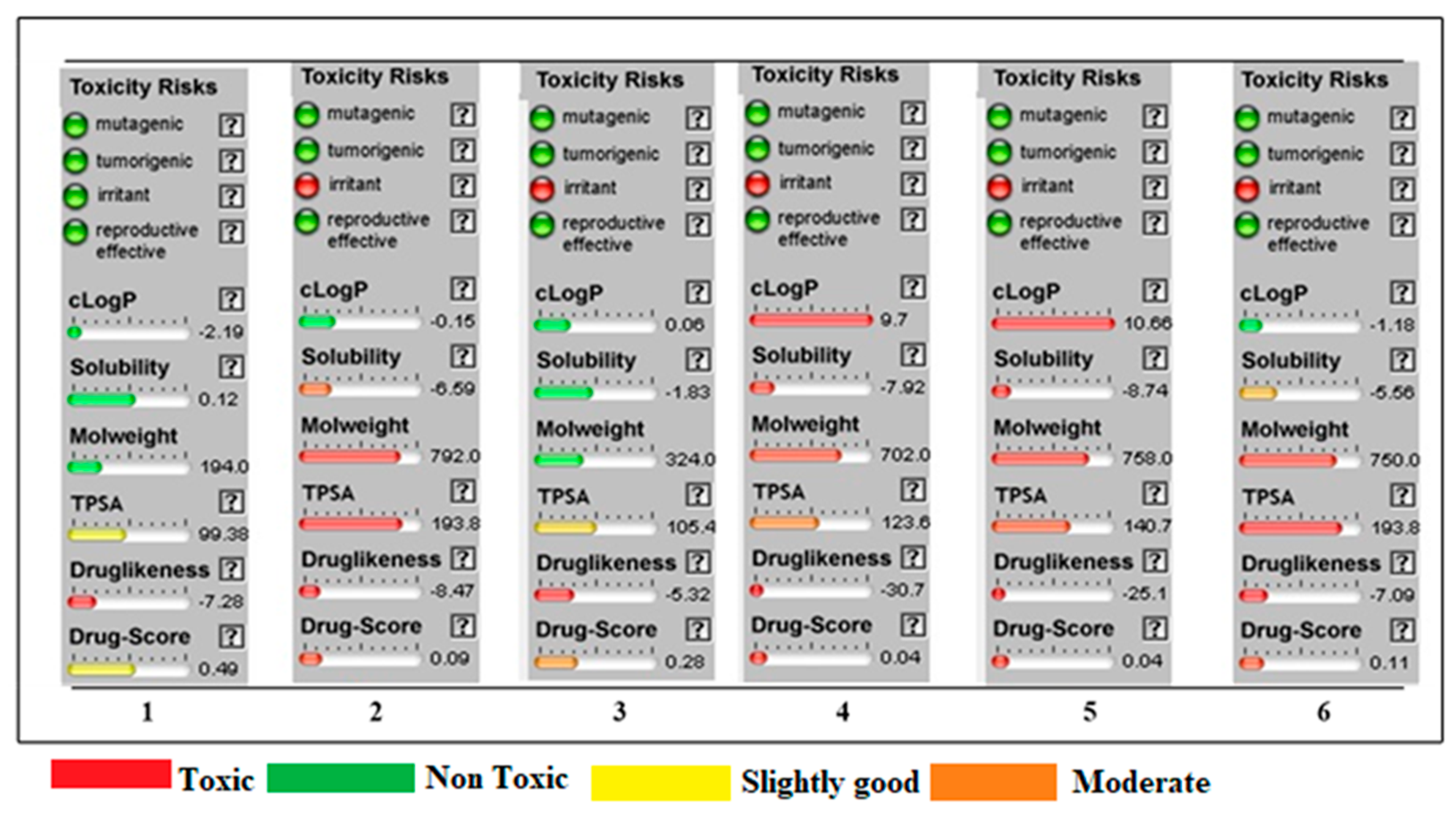
2.13. OSIRIS Data
3. Discussion
4. Materials and Methods
4.1. Material and Equipment
4.2. Synthesis
- Methyl 6-O-cinnamoyl-α-d-mannopyranoside (2)
General Procedure for the Synthesis of Cinnamoyl Derivatives 3–6
- Methyl 2,3,4-tri-O-heptanoyl-6-O-cinnamoyl-α-d-mannopyranoside (3)
- Methyl 6-O-cinnamoyl-2,3,4-tri-O-octanoyl-α-d-mannopyranoside (4)
- Methyl 2,3,4-tri-O-p-toluenesulfonyl-6-O-cinnamoyl-α-d-mannopyranoside (5)
- Methyl 6-O-cinnamoyl-2,3,4-tri-O-benzenesulfonyl-α-d-mannopyranoside (6)
4.3. In Vitro Antimicrobial Activity Test
4.3.1. Test Microorganisms
4.3.2. Antibacterial Activity Evaluation
4.3.3. Determination of MIC and MBC
4.3.4. Evaluation of Mycelial Growth
4.4. Cytotoxic Activity Evaluation
4.5. Structure Activity Relationship (SAR)
4.6. Molecular Docking Studies
4.6.1. Selection and Preparation of the Receptor
4.6.2. Ligand Preparation
4.6.3. Molecular Docking Parameters of the Synthesized Compounds Interacting with 6VMZ
4.7. Molecular Dynamic Simulation
4.8. MM/PBSA Analysis
4.9. Frontier Molecular Orbital (FMO) Analysis
4.10. Molecular Electrostatic Potential (MEP) Investigation
4.11. OSIRIS Calculation
4.12. Pharmacokinetic Prediction
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| DFT | Density functional theory |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
| MD | Molecular dynamics |
| MEP | Molecular electrostatic potential |
| PASS | Prediction of substance activity spectra |
| SAR | Structure–activity relationship |
| FTIR | Fourier-transform infrared |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| SASA | Solvent-accessible surface area |
| MM/PBSA | Molecular mechanics/Poisson–Boltzmann surface area |
| FMO | Frontier molecular orbital |
References
- Porras, G.; Chassagne, F.; Lyles, J.T.; Marquez, L.; Dettweiler, M.; Salam, A.M.; Samarakoon, T.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. Ethnobotany and the role of plant natural products in antibiotic drug discovery. Chem. Rev. 2021, 121, 3495–3560. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Sulaiman, T.; Al-Ahmed, S.H.; Buhaliqah, Z.A.; Buhaliqah, A.A.; AlYuosof, B.; Alfaresi, M.; Al Fares, M.A.; Alwarthan, S.; Alkathlan, M.S. Potential Strategies to Control the Risk of Antifungal Resistance in Humans: A Comprehensive Review. Antibiotics 2023, 12, 608. [Google Scholar] [CrossRef]
- Khaja, U.M.; Bhat, A.H.; Ahmed, M.; Ali, A.; Ganie, S.A. Pharmacogenomics in Viral Diseases. In Pharmacogenomics; Elsevier: Amsterdam, The Netherlands, 2023; pp. 247–269. [Google Scholar]
