The Conformational Change of the L3 Loop Affects the Structural Changes in the Substrate Binding Pocket Entrance of β-Glucosidase
Abstract
:1. Introduction
2. Results
2.1. Structure Determination
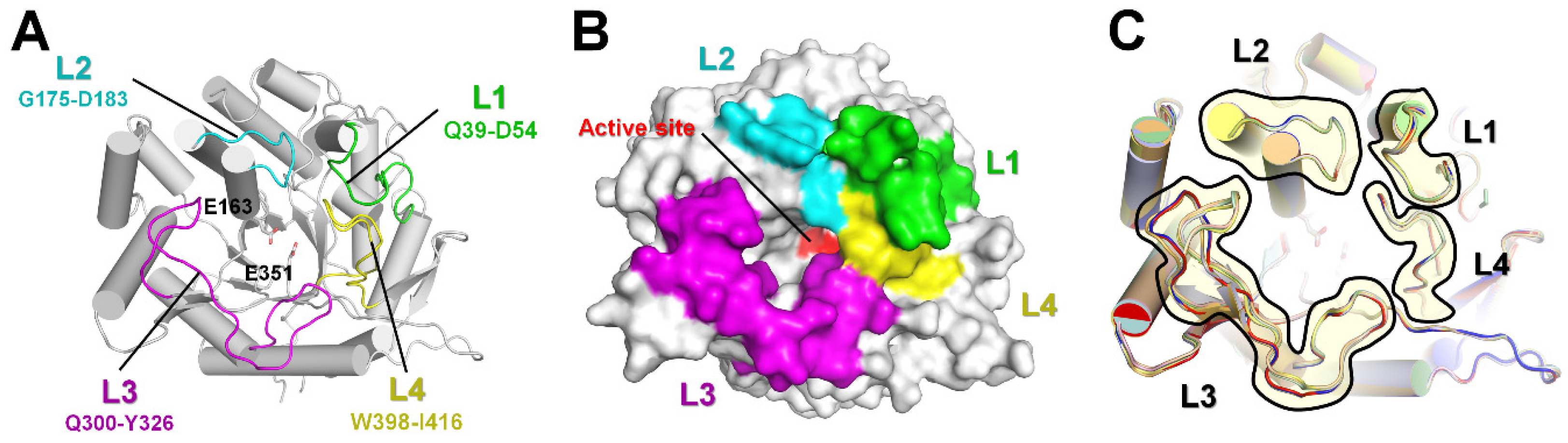
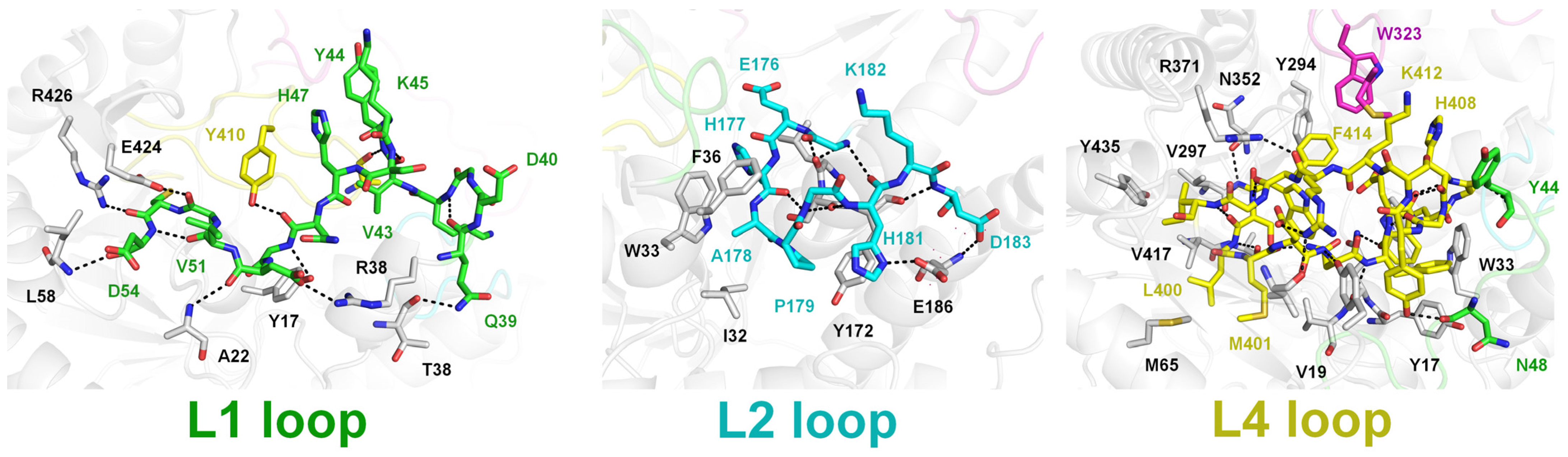
2.2. Structural Dissection of Loops above the TsaBgl Substrate-Binding Pocket
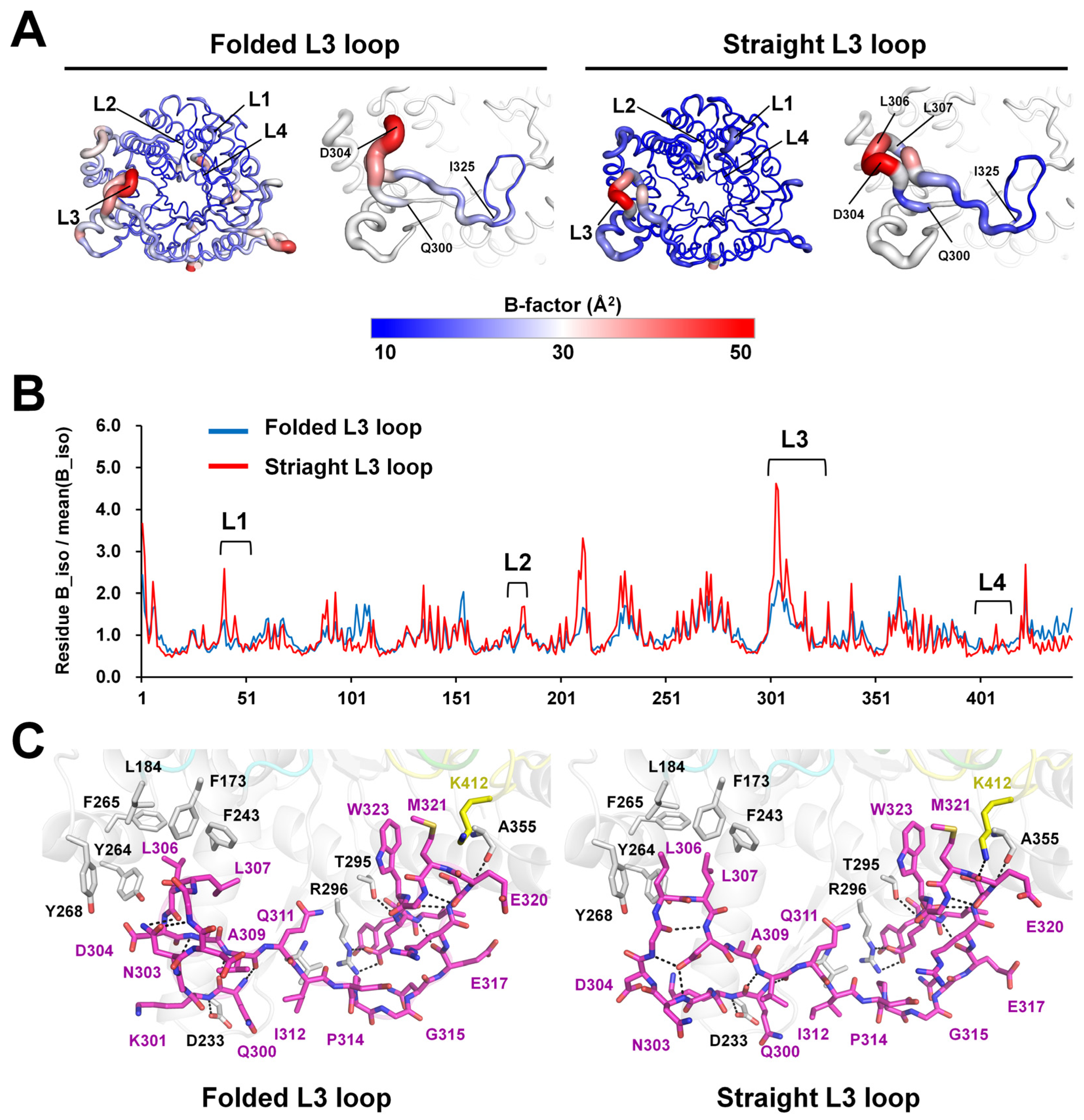
2.3. L3 Loop Conformation-Induced Structural Differences
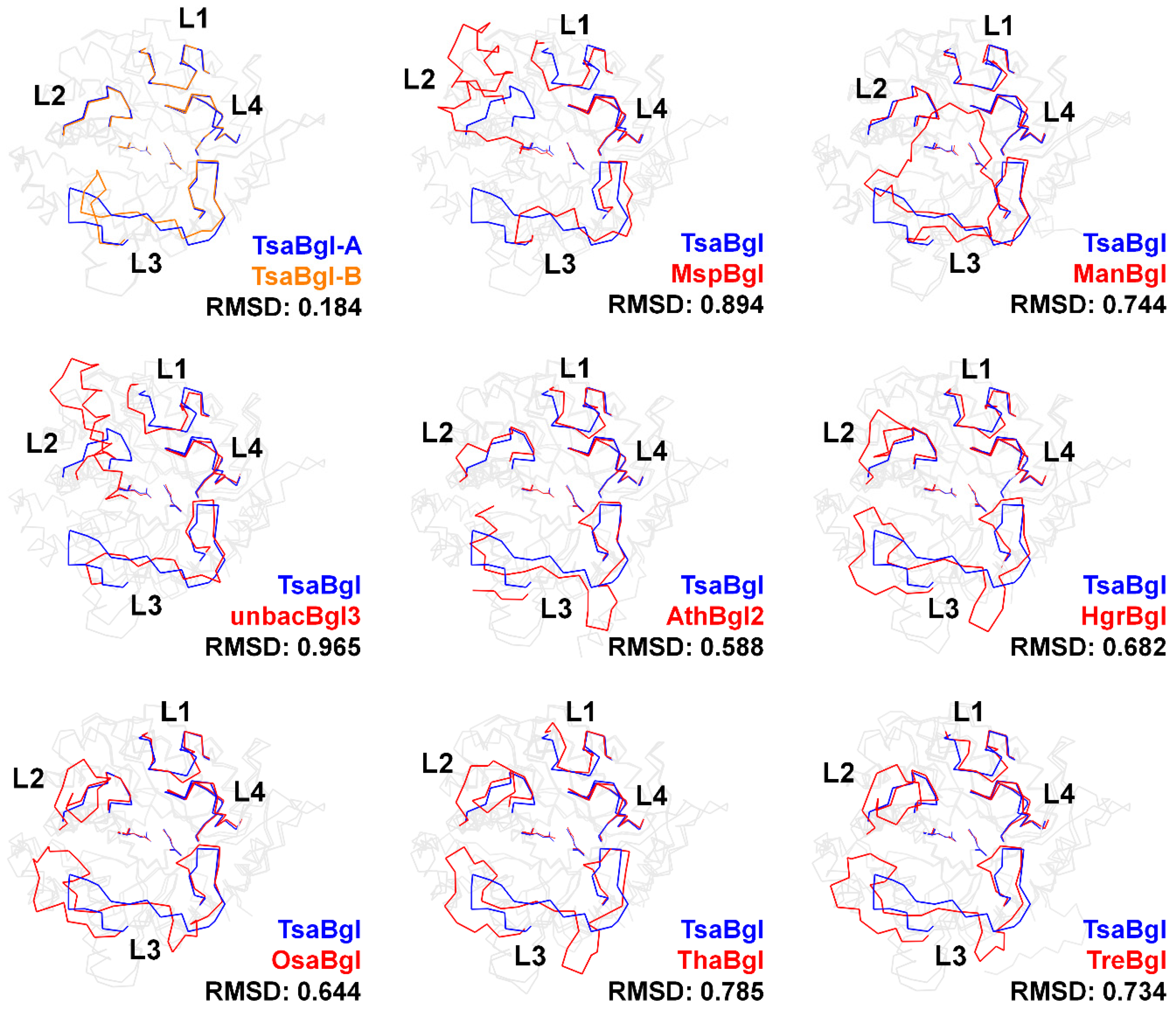
2.4. Analysis of the Loop Region in Bgl Family Enzymes
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Crystallization
4.3. X-ray Diffraction Data Collection
4.4. Structure Determination
4.5. Analysis of the Bgl Loop Region
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ouyang, B.; Wang, G.; Zhang, N.; Zuo, J.; Huang, Y.; Zhao, X. Recent Advances in β-Glucosidase Sequence and Structure Engineering: A Brief Review. Molecules 2023, 28, 4990. [Google Scholar] [CrossRef]
- Goswami, S.; Gupta, N.; Datta, S. Using the β-glucosidase catalyzed reaction product glucose to improve the ionic liquid tolerance of β-glucosidases. Biotechnol. Biofuels 2016, 9, 72. [Google Scholar] [CrossRef]
