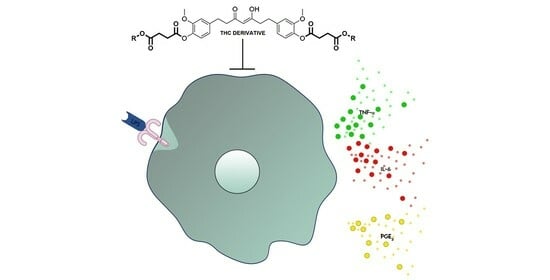
Tetrahydrocurcumin Derivatives Enhanced the Anti-Inflammatory Activity of Curcumin: Synthesis, Biological Evaluation, and Structure–Activity Relationship Analysis
Abstract
1. Introduction
2. Results
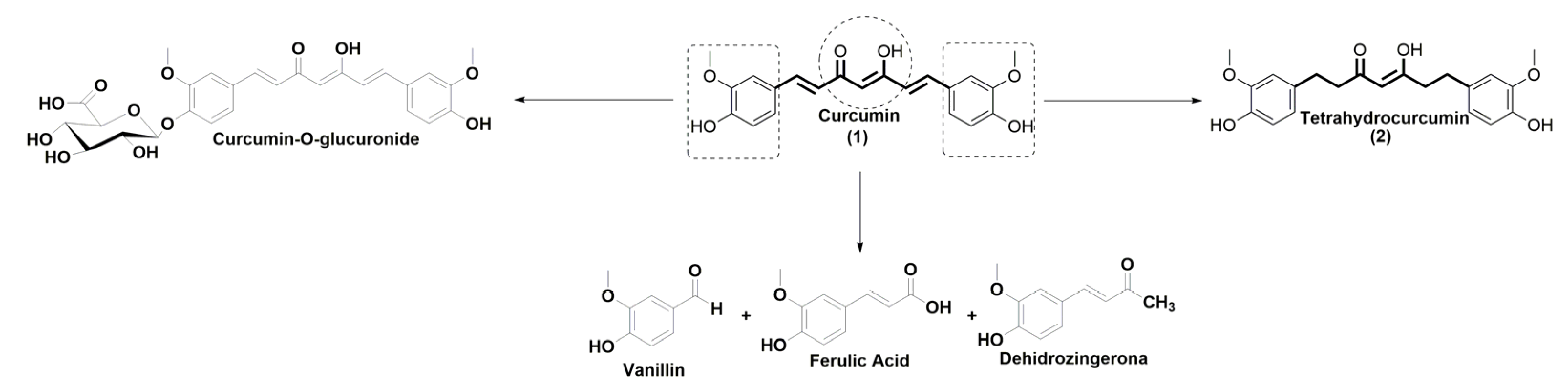
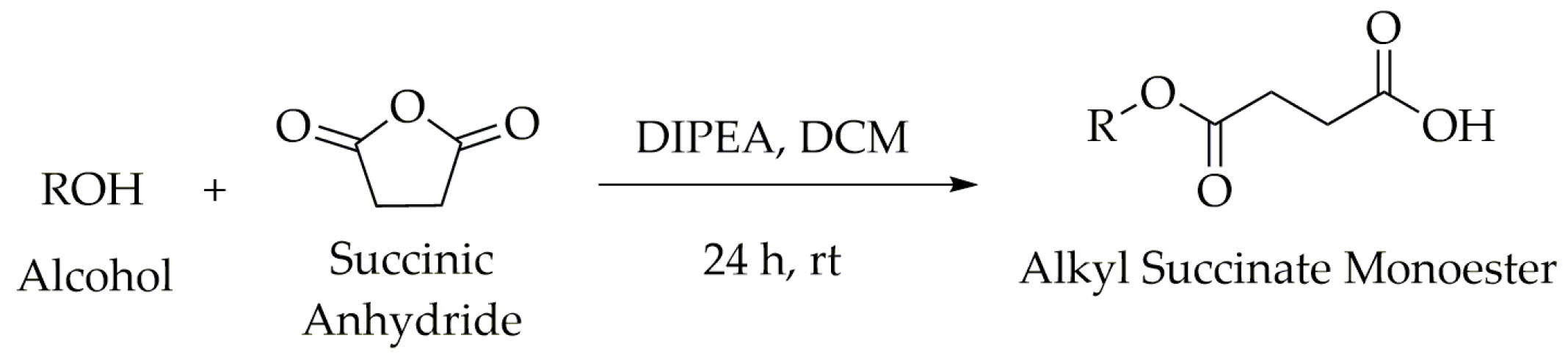
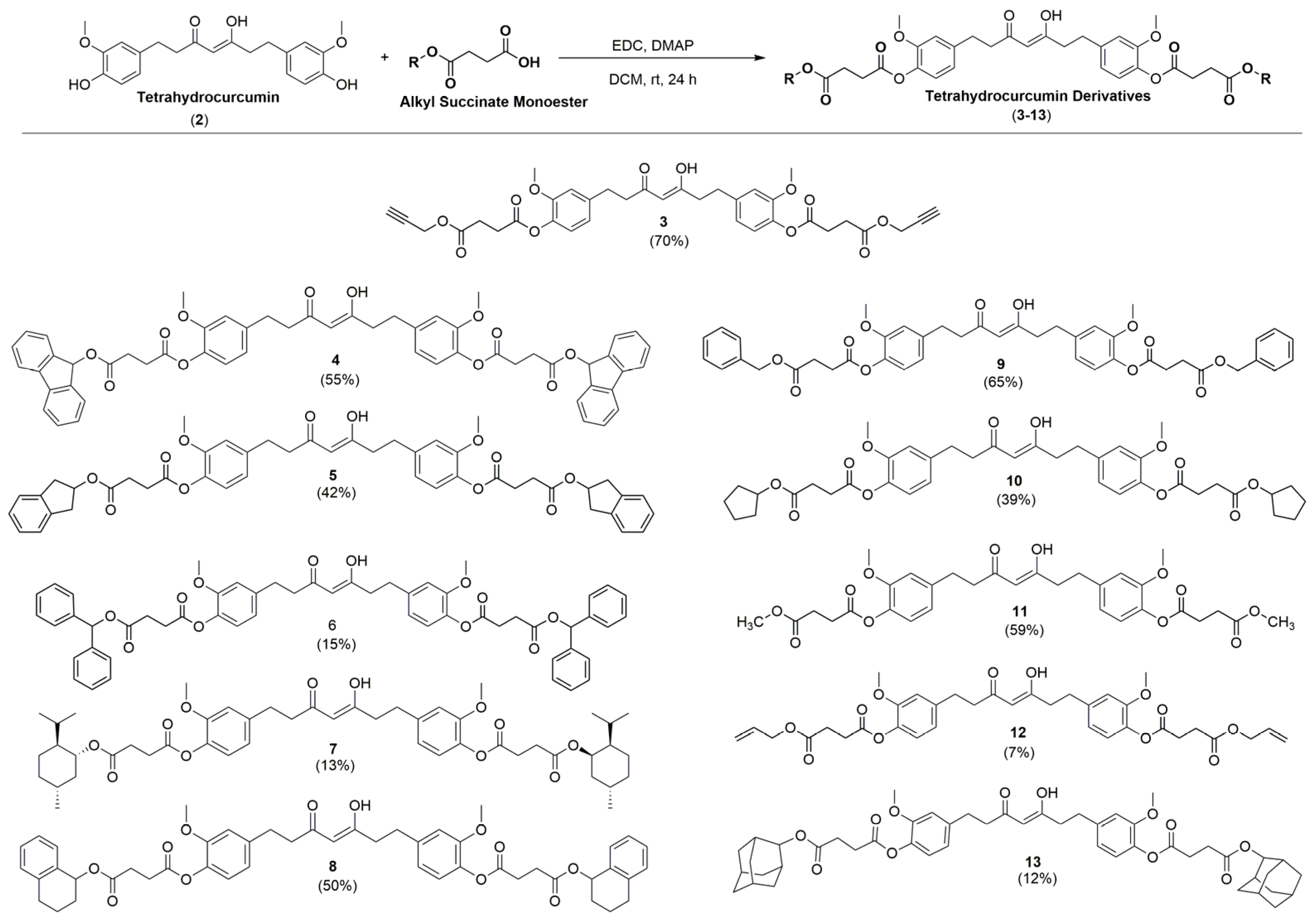
2.1. Synthesis of the Novel Tetrahydrocurcumin Derivatives
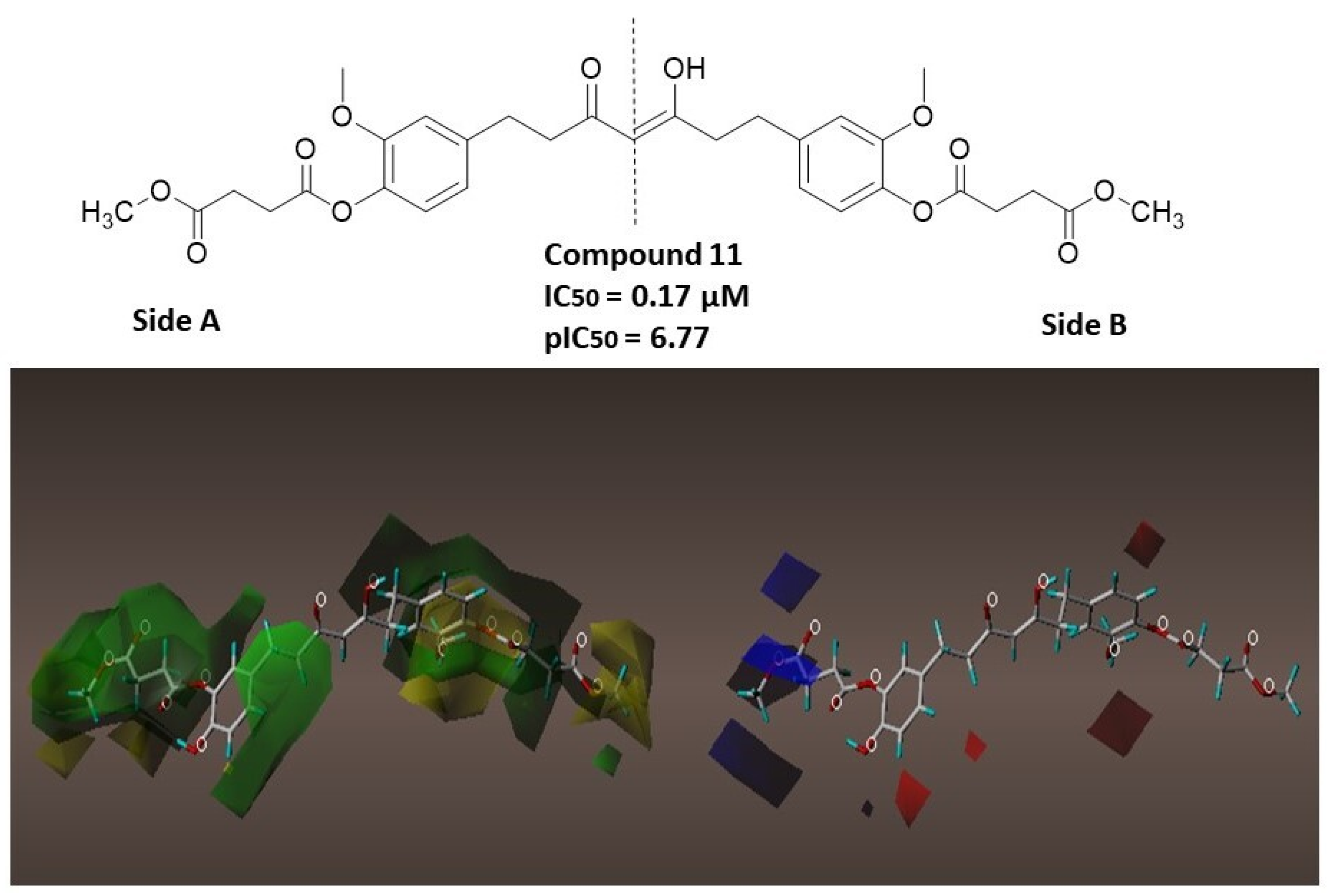
2.2. Anti-Inflammatory Activities of the Tetrahydrocurcumin Derivatives In Vitro
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.2. General Procedure 1 (GP1) for the Synthesis of Alkyl Succinate Monoesters (S1–S11)
4.2.1. Synthesis of 4-oxo-4-(prop-2-yn-1-yloxy)butanoic Acid (S2)
4.2.2. Synthesis of 4-(Benzyloxy)-4-oxobutanoic Acid (S4)
4.2.3. Synthesis of 4-(Cyclopentyloxy)-4-oxobutanoic Acid (S10)
4.2.4. Synthesis of 4-(Allyloxy)-4-oxobutanoic Acid (S11)
4.3. General Procedure 2 (GP2) for the Synthesis of Tetrahydrocurcumin Derivatives
4.3.1. Synthesis of (Z)-O,O′-((3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) di(prop-2-yn-1-yl) Disuccinate (3)
4.3.2. Synthesis of (Z)-di(9H-Fluoren-9-yl)O,O′-((3-hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (4)
4.3.3. Synthesis of (Z)-bis(2,3-Dihydro-1H-inden-2-yl) O,O′-((3-hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (5)
4.3.4. Synthesis of (Z)-Dibenzhydryl O,O′-((3-hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (6)
4.3.5. Synthesis of O,O′-(((Z)-3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) bis((1R,2S,5R)-2-Isopropyl-5-methylcyclohexyl) Disuccinate (7)