- Hutchinson, E.C. Influenza virus. Trends Microbiol. 2018, 26, 809–810. [Google Scholar] [CrossRef]
- Watson, J.M.; Francis, J.N.; Mesens, S.; Faiman, G.A.; Makin, J.; Patriarca, P.; Treanor, J.J.; Georges, B.; Bunce, C.J. Characterization of a wild-type influenza (A/H1N1) virus strain as an experimental challenge agent in humans. Virol. J. 2015, 12, 13. [Google Scholar] [CrossRef]
- James, S.H.; Whitley, R.J. Influenza Viruses. In Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1465–1471.e1. [Google Scholar]
- Nypaver, C.; Dehlinger, C.; Carter, C. Influenza and influenza vaccine: A review. J. Midwifery Women’s Health 2021, 66, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.A. Nurse Practitioner Certification Examination and Practice Preparation; FA Davis: Philadelphia, PA, USA, 2017. [Google Scholar]
- Cheng, C.; Dong, J.; Yao, L.; Chen, A.; Jia, R.; Huan, L.; Guo, J.; Shu, Y.; Zhang, Z. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008, 366, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Gaitonde, D.Y.; Moore, F.C.; Morgan, M.K. Influenza: Diagnosis and treatment. Am. Fam. Physician 2019, 100, 751–758. [Google Scholar] [PubMed]
- Jones-Gray, E.; Robinson, E.J.; Kucharski, A.J.; Fox, A.; Sullivan, S.G. Does repeated influenza vaccination attenuate effectiveness? A systematic review and meta-analysis. Lancet Respir. Med. 2022, 11, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Kotalik, J. Preparing for an influenza pandemic: Ethical issues. Bioethics 2005, 19, 422–431. [Google Scholar] [CrossRef]
- Ceriotti, F.; Zakowski, J.; Sine, H.; Altaie, S.; Horowitz, G.; Pesce, A.; Boyd, J.; Horn, P.; Gard, U.; Horowitz, G. Clinical and Laboratory Standards Institute (CLSI). 2012. Available online: https://www.scienceopen.com/document?vid=1d021ccc-8583-4e6b-a47c-8735734df154 (accessed on 30 December 2022).
- ALrajhi, M.; Al-Rasheedi, M.; Eltom, S.E.M.; Alhazmi, Y.; Mustafa, M.M.; Ali, A.M. Antibacterial activity of date palm cake extracts (Phoenix dactylifera). Cogent Food Agric. 2019, 5, 1625479. [Google Scholar] [CrossRef]
- Mikolo, B.; Moyen, R.; Baloki, N.T.; Nguimbi, E. Optimization by mixture design of the antimicrobial activities of five selected essential oils. J. Med. Plant Res. 2020, 14, 570–578. [Google Scholar] [CrossRef]
- Maowa, J.; Alam, A.; Rana, K.M.; Hosen, A.; Dey, S.; Hasan, I.; Fujii, Y.; Ozeki, Y.; Kawsar, S.M.A. Synthesis, characterization, synergistic antimicrobial properties and molecular docking of sugar modified uridine derivatives. Ovidius. Univ. Ann. Chem. 2021, 32, 6–21. [Google Scholar] [CrossRef]
