Helium Cold Atmospheric Plasma Causes Morphological and Biochemical Alterations in Candida albicans Cells
Abstract
:1. Introduction
2. Results
2.1. Antifungal Activity of CAP on Candida albicans and Determination of the Minimum Inhibitory Concentration (MIC) of Caspofungin
2.2. Morphological and Topographical Evaluation of Candida albicans Cells Exposed to CAP by Atomic Force Microscopy (AFM)
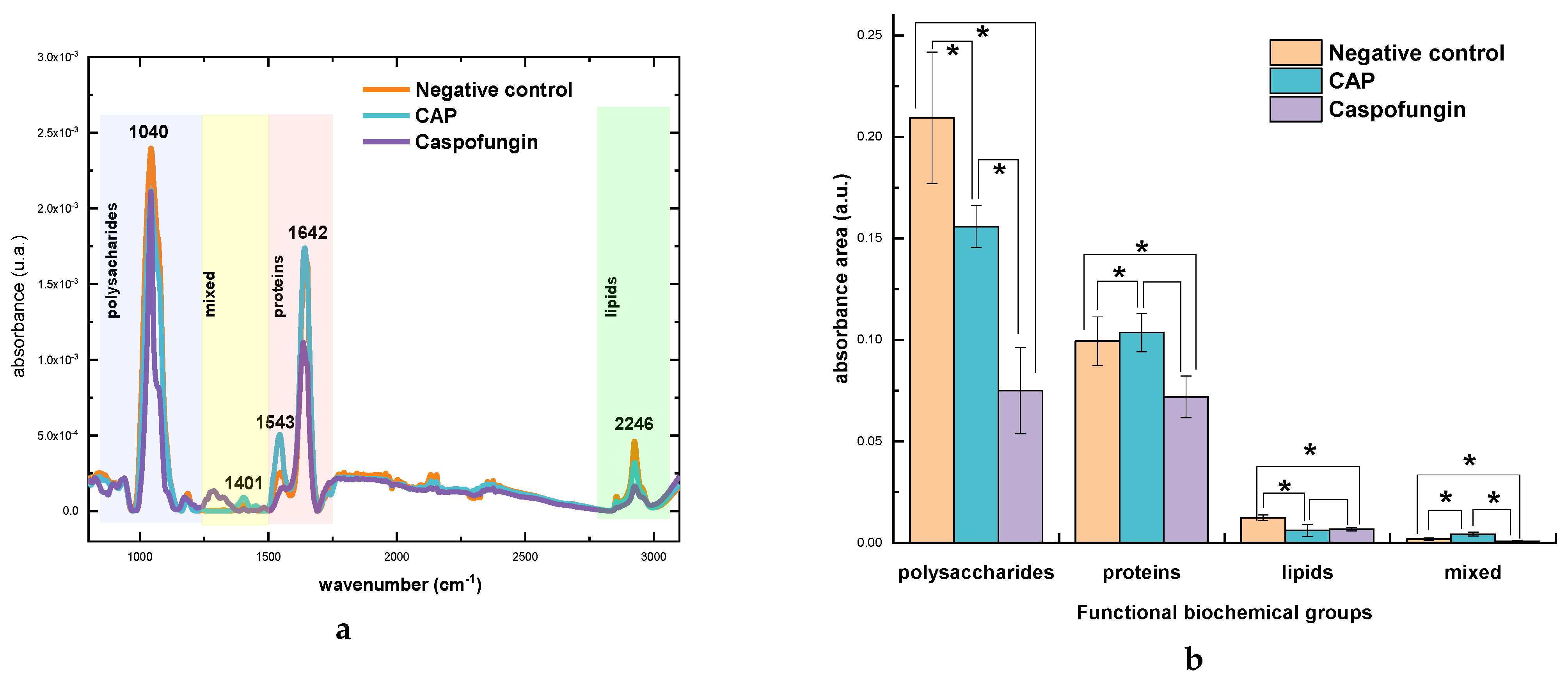
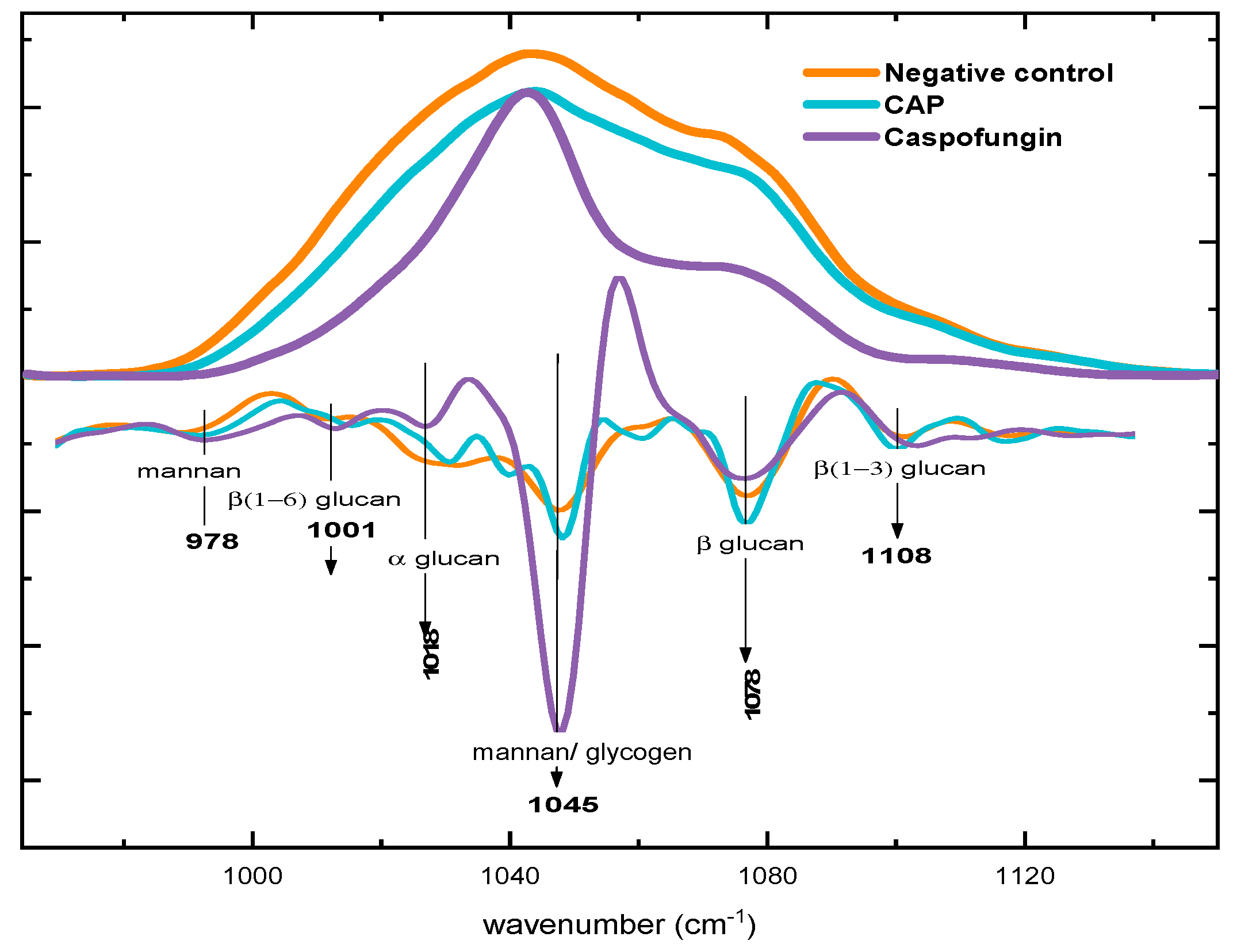
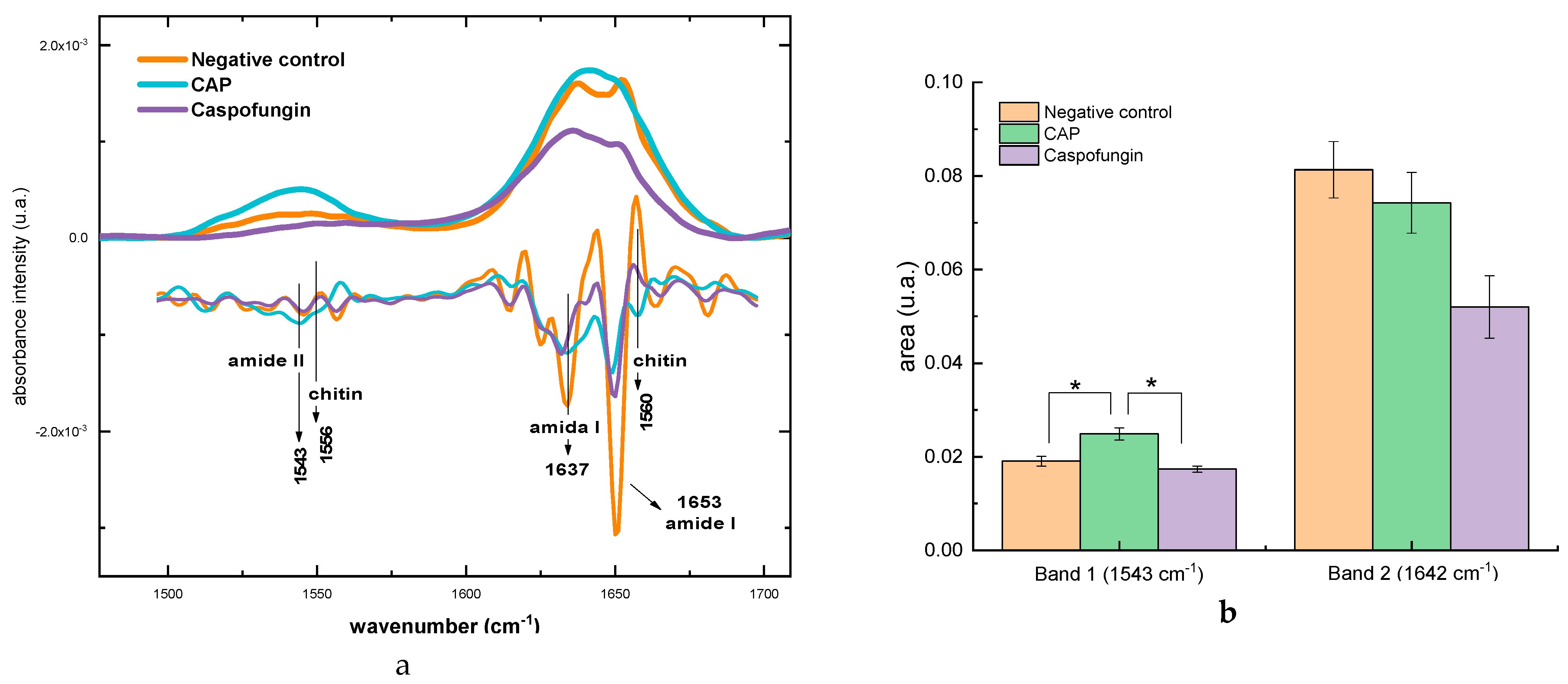
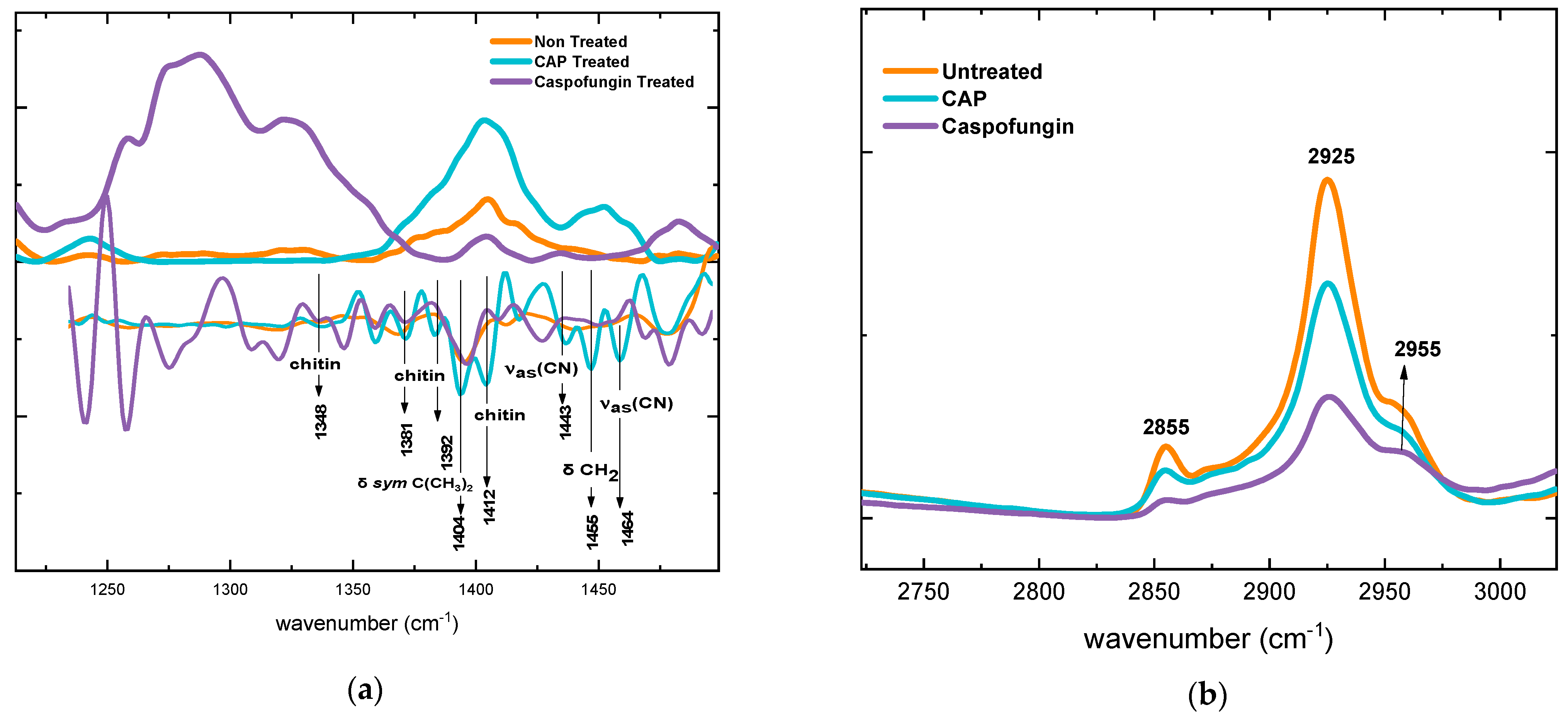
2.3. Biochemical Evaluation of Candida albicans Cell Wall Exposed to CAP by Fourier Transform Infrared Spectroscopy (FTIR)
2.3.1. General Comparison
2.3.2. Alterations in Polysaccharide Content
2.3.3. Alterations in Protein Content
2.3.4. Alterations in the Mixed Region
2.3.5. Alterations in the Lipid Content
3. Materials and Methods
3.1. Plasma Source
3.2. Fungal Strain and Growth Conditions
3.3. Antifungal Activity of CAP
3.4. Determination of Minimum Inhibitory Concentration (MIC) of Caspofungin
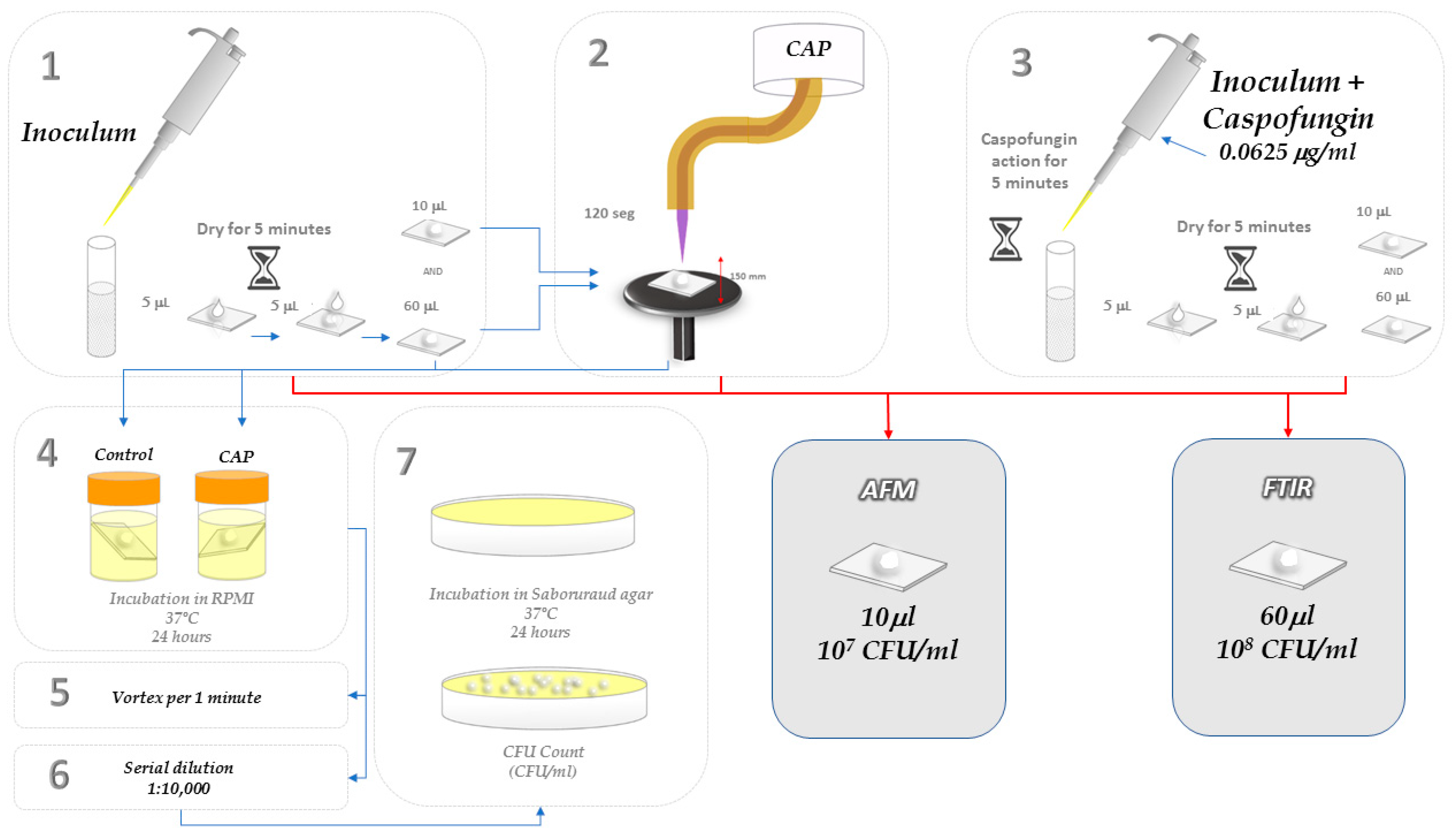
3.5. Morphological and Topographic Analyses of Candida albicans Cells Exposed to CAP by Atomic Force Microscopy (AFM)
3.6. Biochemical Analyses of C. albicans Cells Exposed to CAP by Infrared Spectroscopy with Fourier Transform (FT-IR)
3.7. Statistical Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hasim, S.; Coleman, J.J. Targeting the Fungal Cell Wall: Current Therapies and Implications for Development of Alternative Antifungal Agents. Future Med. Chem. 2019, 11, 869–883. [Google Scholar] [CrossRef]
- Thompson, A.; Griffiths, J.S.; Walker, L.; da Fonseca, D.M.; Lee, K.K.; Taylor, P.R.; Gow, N.A.R.; Orr, S.J. Dependence on Dectin-1 Varies with Multiple Candida Species. Front. Microbiol. 2019, 10, 1800. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, M.F.; Istel, F.; Jenull, S.; Walker, L.A.; Gow, N.; Lion, T. Quantitative Analysis of Candida Cell Wall Components by Flow Cytometry with Triple-Fluorescence Staining. J. Microbiol. Mod. Technol. 2017, 2, 1–9. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Almeida Nogueira, M.F.