- Sternberg, D.; Vuayakumar, P.; Reese, E.T. β-Glucosidase: Microbial production and effect on enzymatic hydrolysis of cellulose. Can. J. Microbiol. 1977, 23, 139–147. [Google Scholar] [CrossRef]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose tolerant and glucose stimulated β-glucosidases—A review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech. 2016, 6, 3. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, R.; Yang, X.; Zhang, Z.; Song, S.; Miao, Y.; Shen, Q. Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microb. Cell Factories 2012, 11, 25. [Google Scholar] [CrossRef]
- Lee, S.-M.; Jin, L.H.; Kim, J.H.; Han, S.O.; Na, H.B.; Hyeon, T.; Koo, Y.-M.; Kim, J.; Lee, J.-H. β-Glucosidase coating on polymer nanofibers for improved cellulosic ethanol production. Bioprocess Biosyst. Eng. 2009, 33, 141–147. [Google Scholar] [CrossRef]
- Konar, S.; Sinha, S.K.; Datta, S.; Ghorai, P.K. Probing the Effect of Glucose on the Activity and Stability of β-Glucosidase: An All-Atom Molecular Dynamics Simulation Investigation. ACS Omega 2019, 4, 11189–11196. [Google Scholar] [CrossRef]
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421. [Google Scholar] [CrossRef]
- Yanase, H.; Nozaki, K.; Okamoto, K. Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnol. Lett. 2005, 27, 259–263. [Google Scholar] [CrossRef]
- Sengupta, S.; Datta, M.; Datta, S. β-Glucosidase: Structure, function and industrial applications. In Glycoside Hydrolases; Goyal, A., Sharma, K., Eds.; Elsevier: Amsterdam, Netherlands, 2023; pp. 97–120. [Google Scholar] [CrossRef]