- Afroz, P.; Vijey, A.M.; Gopinath, P. Molecular dynamics simulation approach of hybrid chalcone–thiazole complex derivatives for DNA gyrase B inhibition: Lead generation. RSC Adv. 2023, 13, 24291–24308. [Google Scholar]
- Li, W.R.; Xie, X.B.; Shi, Q.S.; Zeng, H.Y.; Ou-Yang, Y.S.; Chen, Y.B. Antibacterial Activity and Mechanism of Silver Nanoparticles on Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 85, 1115–1122. [Google Scholar] [CrossRef]
- Rana, K.M.; Maowa, J.; Alam, A.; Hosen, A.; Dey, S.; Hasan, I.; Fujii, Y.; Ozeki, Y.; Kawsar, S.M.A. In silico DFT study, molecular docking, and ADMET predictions of cytidine analogs with antimicrobial and anticancer properties. Silico Pharmacol. 2021, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Kholmirzaeva, K.; Shavakat, K. Study of Structure New Organic Compound with Nmr Spectroscopy. Int. J. Innov. Eng. Res. Tech. 2020, 7, 52–55. [Google Scholar]
- Klekota, J.; Roth, F.P. Chemical Substructures that Enrich for Biological Activity. Bioinformatics 2008, 24, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Jumina; Nurmala, A.; Fitria, A.; Pranowo, D.; Sholikhah, E.N.; Kurniawan, Y.S.; Kuswandi, B. Monomyristin and Monopalmitin Derivatives: Synthesis and Evaluation as Potential Antibacterial and Antifungal Agents. Molecules 2018, 23, 3141. [Google Scholar] [CrossRef] [PubMed]
- Janowska, S.; Andrzejczuk, S.; Gawryś, P.; Wujec, M. Synthesis and Antimicrobial Activity of New Mannich Bases with Piperazine Moiety. Molecules 2023, 28, 5562. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Dhanjal, J.K.; Kaul, S.C.; Wadhwa, R.; Sundar, D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease M(pro) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. 2020, 75, 3842–3854. [Google Scholar]
- Bär, W.; Bäde-Schumann, U.; Krebs, A.; Cromme, L. Rapid method for detection of minimal bactericidal concentration of antibiotics. J. Microbiol. Methods 2009, 77, 85–89. [Google Scholar] [CrossRef]
- Ettahiri, W.; Salim, R.; Adardour, M.; Ech-chihbi, E.; Yunusa, I.; Alanazi, M.M.; Lahmidi, S.; Barnossi, A.E.; Merzouki, O.; Iraqi Housseini, A.; et al. Synthesis, Characterization, Antibacterial, Antifungal and Anticorrosion Activities of 1,2,4-Triazolo [1,5-a]quinazolinone. Molecules 2023, 28, 5340. [Google Scholar] [CrossRef]
- Farhana, Y.; Amin, M.R.; Hosen, M.A.; Bulbul, M.Z.H.; Dey, S.; Kawsar, S.M.A. Monosaccharide Derivatives: Synthesis, Antimicrobial, PASS, Antiviral, and Molecular Docking studies Against SARS-CoV-2 Mpro Inhibitors. J. Cellul. Chem. Technol. 2021, 55, 477–499. [Google Scholar]