4.3.6. Synthesis of (Z)-O,O′-((3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) bis(1,2,3,4-Tetrahydronaphthalen-1-yl) Disuccinate (8)
4.3.7. Synthesis of (Z)-Dibenzyl O,O′-((3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (9)
4.3.8. Synthesis of (Z)-Dicyclopentyl O,O′-((3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (10)
4.3.9. Synthesis of (Z)-O,O′-((3-Hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) dimethyl Disuccinate (11)
4.3.10. Synthesis of (Z)-Diallyl O,O′-((3-hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (12)
4.3.11. Synthesis of di((1r,3r,5r,7r)-Adamantan-2-yl) O,O′-(((Z)-3-hydroxy-5-oxohept-3-ene-1,7-diyl)bis(2-methoxy-4,1-phenylene)) Disuccinate (13)
4.4. Mice
4.5. Macrophage Culture and Mediator Measurement
4.6. Cytotoxicity Assay
4.7. Statistical Analysis
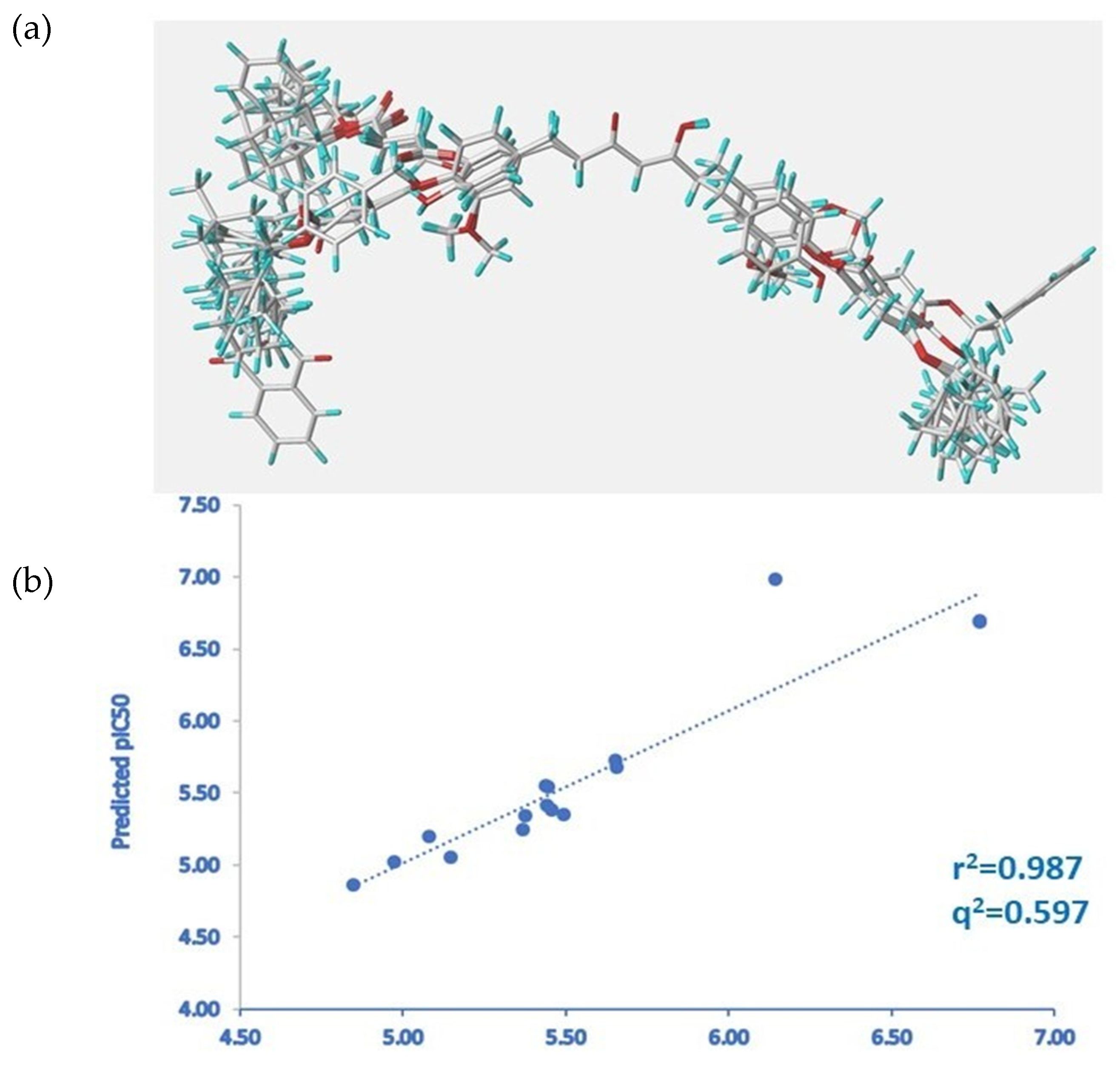
4.8. Three-Dimensional QSAR Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Priyadarsini, K.I. The chemistry of curcumin: From extraction to therapeutic agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef]
- Feng, L.; Li, Y.; Song, Z.-F.; Huai, Q.-Y.; Li, H.-J. Synthesis and biological evaluation of curcuminoid derivatives. Chem. Pharm. Bull. 2015, 63, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, K.S.; Negi, P.S.; Srinivas, P. Curcumin-amino acid conjugates: Synthesis, antioxidant and antimutagenic attributes. Food Chem. 2010, 120, 523–530. [Google Scholar] [CrossRef]