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M.; et al. Fungal Model Systems and the Elucidation of Pathogenicity Determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.N.; Yau, R.; Lasco, T.M.; Weston, J.; Salazar, M.; Palmer, H.R.; Garey, K.W. Impact of Prior Inappropriate Fluconazole Dosing on Isolation of Fluconazole-Nonsusceptible Candida Species in Hospitalized Patients with Candidemia. Antimicrob. Agents Chemother. 2012, 56, 3239–3243. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Huang, X.; Liu, Y.; Ni, T.; Li, L.; Yan, L.; An, M.; Zhang, D.; Jiang, Y. 11g, a Potent Antifungal Candidate, Enhances Candida albicans Immunogenicity by Unmasking β-Glucan in Fungal Cell Wall. Front. Microbiol. 2020, 11, 1324. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Lu, C.; Gong, Y.; Li, M.; Sun, S. Promising Antifungal Targets against Candida albicans Based on Ion Homeostasis. Front. Cell. Infect. Microbiol. 2018, 8, 286. [Google Scholar] [CrossRef]
- Bersani, I.; Piersigilli, F.; Goffredo, B.M.; Santisi, A.; Cairoli, S.; Ronchetti, M.P.; Auriti, C. Antifungal Drugs for Invasive Candida Infections (ICI) in Neonates: Future Perspectives. Front. Pediatr. 2019, 7, 375. [Google Scholar] [CrossRef]
- Aldardeer, N.F.; Albar, H.; Al-Attas, M.; Eldali, A.; Qutub, M.; Hassanien, A.; Alraddadi, B. Antifungal Resistance in Patients with Candidaemia: A Retrospective Cohort Study. BMC Infect. Dis. 2020, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Costa-de-Oliveira, S.; Rodrigues, A.G. Candida albicans Antifungal Resistance and Tolerance in Bloodstream Infections: The Triad Yeast-Host-Antifungal. Microorganisms 2020, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Flynn, P.B.; Gilmore, B.F. Understanding Plasma Biofilm Interactions for Controlling Infection and Virulence. J. Phys. D Appl. Phys. 2018, 51, 263001. [Google Scholar] [CrossRef]
- Franco, O.L.; Parachin, N.S.; Monteiro, C.A.; Br, C.M.; Moffa, E.B.; Terças, A.L.G.; Marques, S.G.; Alves, M.B.; De Azevedo, C.M.P.S.; Siqueira, W.L. Antifungal Drug Susceptibility of Candida Species Isolated from HIV-Positive Patients Recruited at a Public Hospital in São Luís, Maranhão, Brazil. Front. Microbiol. 2017, 8, 298. [Google Scholar] [CrossRef]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond. Chem. Rev. 2020, 121, 3390–3411. [Google Scholar] [CrossRef]