- Sharma, J.; Kumar, V.; Prasad, R.; Gaur, N.A. Engineering of Saccharomyces cerevisiae as a consolidated bioprocessing host to produce cellulosic ethanol: Recent advancements and current challenges. Biotechnol. Adv. 2022, 56, 107925. [Google Scholar] [CrossRef]
- Nam, K.H.; Kim, S.J.; Kim, M.Y.; Kim, J.H.; Yeo, Y.S.; Lee, C.M.; Jun, H.K.; Hwang, K.Y. Crystal structure of engineered β-glucosidase from a soil metagenome. Proteins 2008, 73, 788–793. [Google Scholar] [CrossRef]
- Godse, R.; Bawane, H.; Tripathi, J.; Kulkarni, R. Unconventional β-Glucosidases: A Promising Biocatalyst for Industrial Biotechnology. Appl. Biochem. Biotechnol. 2021, 193, 2993–3016. [Google Scholar] [CrossRef] [PubMed]
- Czjzek, M.; Cicek, M.; Zamboni, V.; Bevan, D.R.; Henrissat, B.; Esen, A. The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 13555–13560. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Sung, M.W.; Hwang, K.Y. Structural insights into the substrate recognition properties of β-glucosidase. Biochem. Biophys. Res. Commun. 2010, 391, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.; Bornscheuer, U.T.; Nam, K.H. Biochemical and Structural Analysis of a Glucose-Tolerant β-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum. Molecules 2022, 27, 290–305. [Google Scholar] [CrossRef]