- Rabasseda, X. Brivudine: A Herpes Virostatic with Rapid Antiviral Activity and Once-Daily Dosing. Drugs Today. 2003, 39, 359–371. [Google Scholar] [CrossRef]
- Martínez-Culebras, P.V.; Gandía, M.; Boronat, A.; Marcos, J.F.; Manzanares, P. Differential susceptibility of mycotoxin-producing fungi to distinct antifungal proteins (AFPs). Food Microbiol. 2021, 97, 103760. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Nobmann, P.; Henehan, G.; Bourke, P.; Dunne, J. Synthesis and Antimicrobial Evaluation of Carbohydrate and Polyhydroxylated Non-Carbohydrate Fatty Acid Ester and Ether Derivatives. Carbohydr. Res. 2008, 343, 2557–2566. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Lee, A.R.; Kim, J.H. Antifungal effect of nanoparticles against COVID-19 linked black fungus: A perspective on biomedical applications. Int. J. Mol. Sci. 2022, 23, 12526. [Google Scholar] [CrossRef]
- Kawsar, S.M.A.; Huq, E.; Nahar, N. Cytotoxicity Assessment of the Aerial Parts of Macrotyloma uniflorum Linn. Int. J. Pharmacol. 2008, 4, 297–300. [Google Scholar] [CrossRef]
- Marinescu, M.; Popa, C.V. Pyridine Compounds with Antimicrobial and Antiviral Activities. Int J Mol Sci. 2022, 23, 5659. [Google Scholar] [CrossRef] [PubMed]
- Kumaresan, S.; Senthilkumar, V.; Stephen, A.; Balakumar, B.S. GC–MS analysis and pass-assisted prediction of biological activity spectra of extract of Phomopsis sp. isolated from Andrographis paniculata. World J. Pharm. Res. 2015, 4, 1035–1053. [Google Scholar]
- Saleh, S.S.; Salihi, S.S.A.; Mohammed, I.A. Biological activity Study for some heterocyclic compounds and their impact on the gram-positive and negative bacteria. Energy Procedia 2019, 157, 296–306. [Google Scholar] [CrossRef]
- Liao, S.; Zhang, Y.; Pan, X.; Zhu, F.; Jiang, C.; Liu, Q.; Cheng, Z.; Dai, G.; Wu, G.; Wang, L.; et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int. J. Nanomed 2019, 14, 1469–1487. [Google Scholar] [CrossRef]
- Judge, V.; Narasimhan, B.; Ahuja, M.; Sriram, D.; Yogeeswari, P.; Clercq, E.D.; Pannecouque, C.; Balzarini, J. Synthesis, antimycobacterial, antiviral, antimicrobial activities, and QSAR studies of isonicotinic acid-1-(substituted phenyl)-ethylidene/cycloheptylidene hydrazides. Med. Chem. Res. 2012, 21, 1935–1952. [Google Scholar] [CrossRef]
