Anti-Inflammatory Ergosteroid Derivatives from the Coral-Associated Fungi Penicillium oxalicum HL-44
Abstract
:1. Introduction
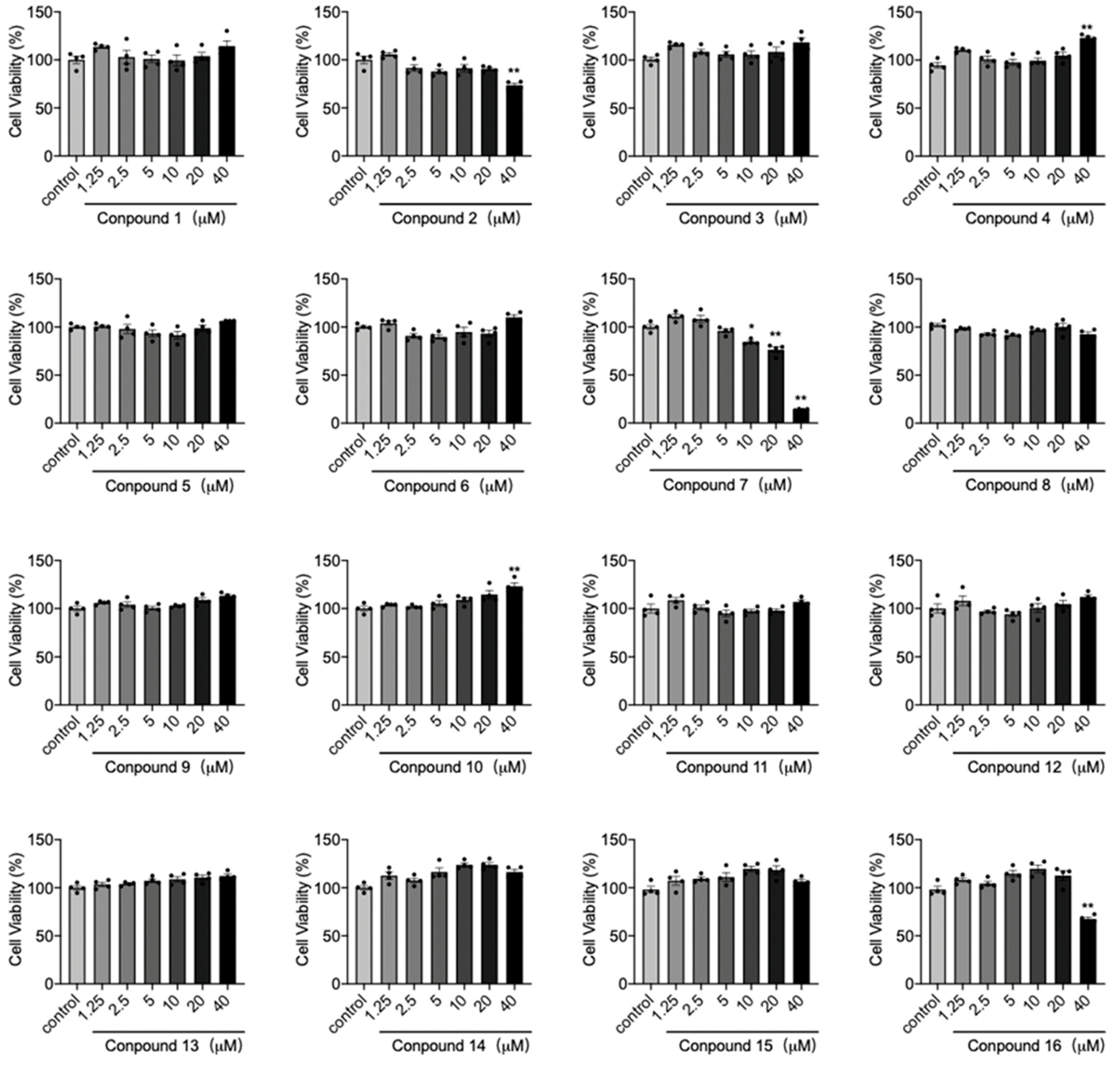
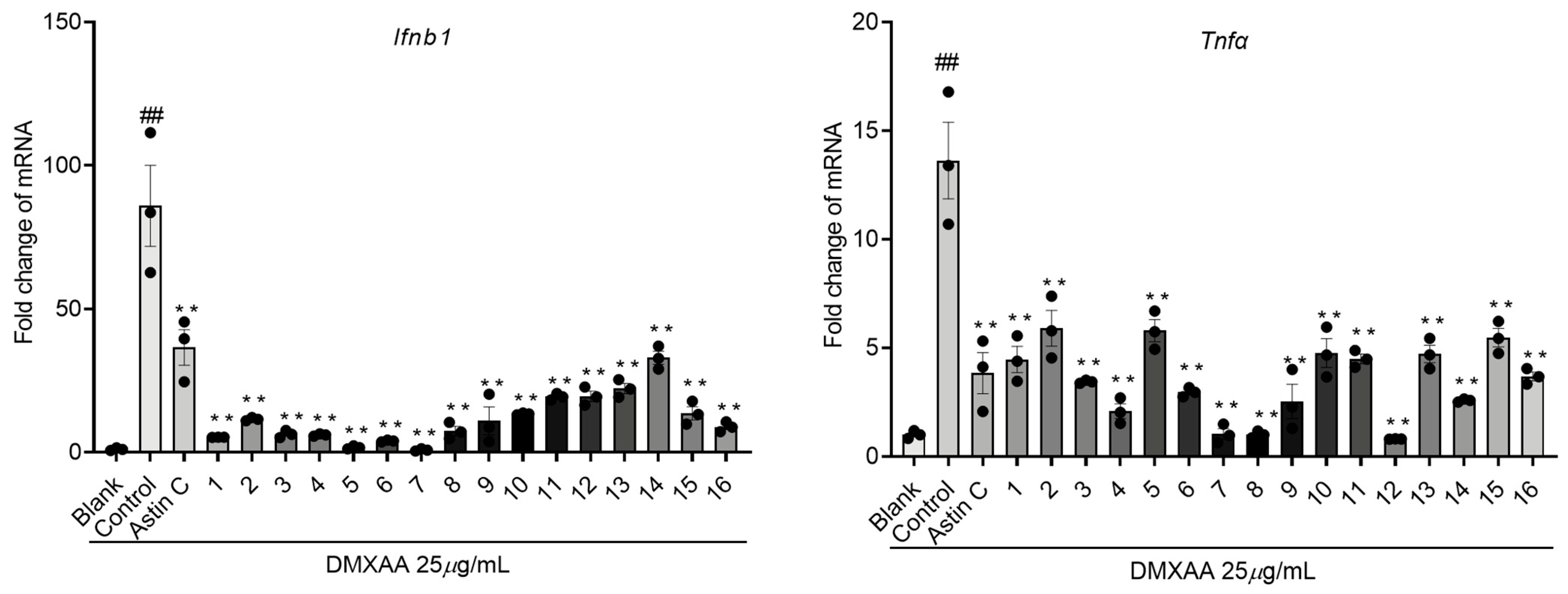
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Screening Culture Medium and Cultivation
3.4. Extraction and Isolation
3.5. Cell Viability Detection
3.6. Quantitative Real-Time PCR (qPCR)
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chen, Y.; Chan, W.Y. Marine natural products with anti-inflammatory activity. Appl. Microbiol. Biotechnol. 2016, 100, 1645–1666. [Google Scholar] [CrossRef]
- Zhabinskii, V.N.; Drasar, P.; Khripach, V.A. Structure and Biological Activity of Ergostane-Type Steroids from Fungi. Molecules 2022, 27, 2103. [Google Scholar] [CrossRef] [PubMed]
- Kubatova, A.; Hujslova, M.; Frisvad, J.C.; Chudickova, M.; Kolarik, M. Taxonomic revision of the biotechnologically important species Penicillium oxalicum with the description of two new species from acidic and saline soils. Mycol. Prog. 2019, 18, 215–228. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.-M.; Liu, H.; Li, X.; Wang, B.-G. Two new alkaloids from Penicillium oxalicum EN-201, an endophytic fungus derived from the marine mangrove plant Rhizophora stylosa. Phytochem. Lett. 2015, 13, 160–164. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, Y.; Xu, M.-M.; Wei, X.; Yang, C.-B.; Zeng, F.; Duan, J.-A.; Che, C.-T.; Zhou, J.; Zhao, M. Oxalactam A, a Novel Macrolactam with Potent Anti-Rhizoctonia solani Activity from the Endophytic Fungus Penicillium oxalicum. Molecules 2022, 27, 8811. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chao, L.-H.; Sun, J.; Chen, X.-N.; Yao, H.-N.; Zhu, Z.-X.; Dong, D.; Liu, T.; Tu, P.-F.; Li, J. Two new polyketides from the fungus Penicillium oxalicum MHZ153. Nat. Prod. Res. 2019, 33, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Weng, W.; Li, R.; Zhang, Y.; Pan, X.; Jiang, S.; Sun, C.; Zhang, C.; Lu, X. Polyketides isolated from an endophyte Penicillium oxalicum 2021CDF-3 inhibit pancreatic tumor growth. Front. Microbiol. 2022, 13, 1033823. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.-M.; Zhang, P.; Wang, B.-G. A new phenolic enamide and a new meroterpenoid from marine alga-derived endophytic fungus Penicillium oxalicum EN-290. J. Asian Nat. Prod. Res. 2015, 17, 1204–1212. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Li, L.; Li, Y.