- Nami, S.; Aghebati-Maleki, A.; Morovati, H.; Aghebati-Maleki, L. Current Antifungal Drugs and Immunotherapeutic Approaches as Promising Strategies to Treatment of Fungal Diseases. Biomed. Pharmacother. 2019, 110, 857–868. [Google Scholar] [CrossRef]
- Jabra-Rizk, M.A.; Falkler, W.A.; Meiller, T.F. Fungal Biofilms and Drug. Emerg. Infect. Dis. 2004, 10, 10–19. [Google Scholar] [CrossRef]
- Mumtaz, S.; Khan, R.; Rana, J.N.; Javed, R.; Iqbal, M.; Choi, E.H.; Han, I. Review on the Biomedical and Environmental Applications of Nonthermal Plasma. Catalysts 2023, 13, 685. [Google Scholar] [CrossRef]
- Von Woedtke, T.; Schmidt, A.; Bekeschus, S.; Wende, K.; Weltmann, K.D. Plasma Medicine: A Field of Applied Redox Biology. In Vivo 2019, 33, 1011–1026. [Google Scholar] [CrossRef]
- Bourke, P.; Ziuzina, D.; Han, L.; Cullen, P.J.; Gilmore, B.F. Microbiological Interactions with Cold Plasma. J. Appl. Microbiol. 2017, 123, 308–324. [Google Scholar] [CrossRef]
- Winter, T.; Winter, J.; Polak, M.; Kusch, K.; Mäder, U.; Sietmann, R.; Ehlbeck, J.; Van Hijum, S.; Weltmann, K.D.; Hecker, M.; et al. Characterization of the Global Impact of Low Temperature Gas Plasma on Vegetative Microorganisms. Proteomics 2011, 11, 3518–3530. [Google Scholar] [CrossRef]
- Tyczkowska-Sieroń, E.; Kałużewski, T.; Grabiec, M.; Kałużewski, B.; Tyczkowski, J. Genotypic and Phenotypic Changes in Candida albicans as a Result of Cold Plasma Treatment. Int. J. Mol. Sci. 2020, 21, 8100. [Google Scholar] [CrossRef]
- Kostov, K.G.; Nishime, T.M.C.; Machida, M.; Borges, A.C.; Prysiazhnyi, V.; Koga-Ito, C.Y. Study of Cold Atmospheric Plasma Jet at the End of Flexible Plastic Tube for Microbial Decontamination. Plasma Process. Polym. 2015, 12, 1383–1391. [Google Scholar] [CrossRef]
- Kostov, K.G.; Borges, A.C.; Koga-Ito, C.Y.; Nishime, T.M.C.; Prysiazhnyi, V.; Honda, R.Y. Inactivation of Candida albicans by Cold Atmospheric Pressure Plasma Jet. IEEE Trans. Plasma Sci. 2015, 43, 770–775. [Google Scholar] [CrossRef]
- Singh, S.; Halder, A.; Abdul Mohid, S.K.; Bagchi, D.; Sinha, O.; Banerjee, A.; Kumar Sarkar, P.; Bhunia, A.; Ghosh, S.K.; Mitra, A.; et al. Nonthermal Atmospheric Plasma-Induced Cellular Envelope Damage of Staphylococcus aureus and Candida albicans Biofilms: Spectroscopic and Biochemical Investigations. IEEE Trans. Plasma Sci. 2020, 48, 2768–2776. [Google Scholar] [CrossRef]