- Lien, E.J.; Guo, Z.R.; Li, R.L.; Su, C.T. Use of dipole moment as a parameter in drug-receptor interaction and quantitative structure-activity relationship studies. J. Pharm. Sci. 1982, 71, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Leo, A.; Hansch, C.; Church, C. Comparison of parameters currently used in the study of structure-activity relationships. J. Med. Chem. 1969, 12, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Altharawi, A.; Alanazi, M.M.; Alossaimi, M.A.; Alanazi, A.S.; Alqahtani, S.M.; Geesi, M.H.; Riadi, Y. Novel 2-Sulfanylquinazolin-4(3H)-one Derivatives as Multi-Kinase Inhibitors and Apoptosis Inducers: A Synthesis, Biological Evaluation, and Molecular Docking Study. Molecules 2023, 28, 5548. [Google Scholar] [CrossRef] [PubMed]
- Vishvakarma, V.K.; Singh, M.B.; Jain, P.; Kumari, K.; Singh, P. Hunting the main protease of SARS-CoV-2 by plitidepsin: Molecular docking and temperature-dependent molecular dynamics simulations. Amino Acids 2022, 54, 205–213. [Google Scholar] [CrossRef]
- Liu, H.; Long, S.; Rakesh, K.; Zha, G.-F. Structure-activity relationships (SAR) of triazine derivatives: Promising antimicrobial agents. Eur. J. Med. Chem. 2020, 185, 111804. [Google Scholar] [CrossRef]
- Sheikh, J.; Ben Hadda, T. Antibacterial, antifungal and antioxidant activity of some new water-soluble b-diketones. Med. Chem. Res. 2013, 22, 964–975. [Google Scholar] [CrossRef]
- Cui, T.-M.; Altaf, M.; Aldarhami, A.; Bazaid, A.S.; Saeedi, N.H.; Alkayyal, A.A.; Alshabrmi, F.M.; Ali, F.; Aladhadh, M.; Khan, M.Y.; et al. Dihydropyrimidone Derivatives as Thymidine Phosphorylase Inhibitors: Inhibition Kinetics, Cytotoxicity, and Molecular Docking. Molecules 2023, 28, 3634. [Google Scholar] [CrossRef]
- CLSI Documents M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard-Ninth Edition. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012.
- Bhuyan, P.D.; Tamuli, P.; Boruah, P. In-Vitro Efficacy of Certain Essential Oils and Plant Extracts against Three Major Pathogens of Jatropha curcas L. Am. J. Plant Sci. 2015, 6, 362. [Google Scholar] [CrossRef]
- Bauer, A.; Kirby, W.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Hunt, W.A. The Effects of Aliphatic Alcohols on the Biophysical and Biochemical Correlates of Membrane Function. In Biochemical Pharmacology of Ethanol; Springer: Berlin/Heidelberg, Germany, 1975; pp. 195–210. [Google Scholar]
- Kim, Y.-m.; Farrah, S.; Baney, R.H. Structure—Antimicrobial activity relationship for silanols, a new class of disinfectants, compared with alcohols and phenols. Int. J. Antimicrob. Agents 2007, 29, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhang, Y.; Zhu, H. Structure—Activity Relationship Target Prediction Studies of Clindamycin Derivatives with Broad-Spectrum Bacteriostatic Antibacterial Properties. Molecules 2023, 28, 7357. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, J.A.; Catarello, D.P.; Wozniak, J.M.; Skeel, R.D. Langevin Stabilization of Molecular dynamics. J. Chem. Phys. 2001, 114, 2090–2098. [Google Scholar] [CrossRef]