-Q.; Luo, J.-H.; Li, W.; Li, L.-F.; Zheng, C.-J.; Cao, F. Oxalierpenes A and B, unusual indole-diterpenoid derivatives with antiviral activity from a marine-derived strain of the fungus Penicillium oxalicum. J. Nat. Prod. 2022, 85, 1880–1885. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.; Xu, Z.; Bai, Q.; Zhou, X.; Zheng, C.; Bai, M.; Chen, G. Biological secondary metabolites from the Lumnitzera littorea-derived fungus Penicillium oxalicum HLLG-13. Mar. Drugs 2023, 21, 22. [Google Scholar] [CrossRef]
- He, Z.-H.; Xie, C.-L.; Hao, Y.-J.; Xu, L.; Wang, C.-F.; Hu, M.-Y.; Li, S.-J.; Zhong, T.-H.; Yang, X.-W. Solitumergosterol A, a unique 6/6/6/6/5 steroid from the deep-sea-derived Penicillium solitum MCCC 3A00215. Org. Biomol. Chem. 2021, 19, 9369–9372. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Li, X.P.; Zhao, J.; Gao, H.W.; Xu, Q.M.; Wang, J.W. Biotransformation of artemisinic acid to bioactive derivatives by endophytic Penicillium oxalicum B4 from Artemisia annua L. Phytochemistry 2021, 185, 112682. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz, T.M.; Ellena, J.; Porto, A.L.M. Biotransformation of Ethinylestradiol by Whole Cells of Brazilian Marine-Derived Fungus Penicillium oxalicum CBMAI 1996. Mar. Biotechnol. 2020, 22, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Li, R.; Li, J.; He, J.; Cao, Z.Y.; Kurtán, T.; Mándi, A.; Zheng, G.L.; Zhang, W. Alternarin A, a drimane meroterpenoid, suppresses neuronal excitability from the coral-associated fungi Alternaria sp. ZH-15. Org. Lett. 2020, 22, 2995–2998. [Google Scholar] [CrossRef]
- Wang, H.L.; Li, R.; Zhao, M.; Wang, Z.Y.; Tang, H.; Cao, Z.Y.; Zheng, G.L.; Zhang, W. A drimane meroterpenoid borate as a synchronous Ca plus oscillation inhibitor from the coral-associated fungus Alternaria sp. ZH-15. J. Nat. Prod. 2023, 86, 429–433. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.L.; Tang, H.; Su, L.; Zheng, G.L.; Zhang, W. Immunosuppressive 9,10-secosteroids from the gorgonian Verrucella umbraculum collected in the South China Sea. J. Nat. Prod. 2021, 84, 1671–1675. [Google Scholar] [CrossRef]
- Shi, Q.; Huang, Y.; Su, H.; Gao, Y.; Peng, X.; Zhou, L.; Li, X.; Qiu, M. C(28) steroids from the fruiting bodies of Ganoderma resinaceum with potential anti-inflammatory activity. Phytochemistry 2019, 168, 112109. [Google Scholar] [CrossRef]