- He, M.; Duan, J.; Xu, J.; Ma, M.; Chai, B.; He, G.; Gan, L.; Zhang, S.; Duan, X.; Lu, X.; et al. Candida albicans Biofilm Inactivated by Cold Plasma Treatment In Vitro and In Vivo. Plasma Process. Polym. 2020, 17, 1900068. [Google Scholar] [CrossRef]
- Handorf, O.; Weihe, T.; Bekeschus, S.; Graf, A.C.; Schnabel, U.; Riedel, K.; Ehlbeck, J. Nonthermal Plasma Jet Treatment Negatively Affects the Viability and Structure of Candida albicans SC5314 Biofilms. Appl. Environ. Microbiol. 2018, 84, e01163-18. [Google Scholar] [CrossRef] [PubMed]
- Matthes, R.; Duske, K.; Kebede, T.; Pink, C.; Schlüter, R.; von Woedtke, T.; Weltmann, K.-D.; Kocher, T.; Jablonowski, L. Osteoblast Growth after Cleaning of Biofilm Covered Titanium Discs with Air-Polishing and Cold Plasma Running Title: Treated Biofilm and Its Effects on Cells. J. Clin. Periodontol. 2016, 44, 672–680. [Google Scholar] [CrossRef]
- Wang, G.M.; Sun, P.P.; Pan, H.; Ye, G.P.; Sun, K.; Zhang, J.; Pan, J.; Fang, J. Inactivation of Candida albicans Biofilms on Polymethyl Methacrylate and Enhancement of the Drug Susceptibility by Cold Ar/O2 Plasma Jet. Plasma Chem. Plasma Process. 2016, 36, 383–396. [Google Scholar] [CrossRef]
- Fricke, K.; Koban, I.; Tresp, H.; Jablonowski, L.; Schröder, K.; Kramer, A.; Weltmann, K.D.; von Woedtke, T.; Kocher, T. Atmospheric Pressure Plasma: A High-Performance Tool for the Efficient Removal of Biofilms. PLoS ONE 2012, 7, e42539. [Google Scholar] [CrossRef]
- Borges, A.C.; de Morais Gouvêa Lima, G.; Mayumi Castaldelli Nishime, T.; Vidal Lacerda Gontijo, A.; Kostov, K.G.; Koga-Ito, C.Y. Amplitude-Modulated Cold Atmospheric Pressure Plasma Jet for Treatment of Oral Candidiasis: In Vivo Study. PLoS ONE 2018, 13, e0199832. [Google Scholar] [CrossRef]
- Chiodi Borges, A.; Castaldelli Nishime, T.M.; Kostov, K.G.; de Morais Gouvêa Lima, G.; Vidal Lacerda Gontijo, A.; Nóbrega Martins Marchesotti de Carvalho, J.; Yzumi Honda, R.; Koga-Ito, C.Y. Cold Atmospheric Pressure Plasma Jet Modulates Candida albicans Virulence Traits. Clin. Plasma Med. 2017, 7–8, 9–15. [Google Scholar] [CrossRef]