- Joshi, B.C.; Juyal, V.; Sah, A.N.; Saha, S. Computational Investigation of Geniposidic Acid as an Anticancer Agent Using Molecular Docking, Molecular Dynamic Simulation, DFT Calculation, and OSIRIS-Molinspiration Profiling. Phys. Chem. Res. 2023, 11, 801–823. [Google Scholar]
- Kumari, R.; Kumar, R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- He, X.; Liu, S.; Lee, T.-S.; Ji, B.; Man, V.H.; York, D.M.; Wang, J. Fast, Accurate, and Reliable Protocols for Routine Calculations of Protein–Ligand Binding Affinities in Drug Design Projects Using AMBER GPU-TI with ff14SB/GAFF. ACS Omega 2020, 5, 4611–4619. [Google Scholar] [CrossRef]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A Fast SHAKE Algorithm to Solve Distance Constraint Equations for Small Molecules in Molecular Dynamics Simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H. The FF14SB Force Field. Amber 2014, 14, 29–31. [Google Scholar]
- Pires, D.E.V.; Blundell, T.L.; Ascher, B.D. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]




| Entry | Chemical Structure |
|---|---|
| 2 | 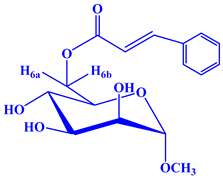 |
| 3 |  |
| 4 | 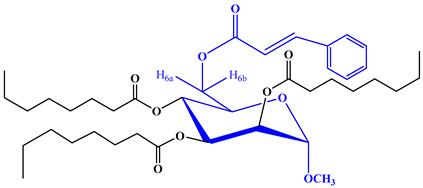 |
| 5 | 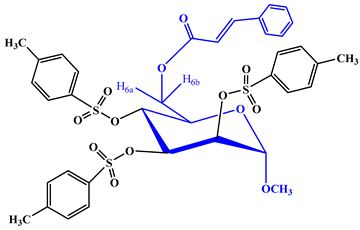 |
| 6 | 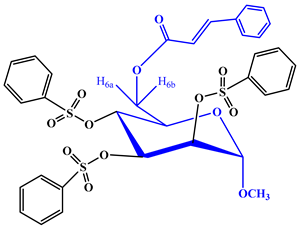 |
| Diameter of Inhibition Zone (mm) | ||||||
|---|---|---|---|---|---|---|
| Entry | B. subtilis (+ve) | S. aureus (+ve) | B. cereus (+ve) | E. coli (−ve) | S.typhi (−ve) | P. aeruginosa (−ve) |
| 1 | NI | NI | NI | NI | NI | NI |
| 2 | NI | 11.75 ± 0.3 | 07.11 ± 0.5 | NI | 12.41 ± 0.3 | NI |
| 3 | NI | NI | NI | 10.25 ± 0.2 | NI | NI |
| 4 | 10.51 ± 0.6 | * 14.10 ± 0.7 | * 15.17 ± 0.4 | 11.53 ± 0.2 | 10.23 ± 0.5 | 09.19 ± 0.6 |
| 5 | 12.15 ± 0.7 | NI | 08.10 ± 0.3 | 9.00 ± 0.2 | NI | 12.36 ± 0.2 |
| 6 | 10.13 ± 0.8 | * 15.12 ± 0.1 | * 15.15 ± 0.3 | 9.00 ± 0.3 | 9.17 ± 0.1 | 8.50 ± 0.2 |
| Azithromycin | ** 18.5 ± 0.3 | ** 17.75 ± 0.3 | ** 17.22 ± 0.3 | ** 17.25 ± 0.1 | ** 18.0 ± 0.2 | ** 18.5 ± 0.3 |
| Entry | % Inhibition of Fungal Mycelial Growth in mm (20 μg/μL) | |
|---|---|---|
| Aspergillus niger | Aspergillus flavus | |
| 1 | NI | NI |
| 2 | NI | * 91.34 ± 1.0 |
| 3 | NI | * 91.12 ± 1.0 |
| 4 | * 96.22 ± 1.1 | * 98.47 ± 1.0 |
| 5 | 57.62 ± 1.1 | * 81.30 ± 1.0 |