- Weng, Y.F.; Lu, J.; Xiang, L.; Matsuura, A.; Zhang, Y.; Huang, Q.M.; Qi, J.H. Ganodermasides C and D, two new anti-aging ergosterols from spores of the medicinal mushroom Ganoderma lucidum. Biosci. Biotechnol. Biochem. 2011, 75, 800–803. [Google Scholar] [CrossRef]
- Kwon, H.C.; Zee, S.D.; Cho, S.Y.; Choi, S.U.; Lee, K.R. Cytotoxic ergosterols from Paecilomyces sp. J300. Arch. Pharmacal Res. 2002, 25, 851–855. [Google Scholar] [CrossRef]
- Liu, X.-H.; Miao, F.-P.; Liang, X.-R.; Ji, N.-Y. Ergosteroid derivatives from an algicolous strain of Aspergillus ustus. Nat. Prod. Res. 2014, 28, 1182–1186. [Google Scholar] [CrossRef]
- Duecker, F.L.; Heinze, R.C.; Mueller, M.; Zhang, S.D.; Heretsch, P. Synthesis of the alleged structures of fortisterol and herbarulide and structural revision of herbarulide. Org. Lett. 2020, 22, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Amagata, T.; Tanaka, M.; Yamada, T.; Doi, M.; Minoura, K.; Ohishi, H.; Yamori, T.; Numata, A. Variation in cytostatic constituents of a sponge-derived Gymnascella dankaliensis by manipulating the carbon source. J. Nat. Prod. 2007, 70, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Fangkrathok, N.; Sripanidkulchai, B.; Umehara, K.; Noguchi, H. Bioactive ergostanoids and a new polyhydroxyoctane from Lentinus polychrous mycelia and their inhibitory effects on E2-enhanced cell proliferation of T47D cells. Nat. Prod. Res. 2013, 27, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Hoque, N.; Afroz, F.; Khatun, F.; Rony, S.R.; Hasan, C.M.; Rana, M.S.; Sohrab, M.H. Physicochemical, pharmacokinetic and cytotoxicity of the compounds Isolated from an endophyte Fusarium oxysporum: In vitro and in silico approaches. Toxins 2022, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fang, Y.; Zhang, M.; Lin, A.; Zhu, A.; Gu, Q.; Zhu, W. Six new ergosterols from the marine-derived fungus Rhizopus sp. Steroids 2008, 73, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zhao, J.-L.; Liu, J.-M.; Zhang, M.; Chen, R.-D.; Xie, K.-B.; Chen, D.-W.; Dai, J.-G. Lanostane triterpenoids and ergostane-type steroids from the cultured mycelia of Ganoderma capense. J. Asian Nat. Prod. Res. 2018, 20, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Hong, K.; Chen, G.-D.; Wang, C.-X.; Tang, J.-S.; Yu, Y.; Jiang, M.-M.; Li, M.-M.; Wang, N.-L.; Yao, X.-S. New oxidized sterols from Aspergillus awamori and the endo-boat conformation adopted by the cyclohexene oxide system. Magn. Reson. Chem. 2010, 48, 38–43. [Google Scholar] [CrossRef]