- Rahimi-Verki, N.; Shapoorzadeh, A.; Razzaghi-Abyaneh, M.; Atyabi, S.M.; Shams-Ghahfarokhi, M.; Jahanshiri, Z.; Gholami-Shabani, M. Cold Atmospheric Plasma Inhibits the Growth of Candida albicans by Affecting Ergosterol Biosynthesis and Suppresses the Fungal Virulence Factors In Vitro. Photodiagn. Photodyn. Ther. 2016, 13, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Van De Veerdonk, F.L.; Brown, A.J.P.; Netea, M.G. Candida albicans Morphogenesis and Host Defence: Discriminating Invasion from Colonization. Nat. Rev. Microbiol. 2012, 10, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Bain, J.M.; Louw, J.; Lewis, L.E.; Okai, B.; Walls, C.A.; Ballou, E.R.; Walker, L.A.; Reid, D.; Munro, C.A.; Brown, A.J.; et al. Candida albicans Hypha Formation and Mannan Masking of β-Glucan Inhibit Macrophage Phagosome Maturation. Eur. J. Med. Res. 2014, 5, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Ne, Z.; Malovíková, A.; Bystrick, S. Preparation and Characterization of Cationic and Amphoteric Mannans from Candida albicans. Carbohydr. Polym. 2016, 149, 1–7. [Google Scholar] [CrossRef]
- Perez-Garcia, L.A. Role of Cell Wall Polysaccharides during Recognition of Candida albicans by the Innate Immune System. J. Glycobiol. 2012, 1. [Google Scholar] [CrossRef]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.R.; Munro, C.A.; Brown, A.J.P. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986-15. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Majumdar, S. Echinocandins in Antifungal Pharmacotherapy. J. Pharm. Pharmacol. 2017, 69, 1635–1660. [Google Scholar] [CrossRef]
- Arroyo, J.; Farka, V.; Sanz, A.B.; Cabib, E. Strengthening the Fungal Cell Wall through Chitin-Glucan Cross-Links: Effects on Morphogenesis and Cell Integrity. Cell Microbiol. 2016, 18, 1239–1250. [Google Scholar] [CrossRef]
- Toubas, D.; Essendoubi, M.; Adt, I.; Pinon, J.-M.; Manfait, M.; Sockalingum, G.D. FTIR Spectroscopy in Medical Mycology: Applications to the Differentiation and Typing of Candida. Anal. Bioanal. Chem. 2007, 387, 1729–1737. [Google Scholar] [CrossRef]
- Adt, I.; Toubas, D.; Pinon, J.M.; Manfait, M.; Sockalingum, G.D. FTIR Spectroscopy as a Potential Tool to Analyse Structural Modifications during Morphogenesis of Candida albicans. Arch. Microbiol. 2006, 185, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Bekiaris, G.; Tagkouli, D.; Koutrotsios, G.; Kalogeropoulos, N.; Zervakis, G.I. Pleurotus Mushrooms Content in Glucans and Ergosterol Assessed by ATR-FTIR Spectroscopy and Multivariate Analysis. Foods 2020, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Quiles, F.; Accoceberry, I.; Couzigou, C.; Francius, G.; Noel, T.; El-Kirat-Chatel, S. AFM Combined to ATR-FTIR Reveals Candida Cell Wall Changes under Caspofungin Treatment. Nanoscale 2017, 9, 13731–13738. [Google Scholar] [CrossRef]