- He, Y.; Yue, Y.; Zheng, X.; Zhang, K.; Chen, S.; Du, Z. Curcumin, inflammation, and chronic diseases: How are they linked? Molecules 2015, 20, 9183–9213. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Dolai, S.; Rizk, S.; Hussain, A.; Tariq, H.; Averick, S.; L’Amoreaux, W.; El Idrissi, A.; Banerjee, P.; Raja, K. Synthesis of monofunctional curcumin derivatives, clicked curcumin dimer, and a PAMAM dendrimer curcumin conjugate for therapeutic applications. Org. Lett. 2007, 9, 5461–5464. [Google Scholar] [CrossRef]
- Esatbeyoglu, T.; Huebbe, P.; Ernst, I.M.A.; Chin, D.; Wagner, A.E.; Rimbach, G. Curcumin-from molecule to biological function. Angew. Chem. Int. Ed. 2012, 51, 5308–5332. [Google Scholar] [CrossRef]
- Jayaraj, R.L.; Elangovan, N.; Manigandan, K.; Singh, S.; Shukla, S. CNB-001 a novel curcumin derivative, guards dopamine neurons in MPTP model of Parkinson’s disease. BioMed Res. Int. 2014, 2014, 236182. [Google Scholar] [CrossRef]
- Gomes, D.d.C.F.; Vilela Alegrio, L.; Edilson Freire de Lima, M.; Leon, L.L.; Araújo, C.A.C. Synthetic Derivatives of Curcumin and their Activity against Leishmania amazonensis. Arzneim. Forsch. Drug Res. 2002, 52, 120–124. [Google Scholar] [CrossRef]
- Banuppriya, G.; Sribalan, R.; Padmini, V.; Shanmugaiah, V. Biological evaluation and molecular docking studies of new curcuminoid derivatives: Synthesis and characterization. Bioorg. Med. Chem. Lett. 2016, 26, 1655–1659. [Google Scholar] [CrossRef]
- Zhao, F.; Gong, Y.; Hu, Y.; Lu, M.; Wang, J.; Dong, J.; Chen, D.; Chen, L.; Fu, F.; Qiu, F. Curcumin and its major metabolites inhibit the inflammatory response induced by lipopolysaccharide: Translocation of nuclear factor-κB as potential target. Mol. Med. Rep. 2015, 11, 3087–3093. [Google Scholar] [CrossRef]