- Miao, L.-L.; Hou, Y.-J.; Fan, H.-X.; Qu, J.; Qi, C.; Liu, Y.; Li, D.-F.; Liu, Z.-P.; Schottel, J.L. Molecular Structural Basis for the Cold Adaptedness of the Psychrophilic β-Glucosidase BglU in Micrococcus antarcticus. Appl. Environ. Microbiol. 2016, 82, 2021–2030. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]

| Data | Data I | Data II | Data III | Data IV |
|---|---|---|---|---|
| Wavelength (Å) | 0.9864 | 0.9864 | 0.9864 | 0.9864 |
| Space group | P1 | P1 | P212121 | P212121 |
| Unit cell (Å) | ||||
| a | 63.414 | 63.213 | 64.970 | 65.066 |
| b | 72.831 | 72.739 | 70.948 | 71.211 |
| c | 97.490 | 97.385 | 98.876 | 99.151 |
| α | 92.495 | 92.484 | 90.000 | 90.000 |
| β | 81.281 | 91.431 | 90.000 | 90.000 |
| γ | 95.128 | 95.215 | 90.000 | 90.000 |
| Molecule/asym. | 4 | 4 | 1 | 1 |
| Resolution (Å) a | 50.00–1.90 (1.93–1.90) | 50.00–2.10 (2.14–2.10) | 50.00–1.50 (1.53–1.50) | 50.00–1.60 (1.63–1.60) |
| Unique reflections | 123,062 (6212) | 90,740 (4342) | 71,920 (3026) | 60,041 (2923) |
| Completeness (%) | 90.7 (91.5) | 90.9 (87.2) | 97.4 (83.5) | 98.8 (97.8) |
| Redundancy | 3.7 (3.4) | 3.7 (3.4) | 4.1 (2.4) | 5.7 (4.6) |
| Mean I/σ(I) | 10.78 (1.90) | 9.02 (1.92) | 15.41 (2.47) | 14.63 (2.15) |
| CC1/2 | 0.974 (0.563) | 0.977 (0.706) | 0.973 (0.771) | 0.981 (0.585) |
| CC* | 0.993 (0.849) | 0.994 (0.910) | 0.993 (0.933) | 0.995 (0.859) |
| Refinement | ||||
| Resolution (Å) | 49.89–1.90 | 49.81–2.10 | 49.44–1.50 | 49.58–1.61 |
| Rwork | 0.1694 | 0.1599 | 0.1546 | 0.1506 |