- Balan, V.; Mihai, C.T.; Cojocaru, F.D.; Uritu, C.M.; Dodi, G.; Botezat, D.; Gardikiotis, I. Vibrational Spectroscopy Fingerprinting in Medicine: From Molecular to Clinical Practice. Materials 2019, 12, 2884. [Google Scholar] [CrossRef]
- Shi, Q.Y.; Câmara, C.R.S.; Schlegel, V. Biochemical Alterations of: Candida albicans during the Phenotypic Transition from Yeast to Hyphae Captured by Fourier Transform Mid-Infrared-Attenuated Reflectance Spectroscopy. Analyst 2018, 143, 5404–5416. [Google Scholar] [CrossRef]
- Oliver, K.V.; Maréchal, A.; Rich, P.R. Effects of the Hydration State on the Mid-Infrared Spectra of Urea and Creatinine in Relation to Urine Analyses. Appl. Spectrosc. 2016, 70, 983–994. [Google Scholar] [CrossRef]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The Estimation of the Bactericidal Power of the Blood. J. Hyg. 1938, 38, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Aparecida Delben, J.; Evelin Zago, C.; Tyhovych, N.; Duarte, S.; Eduardo Vergani, C. Effect of Atmospheric-Pressure Cold Plasma on Pathogenic Oral Biofilms and In Vitro Reconstituted Oral Epithelium. PLoS ONE 2016, 11, e0155427. [Google Scholar] [CrossRef]
- Pawłat, J.; Kwiatkowski, M.; Terebun, P.; Chudzik, B.; Gagoś, M. Candida albicans Inactivation with DBD He/O2 Plasma Jet. In Proceedings of the 2017 International Conference on Electromagnetic Devices and Processes in Environment Protection with Seminar Applications of Superconductors (ELMECO & AoS), Naleczow, Poland, 3–6 December 2017. [Google Scholar]
- El-Kirat-Chatel, S.; Beaussart, A.; Alsteens, D.; Jackson, D.N.; Lipke, P.N.; Dufrêne, Y.F. Nanoscale Analysis of Caspofungin-Induced Cell Surface Remodelling in Candida albicans. Nanoscale 2013, 5, 1105–1115. [Google Scholar] [CrossRef]
- Formosa, C.; Schiavone, M.; Martin-Yken, H.; François, J.M.; Duval, R.E.; Dague, E. Nanoscale Effects of Caspofungin against Two Yeast Species, Saccharomyces Cerevisiae and Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 3498–3506. [Google Scholar] [CrossRef] [PubMed]