- Lozada-García, M.C.; Enríquez, R.G.; Ramírez-Apán, T.O.; Nieto-Camacho, A.; Palacios-Espinosa, J.F.; Custodio-Galván, Z.; Soria-Arteche, O.; Pérez-Villanueva, J. Synthesis of curcuminoids and evaluation of their cytotoxic and antioxidant properties. Molecules 2017, 22, 633. [Google Scholar] [CrossRef] [PubMed]
- Wichitnithad, W.; Nimmannit, U.; Wacharasindhu, S.; Rojsitthisak, P. Synthesis, characterization and biological evaluation of succinate prodrugs of curcuminoids for colon cancer treatment. Molecules 2011, 16, 1888–1900. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Hidaka, K.; Shinohara, A.; Maekawa, T.; Takeda, Y.; Yamaguchi, H. Chemical studies on antioxidant mechanism of curcuminoid: Analysis of radical reaction products from curcumin. J. Agric. Food Chem. 1999, 47, 71–77. [Google Scholar] [CrossRef]
- Rege, S.A.; Varshneya, M.A.; Momin, S.A. A Mini-Review: Comparison between curcumin and tetrahydrocurcumin based on their activities. Croat. J. food Sci. Technol. 2021, 13, 128–132. [Google Scholar] [CrossRef]
- Lakey-Beitia, J.; Berrocal, R.; Rao, K.S.; Durant, A.A. Polyphenols as Therapeutic Molecules in Alzheimer’s Disease through Modulating Amyloid Pathways. Mol. Neurobiol. 2015, 51, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Lu, H.; Chen, F.; Wang, Y.; Fan, W.; Shao, W.; Lu, H.; Lin, B. Tetrahydrocurcumin-induced autophagy via suppression of PI3K/Akt/mTOR in non-small cell lung carcinoma cells. Mol. Med. Rep. 2018, 17, 5964–5969. [Google Scholar] [CrossRef]
- Mahal, A.; Wu, P.; Jiang, Z.H.; Wei, X. Schiff Bases of Tetrahydrocurcumin as Potential Anticancer Agents. ChemistrySelect 2019, 4, 366–369. [Google Scholar] [CrossRef]