| 6 | NI | * 97.02 ± 1.1 |
| Nystatin | ** 65.4 ± 1.0 | ** 64.1 ± 1.0 |
| Entry | Receptors PDB id | Binding Energy (kcal/mol) | Interactive Residues with 4.0 Ǻ |
|---|---|---|---|
| 1. | 6VMZ | (−) 4.3 | ARG 26, ASP 302, GLU 469, and ALA 471 by hydrogen bond interactions; SER 297, ILE 301, and LYS 470 by hydrophobic interactions. |
| 2. | (−) 6.3 | ARG 55 and GLN 311 by hydrogen bond interactions; ALA 284, SER 283, SER 287, TYR 289, LEU 306, ASN 309, and SER 310 by hydrophobic interactions. | |
| 3. | (−) 6.9 | ARG 305 by hydrogen bond interaction; SER 287, GLY 288, TYR 289, GLU 294, LEU 306, GLN 308, ASN 309, and LEU 466 by hydrophobic interactions. | |
| 4. | (−) 7.2 | SER 287 and ARG 305 by hydrogen bond interactions; ALA 284, GLY 288, TYR 289, ASP 290, ARG 293, GLU 294, LEU 306, GLN 308, ASN 309, LEU 466, and SER 467 by hydrophobic interactions. | |
| 5. | (−) 4.7 | ARG 305 by hydrogen bond interaction; GLY 288, TYR 289, ASP 290, ARG 293, GLU 294, LEU 306, GLN 308, ASN 309, LEU 466, and SER 467 by hydrophobic interactions. | |
| 6. | (−) 7.0 | ARG 305 by hydrogen bond interaction; ALA 284, SER 287, TYR 289, GLU 294, LEU 306, GLN 308, ASN 309, and LEU 466 by hydrophobic interactions. |
| Entry | Van der Waal Energy (kJ/mol) | Electrostaticenergy (kJ/mol) | Polar Solvationenergy (kJ/mol) | SASA Energy (kJ/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|---|
| 1 | −3.980 | −2.911 | −16.839 | −0.775 | −24.496 |
| 2 | −20.233 | −8.457 | 14.906 | −3.076 | −16.892 |
| 3 | −0.037 | 0.196 | 17.849 | 0.106 | 17.783 |
| 4 | −0.185 | −158.346 | −39.939 | 0.033 | −198.299 |
| 5 | −0.151 | −1.282 | 24.136 | 0.181 | 22.887 |
| 6 | −104.331 | −23.951 | 113.680 | −13.269 | −27.877 |
| Entry | EHOMO (eV) | ELUMO (eV) | ΔE Gap (eV) | I | A | η | ζ | μ | Ψ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | −7.34 | 0.217 | 7.13 | 7.34 | 0.217 | 3.56 | 0.14 | 3.77 | 1.99 |
| 2 | −6.61 | −2.04 | 4.57 | 6.61 | 2.04 | 2.28 | 0.21 | 4.32 | 4.08 |
| 3 | −6.77 | −2.50 | 4.27 | 6.77 | 2.50 | 2.13 | 0.23 | 4.63 | 5.02 |
| 4 | −6.83 | −2.93 | 3.90 | 6.83 | 1.95 | 2.49 | 0.25 | 4.88 | 6.10 |
| 5 | −6.61 | −2.61 | 4.00 | 6.61 | 2.00 | 2.35 | 0.25 | 4.61 | 5.31 |
| 6 | −6.12 | −2.85 | 3.27 | 6.12 | 2.85 | 1.64 | 0.31 | 4.48 | 6.13 |
| Entry | Water Solubility (log mol/L) | Caco-2 Permeability | Intestinal Absorption | Skin Permeability |
|---|---|---|---|---|
| 2 | −1.673 | 0.31 | 63.236 | −3.437 |
| 3 | −4.937 | 0.784 | 75.372 | −2.729 |
| 4 | −4.408 | 0.748 | 77.499 | −2.733 |
| 5 | −3.822 | −0.216 | 85.269 | −2.735 |