- Francois, J.M.; Formosa, C.; Schiavone, M.; Pillet, F.; Martin-Yken, H.; Dague, E. Use of Atomic Force Microscopy (AFM) to Explore Cell Wall Properties and Response to Stress in the Yeast Saccharomyces Cerevisiae. Curr. Genet. 2013, 59, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Nastuta, A.V.; Topala, I.; Grigoras, C.; Pohoata, V.; Popa, G. Stimulation of Wound Healing by Helium Atmospheric Pressure Plasma Treatment. J. Phys. D Appl. Phys. 2011, 44, 105204. [Google Scholar] [CrossRef]
- Ryan, T.P.; Stalder, K.R.; Woloszko, J. Antimicrobial Outcomes in Plasma Medicine. In Proceedings of the Energy-Based Treatment of Tissue and Assessment VIII, San Francisco, CA, USA, 7–12 February 2015; SPIE: Bellingham, WA, USA, 2015; Volume 9326, p. 932606. [Google Scholar]
- Pemmaraju, S.C.; Padmapriya, K.; Pruthi, P.A.; Prasad, R.; Pruthi, V. Impact of Oxidative and Osmotic Stresses on Candida albicans Biofilm Formation. Biofouling 2016, 32, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Karkowska-Kuleta, J.; Satala, D.; Bochenska, O.; Rapala-Kozik, M.; Kozik, A. Moonlighting Proteins Are Variably Exposed at the Cell Surfaces of Candida glabrata, Candida parapsilosis and Candida tropicalis under Certain Growth Conditions. BMC Microbiol. 2019, 19, 149. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Fujarte, I.; López-Romero, E.; Cuéllar-Cruz, M. Moonlight-like Proteins of the Cell Wall Protect Sessile Cells of Candida from Oxidative Stress. Microb. Pathog. 2016, 90, 22–33. [Google Scholar] [CrossRef]
- Medrano-Díaz, C.L.; Vega-González, A.; Ruiz-Baca, E.; Moreno, A.; Cuéllar-Cruz, M. Moonlighting Proteins Induce Protection in a Mouse Model against Candida Species. Microb. Pathog. 2018, 124, 21–29. [Google Scholar] [CrossRef]
- Ramírez-Quijas, M.D.; López-Romero, E.; Cuéllar-Cruz, M. Proteomic Analysis of Cell Wall in Four Pathogenic Species of Candida Exposed to Oxidative Stress. Microb. Pathog. 2015, 87, 1–12. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Victoria Elorza, M.; Valentín, E.; Sentandreu, R. Molecular Organization of the Cell Wall of Candida albicans and Its Relation to Pathogenicity. FEMS Yeast Res. 2006, 6, 14–29. [Google Scholar] [CrossRef]