- Mahal, A.; Wu, P.; Jiang, Z.H.; Wei, X. Synthesis and Cytotoxic Activity of Novel Tetrahydrocurcumin Derivatives Bearing Pyrazole Moiety. Nat. Prod. Bioprospect. 2017, 7, 461–469. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Li, S.; Wang, Q.; Wang, H.; Xu, M.; An, Y. Tetrahydrocurcumin protects against sepsis-induced acute kidney injury via the SIRT1 pathway. Ren. Fail. 2021, 43, 1028–1040. [Google Scholar] [CrossRef]
- Karthikesan, K.; Pari, L.; Menon, V.P. Protective effect of tetrahydrocurcumin and chlorogenic acid against streptozotocin-nicotinamide generated oxidative stress induced diabetes. J. Funct. Foods 2010, 2, 134–142. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.R.; Kim, J.C.; Lee, E.S.; Chung, C.H.; Lee, E.Y.; Chung, B.Y. Tetrahydrocurcumin Ameliorates Skin Inflammation by Modulating Autophagy in High-Fat Diet-Induced Obese Mice. BioMed Res. Int. 2021, 2021, 6621027. [Google Scholar] [CrossRef]
- Plyduang, T.; Lomlim, L.; Yuenyongsawad, S.; Wiwattanapatapee, R. Carboxymethylcellulose-tetrahydrocurcumin conjugates for colon-specific delivery of a novel anti-cancer agent, 4-amino tetrahydrocurcumin. Eur. J. Pharm. Biopharm. 2014, 88, 351–360. [Google Scholar] [CrossRef]
- Manjunatha, J.R.; Bettadaiah, B.K.; Negi, P.S.; Srinivas, P. Synthesis of amino acid conjugates of tetrahydrocurcumin and evaluation of their antibacterial and anti-mutagenic properties. Food Chem. 2013, 139, 332–338. [Google Scholar] [CrossRef]
- Lakey-Beitia, J.; Gonzalez, Y.; Doens, D.; Stephens, D.E.; Santamaria, R.; Murillo, E.; Gutierrez, M.; Fernandez, P.L.; Rao, K.S.; Larionov, O.V.; et al. Assessment of Novel Curcumin Derivatives as Potent Inhibitors of Inflammation and Amyloid-Beta Aggregation in Alzheimer Disease. J. Alzheimers Dis. 2017, 2017, S59–S68. [Google Scholar] [CrossRef] [PubMed]