| 6 | −3.781 | −0.439 | 84.047 | −2.735 |
| Entry | Metabolism | Distribution | Execration | ||
|---|---|---|---|---|---|
| CYP2C19 Inhibitior | Vdss (log L/kg) | BBB Permeability | CNS Permeability | Total Clearance | |
| 2 | No | −0.668 | −0.848 | −3.643 | 0.473 |
| 3 | No | −0.404 | −1.858 | −2.81 | 1.559 |
| 4 | No | −0.522 | −1.927 | −2.694 | 1.609 |
| 5 | No | −0.343 | −2.814 | −3.575 | 0.175 |
| 6 | No | −0.446 | −2.778 | −3.726 | 0.384 |
| Entry | Toxicity Risk | Clog P | Solubility | M. Weight | TPSA | Drug Likeness | Drug Score | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mutagenic | Tumorigenic | Irritant | Reproductive effective | −2.19 | 0.12 | 194 | 99.38 | −7.28 | 0.49 |
| 2 | −ve | −ve | −ve | −ve | −0.15 | −6.59 | 792 | 193.8 | −8.47 | 0.09 |
| 3 | −ve | −ve | +ve | −ve | 0.06 | −1.83 | 324 | 105.4 | −5.32 | 0.28 |
| 4 | −ve | −ve | +ve | −ve | 9.7 | −7.92 | 702 | 123.6 | −30.7 | 0.04 |
| 5 | −ve | −ve | +ve | −ve | 10.66 | −8.74 | 758 | 140.7 | −25.1 | 0.04 |
| 6 | −ve | −ve | +ve | −ve | −1.18 | −5.56 | 724 | 193.8 | −7.09 | 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, S.; Alhatlani, B.Y.; Abdallah, E.M.; Saha, S.; Ferdous, J.; Hossain, M.E.; Ali, F.; Kawsar, S.M.A. Exploring Cinnamoyl-Substituted Mannopyranosides: Synthesis, Evaluation of Antimicrobial Properties, and Molecular Docking Studies Targeting H5N1 Influenza A Virus. Molecules 2023, 28, 8001. https://doi.org/10.3390/molecules28248001
Akter S, Alhatlani BY, Abdallah EM, Saha S, Ferdous J, Hossain ME, Ali F, Kawsar SMA. Exploring Cinnamoyl-Substituted Mannopyranosides: Synthesis, Evaluation of Antimicrobial Properties, and Molecular Docking Studies Targeting H5N1 Influenza A Virus. Molecules. 2023; 28(24):8001. https://doi.org/10.3390/molecules28248001
Chicago/Turabian StyleAkter, Sabina, Bader Y. Alhatlani, Emad M. Abdallah, Supriyo Saha, Jannatul Ferdous, Md Emdad Hossain, Ferdausi Ali, and Sarkar M. A. Kawsar. 2023. "Exploring Cinnamoyl-Substituted Mannopyranosides: Synthesis, Evaluation of Antimicrobial Properties, and Molecular Docking Studies Targeting H5N1 Influenza A Virus" Molecules 28, no. 24: 8001. https://doi.org/10.3390/molecules28248001
APA StyleAkter, S., Alhatlani, B. Y., Abdallah, E. M., Saha, S., Ferdous, J., Hossain, M. E., Ali, F., & Kawsar, S. M. A. (2023). Exploring Cinnamoyl-Substituted Mannopyranosides: Synthesis, Evaluation of Antimicrobial Properties, and Molecular Docking Studies Targeting H5N1 Influenza A Virus. Molecules, 28(24), 8001. https://doi.org/10.3390/molecules28248001