- Nishime, T.M.C.; Wagner, R.; Kostov, K.G. Study of Modified Area of Polymer Samples Exposed to a He Atmospheric Pressure Plasma Jet Using Different Treatment Conditions. Polymers 2020, 12, 1028. [Google Scholar] [CrossRef]
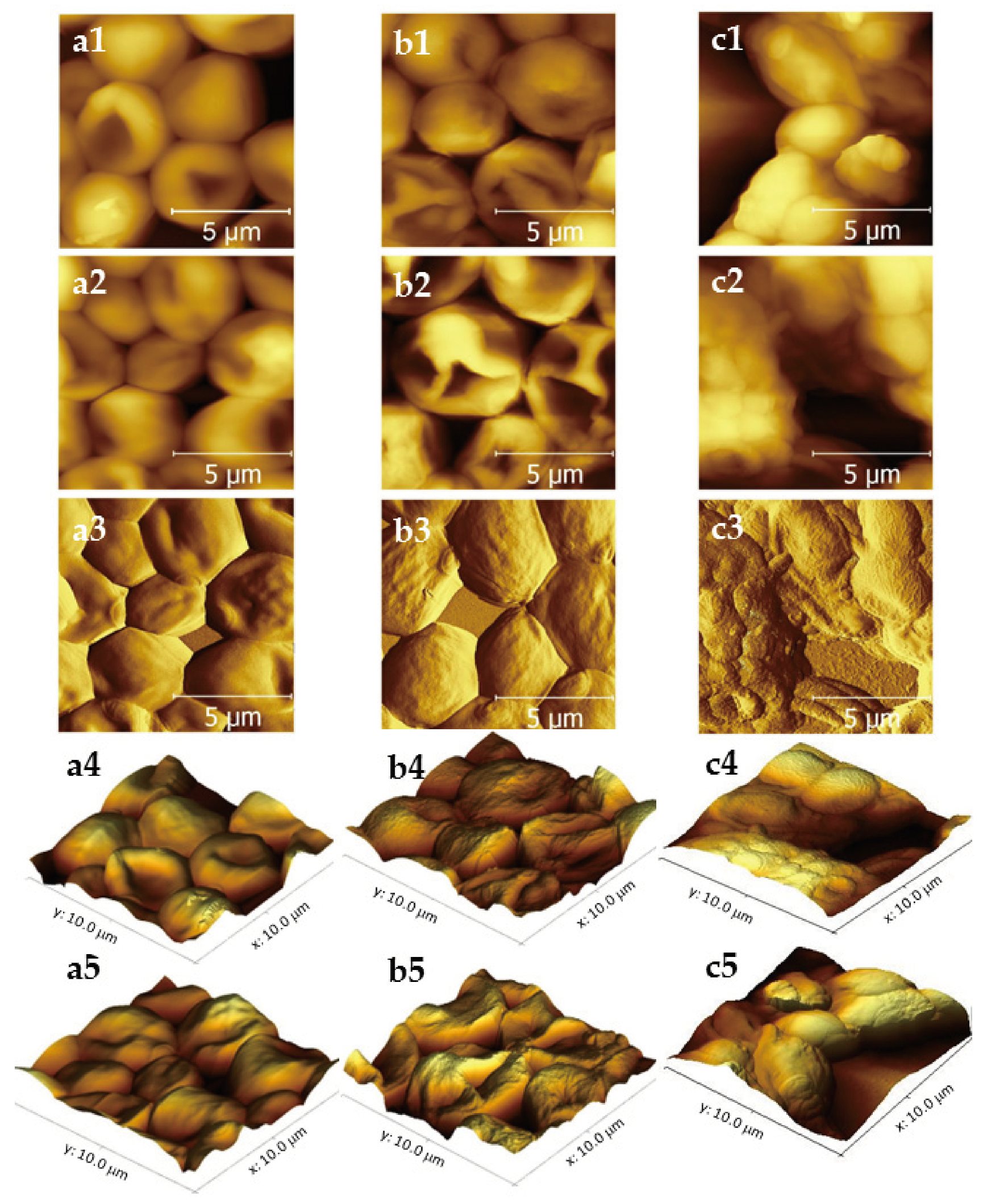
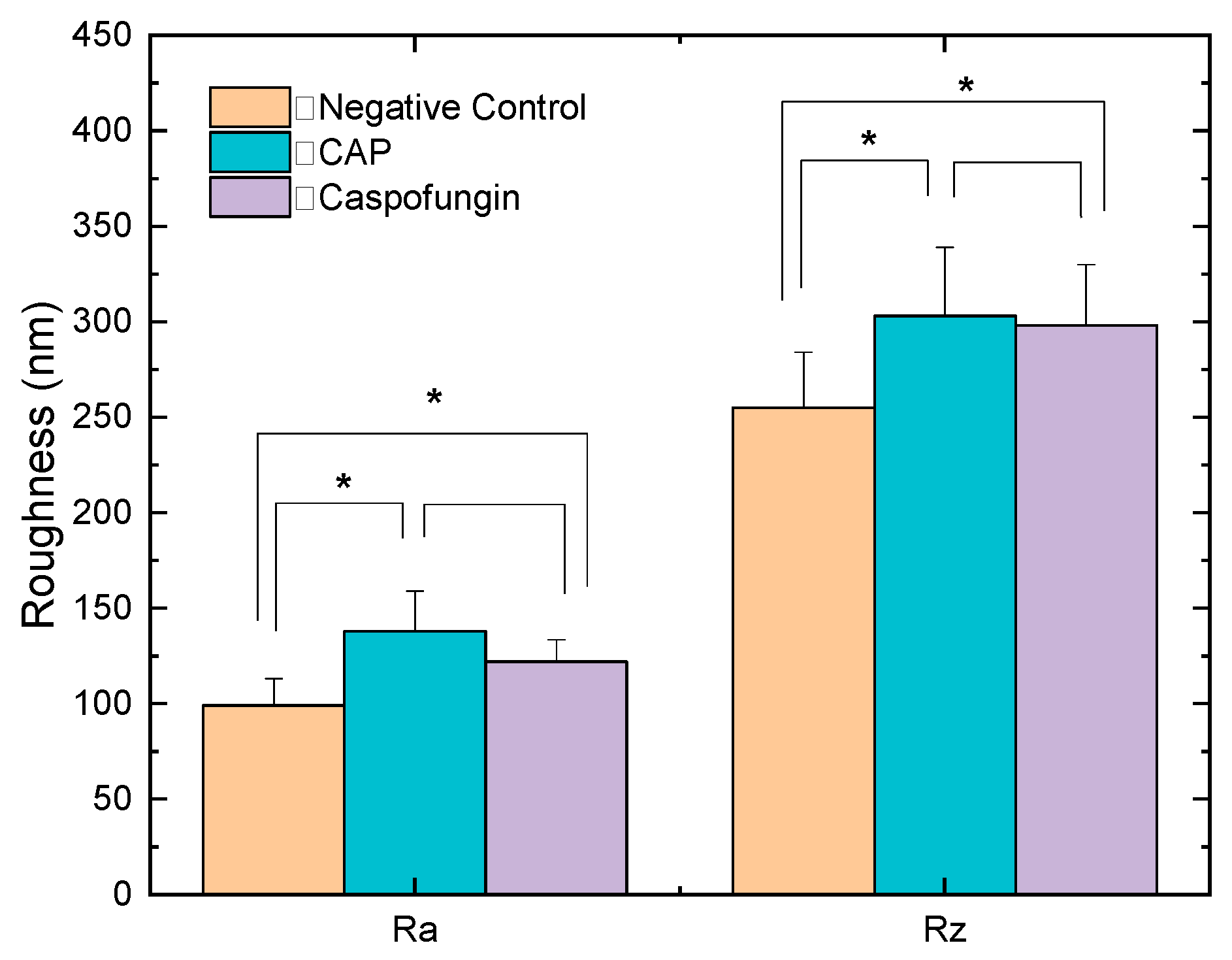
| Groups | Cell Size (μm) | Ra Roughness (nm) | Rz Roughness (nm) | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| CAP | 3.41 | 0.3827 | 138 * | 21 | 303 * | 36 |
| Caspofungin | 2.53 | 0.2998 | 122 * | 11.5 | 298 * | 32 |
| Negative control | 3.94 | 0.1067 | 99 | 14 | 255 | 29 |
| Functional Groups | CAP | Caspofungin | Negative Control | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Polysaccharides | 0.156 * | 0.01034 | 0.075 * | 0.02134 | 0.209 | 0.03235 |
| Proteins | 0.104 * | 0.10358 | 0.072 * | 0.01029 | 0.099 | 0.01203 |
| Lipids | 0.006 * | 0.00617 | 0.007 * | 8.68001 × 10−4 | 0.012 | 0.00133 |
| Mixed region | 0.004 * | 0.00429 | 0.0007 * | 6.22024 × 10−4 | 0.002 | 6.55526 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rovetta-Nogueira, S.d.M.; Borges, A.C.; Oliveira Filho, M.d.; Nishime, T.M.C.; Hein, L.R.d.O.; Kostov, K.G.; Koga-Ito, C.Y. Helium Cold Atmospheric Plasma Causes Morphological and Biochemical Alterations in Candida albicans Cells. Molecules 2023, 28, 7919. https://doi.org/10.3390/molecules28237919
Rovetta-Nogueira SdM, Borges AC, Oliveira Filho Md, Nishime TMC, Hein LRdO, Kostov KG, Koga-Ito CY. Helium Cold Atmospheric Plasma Causes Morphological and Biochemical Alterations in Candida albicans Cells. Molecules. 2023; 28(23):7919. https://doi.org/10.3390/molecules28237919
Chicago/Turabian StyleRovetta-Nogueira, Sabrina de Moura, Aline Chiodi Borges, Maurício de Oliveira Filho, Thalita Mayumi Castaldelli Nishime, Luis Rogerio de Oliveira Hein, Konstantin Georgiev Kostov, and Cristiane Yumi Koga-Ito. 2023. "Helium Cold Atmospheric Plasma Causes Morphological and Biochemical Alterations in Candida albicans Cells" Molecules 28, no. 23: 7919. https://doi.org/10.3390/molecules28237919
APA StyleRovetta-Nogueira, S. d. M., Borges, A. C., Oliveira Filho, M. d., Nishime, T. M. C., Hein, L. R. d. O., Kostov, K. G., & Koga-Ito, C. Y. (2023). Helium Cold Atmospheric Plasma Causes Morphological and Biochemical Alterations in Candida albicans Cells. Molecules, 28(23), 7919. https://doi.org/10.3390/molecules28237919