- González, Y.; Mojica-Flores, R.; Moreno-Labrador, D.; Cubilla-Rios, L.; Rao, K.S.J.; Fernández, P.L.; Larionov, O.V.; Lakey-Beitia, J. Polyphenols with Anti-Inflammatory Properties: Synthesis and Biological Activity of Novel Curcumin Derivatives. Int. J. Mol. Sci. 2023, 24, 3691. [Google Scholar] [CrossRef] [PubMed]
- Payton, F.; Sandusky, P.; Wl, A. NMR study of the solution structure of curcumin. J. Nat. Prod. 2014, 70, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.T.; Herschman, H.R. Ligand-induced prostaglandin synthesis requires expression of the TIS10/PGS-2 prostaglandin synthase gene in murine fibroblasts and macrophages. J. Biol. Chem. 1994, 269, 15473–15480. [Google Scholar] [CrossRef]
- Tropsha, A.; Golbraikh, A. Predictive QSAR Modeling Workflow, Model Applicability Domains, and Virtual Screening. Curr. Pharm. Des. 2007, 13, 3494–3504. [Google Scholar] [CrossRef] [PubMed]
- Sarikavak, K.; Sevin, F. Structural and Spectral (IR, NMR and UV/Visible) Properties of Newly Designed Boronic Acid Derivatives Containing DO3A Sensitive to Uranyl Ion: A DFT and TD-DFT Study. Comput. Chem. 2017, 5, 145–158. [Google Scholar] [CrossRef][Green Version]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09, Revision A. 02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative Molecular Field Analysis (CoMFA). 1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.D.; Bunce, J.D.; Patterson, D.E.; Frank, I.E. Crossvalidation, Bootstrapping, and Partial Least Squares Compared with Multiple Regression in Conventional QSAR Studies. Quant. Struct. Relatsh. 1988, 7, 18–25. [Google Scholar] [CrossRef]