| Rfree | 0.1952 | 0.2019 | 0.1712 | 0.1762 |
| RMS deviations | ||||
| Bonds (Å) | 0.003 | 0.004 | 0.003 | 0.009 |
| Angles (degree) | 0.632 | 0.670 | 0.648 | 0.963 |
| B factor (Å2) | ||||
| Protein | 21.99 | 23.91 | 12.28 | 13.40 |
| Water | 31.42 | 31.42 | 27.10 | 27.97 |
| Ramachandran plot (%) | ||||
| Most favored | 96.99 | 96.48 | 97.74 | 97.96 |
| Allowed | 2.90 | 3.46 | 2.26 | 2.04 |
| Outliers | 0.11 | 0.06 | 0.0 | 0.0 |
| PDB code | 8WFT | 8WFU | 8WFV | 8WFW |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. The Conformational Change of the L3 Loop Affects the Structural Changes in the Substrate Binding Pocket Entrance of β-Glucosidase. Molecules 2023, 28, 7807. https://doi.org/10.3390/molecules28237807
Nam KH. The Conformational Change of the L3 Loop Affects the Structural Changes in the Substrate Binding Pocket Entrance of β-Glucosidase. Molecules. 2023; 28(23):7807. https://doi.org/10.3390/molecules28237807
Chicago/Turabian StyleNam, Ki Hyun. 2023. "The Conformational Change of the L3 Loop Affects the Structural Changes in the Substrate Binding Pocket Entrance of β-Glucosidase" Molecules 28, no. 23: 7807. https://doi.org/10.3390/molecules28237807
APA StyleNam, K. H. (2023). The Conformational Change of the L3 Loop Affects the Structural Changes in the Substrate Binding Pocket Entrance of β-Glucosidase. Molecules, 28(23), 7807. https://doi.org/10.3390/molecules28237807