- Bush, B.L.; Nachbar, R.B. Sample-distance partial least squares: PLS optimized for many variables, with application to CoMFA. J. Comput. Aided. Mol. Des. 1993, 7, 587–619. [Google Scholar] [CrossRef] [PubMed]
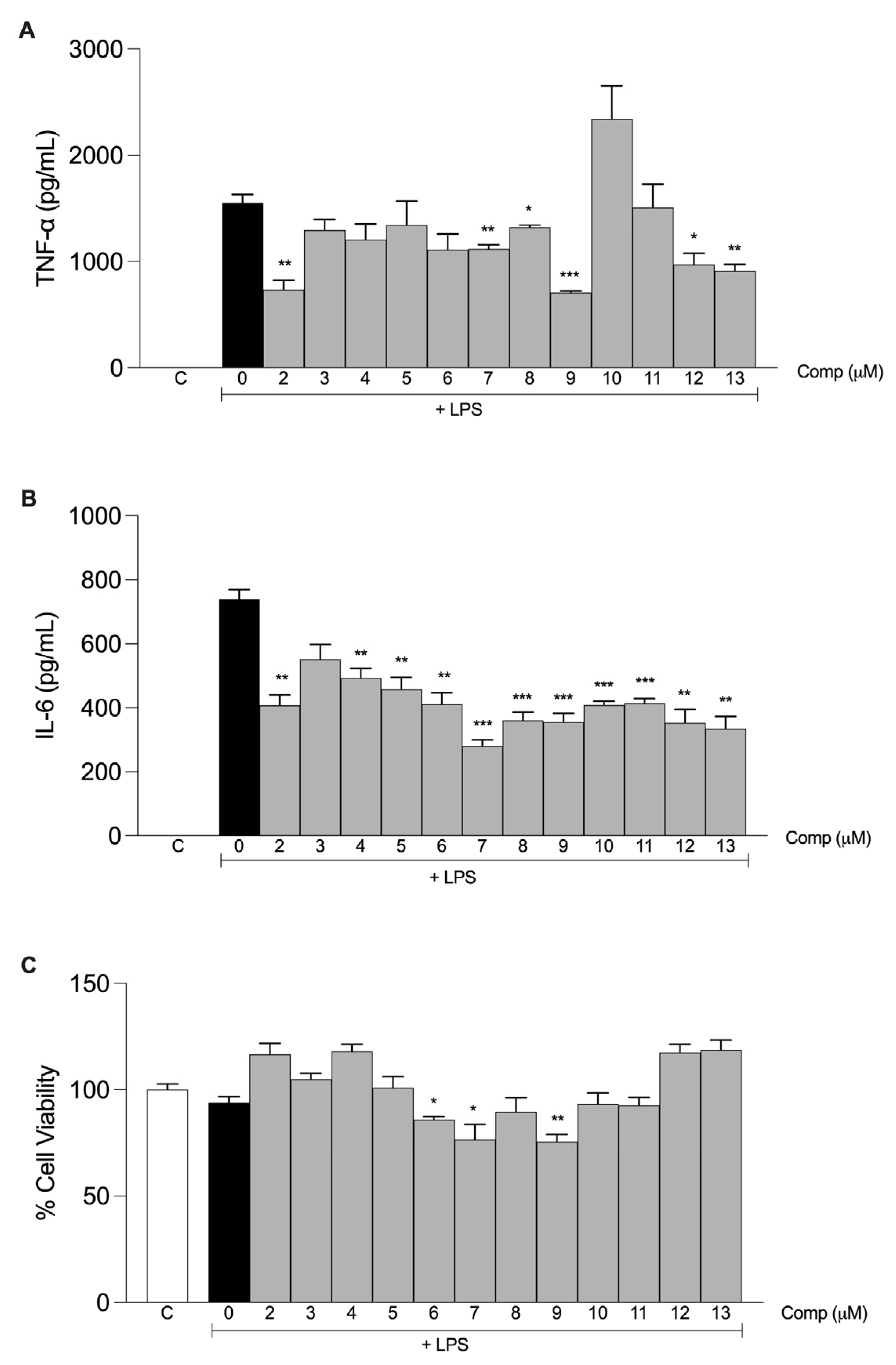
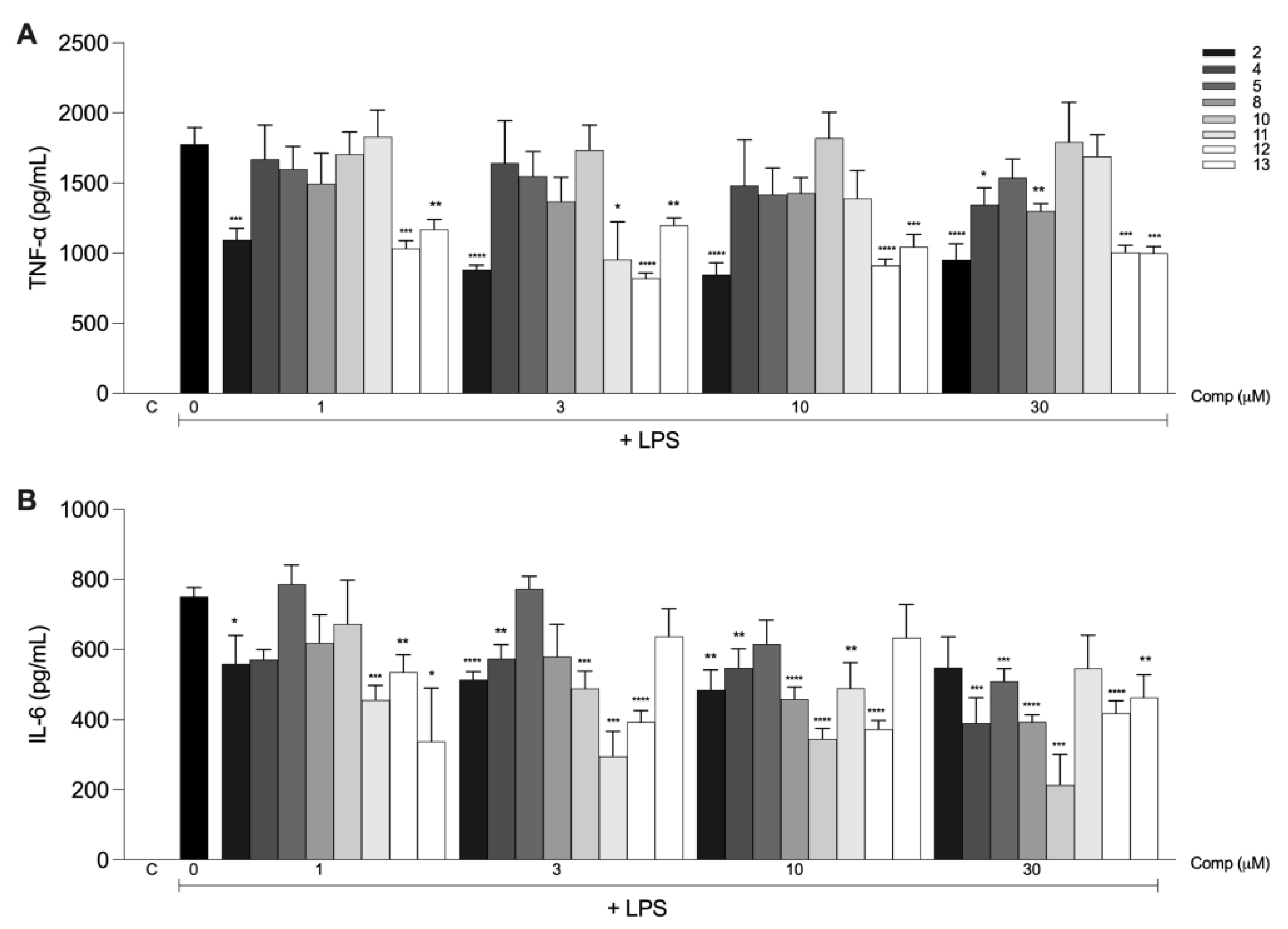
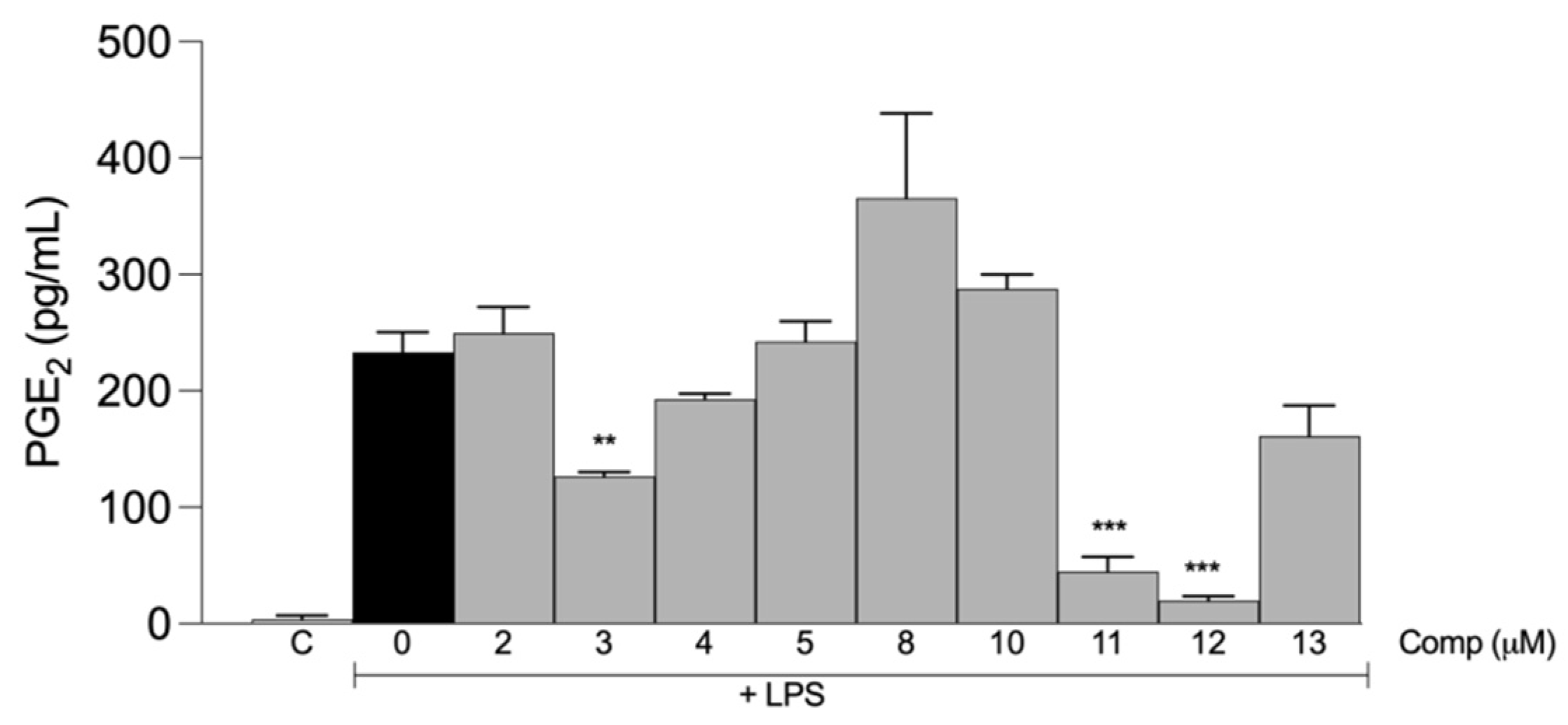
| Compound | IC50 ± S.D. (μM) TNF-α | IC50 ± S.D. (μM) IL-6 |
|---|---|---|
| 2 | 0.18 ± 0.18 | 0.17 ± 0.20 |
| 4 | N.A. | 4.28 ± 4.88 |
| 5 | N.A. | 9.13 ± 5.90 |
| 8 | 3.21 ± 4.52 | 3.48 ± 4.39 |
| 10 | N.A. | 3.66 ± 4.21 |
| 11 | N.A. | 0.17 ± 0.21 |
| 12 | 0.70 ± 0.10 | 0.72 ± 0.38 |
| 13 | 0.35 ± 0.047 | 1.83 ± 2.55 |
| Model | q2 Cross-Validated r2 | r2 Non-Cross-Validated r2 | SE Standard Error | ONC NUMBER of Optimal Components | F Values | %E Contribution of Electrostatic Field | %S Contribution of Steric Field |
|---|---|---|---|---|---|---|---|
| STD standard scaling | 0.424 | 0.973 | 0.108 | 4 | 97.620 | 35 | 65 |
| RF region focusing | 0.597 | 0.987 | 0.1077 | 4 | 204.088 | 42.7 | 0.573 |
| Compound | Experimental pIC50 | Predicted pIC50 | Residues |
|---|---|---|---|
| 2 | 6.77 | 6.69 | 0.08 |
| 4 | 5.37 | 5.25 | 0.12 |
| 8 | 5.46 | 5.38 | 0.07 |
| 10 | 5.44 | 5.55 | −0.12 |
| 11 | 6.77 | 6.69 | 0.08 |
| 12 | 6.14 | 6.98 | −0.84 |
| 14 | 5.65 | 5.72 | −0.07 |
| 15 | 5.44 | 5.54 | −0.10 |
| 16 | 5.66 | 5.68 | −0.03 |
| 18 | 4.85 | 4.87 | −0.02 |
| 19 | 5.08 | 5.20 | −0.12 |
| 20 | 5.49 | 5.35 | 0.14 |
| 21 | 4.97 | 5.02 | −0.05 |
| 24 | 5.44 | 5.41 | 0.03 |
| 26 | 5.38 | 5.35 | 0.03 |
| 27 | 5.15 | 5.06 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, Y.; Mojica-Flores, R.; Moreno-Labrador, D.; Pecchio, M.; Rao, K.S.J.; Ahumedo-Monterrosa, M.; Fernández, P.L.; Larionov, O.V.; Lakey-Beitia, J. Tetrahydrocurcumin Derivatives Enhanced the Anti-Inflammatory Activity of Curcumin: Synthesis, Biological Evaluation, and Structure–Activity Relationship Analysis. Molecules 2023, 28, 7787. https://doi.org/10.3390/molecules28237787
González Y, Mojica-Flores R, Moreno-Labrador D, Pecchio M, Rao KSJ, Ahumedo-Monterrosa M, Fernández PL, Larionov OV, Lakey-Beitia J. Tetrahydrocurcumin Derivatives Enhanced the Anti-Inflammatory Activity of Curcumin: Synthesis, Biological Evaluation, and Structure–Activity Relationship Analysis. Molecules. 2023; 28(23):7787. https://doi.org/10.3390/molecules28237787
Chicago/Turabian StyleGonzález, Yisett, Randy Mojica-Flores, Dilan Moreno-Labrador, Marisín Pecchio, K. S. Jagannatha Rao, Maicol Ahumedo-Monterrosa, Patricia L. Fernández, Oleg V. Larionov, and Johant Lakey-Beitia. 2023. "Tetrahydrocurcumin Derivatives Enhanced the Anti-Inflammatory Activity of Curcumin: Synthesis, Biological Evaluation, and Structure–Activity Relationship Analysis" Molecules 28, no. 23: 7787. https://doi.org/10.3390/molecules28237787
APA StyleGonzález, Y., Mojica-Flores, R., Moreno-Labrador, D., Pecchio, M., Rao, K. S. J., Ahumedo-Monterrosa, M., Fernández, P. L., Larionov, O. V., & Lakey-Beitia, J. (2023). Tetrahydrocurcumin Derivatives Enhanced the Anti-Inflammatory Activity of Curcumin: Synthesis, Biological Evaluation, and Structure–Activity Relationship Analysis. Molecules, 28(23), 7787. https://doi.org/10.3390/molecules28237787