- Zhang, W.; Draeger, S.; Schulz, B.; Krohn, K. Ring B aromatic steroids from an endophytic fungus, Colletotrichum sp. Nat. Prod. Commun. 2009, 4, 1449–1454. [Google Scholar] [CrossRef]
- Kim, K.H.; Choi, S.U.; Park, K.M.; Seok, S.J.; Lee, K.R. Cytotoxic constituents of Amanita subjunquillea. Arch. Pharmacal Res. 2008, 31, 579–586. [Google Scholar] [CrossRef]
- Du, Z.-Z.; Shen, Y.-M. A rare new cleistanthane diterpene from the pericarp of Trewia nudiflora. Helv. Chim. Acta 2006, 89, 2841–2845. [Google Scholar] [CrossRef]
- Yajima, A.; Kagohara, Y.; Shikai, K.; Katsuta, R.; Nukada, T. Synthesis of two osteoclast-forming suppressors, demethylincisterol A(3) and chaxine A. Tetrahedron 2012, 68, 1729–1735. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, O.; Peng, C.; Su, H.G.; Miao, L.L.; Liu, L.S.; Xiong, L. Polyoxygenated ergosteroids from the macrofungus Omphalia lapidescens and the structure-cytotoxicity relationship in a human gastric cancer cell line. Phytochem. Lett. 2018, 25, 99–104. [Google Scholar] [CrossRef]
- Wright, J.L.C.; McInnes, A.G.; Shimizu, S.; Smith, D.G.; Walter, J.A.; Idler, D.; Khalil, W. Identification of C-24 alkyl epimers of marine sterols by C-13 nuclear magnetic-resonance spectroscopy. Can. J. Chem. 1978, 56, 1898–1903. [Google Scholar] [CrossRef]
- Gao, H.; Hong, K.; Zhang, X.; Liu, H.-W.; Wang, N.-L.; Zhuang, L.; Yao, X.-S. New steryl esters of fatty acids from the mangrove fungus Aspergillus awamori. Helv. Chim. Acta 2007, 90, 1165–1178. [Google Scholar] [CrossRef]
- Mahato, S.B.; Banerjee, S.; Podder, S. Steroid transformations by microorganisms-III. Phytochemistry 1989, 28, 7–40. [Google Scholar] [CrossRef]
- Schlosser, D.; Schmauder, H.P. 15-alpha-hydroxylation of 13-ethyl-gon-4-ene-3,17-dione using a hyphal fungus immobilized in calcium alginate gel beads. J. Basic Microbiol. 1991, 31, 385–390. [Google Scholar] [CrossRef]
- Irrgang, S.; Schlosser, D.; Fritsche, W. Involvement of cytochrome P-450 in the 15 alpha-hydroxylation of 13-ethyl-gon-4-ene-3,17-dione by Penicillium raistrickii. J. Steroid Biochem. Mol. Biol. 1997, 60, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.H.; Wang, X.R.; Zhang, Z.H.; Wang, S.; Li, K.; Lu, F.P.; Qin, H.M. 15 alpha-hydroxylation of D-ethylgonendione by Penicillium raistrickii in deep eutectic solvents DESs containing system. Biochem. Eng. J. 2020, 164, 7781–7789. [Google Scholar] [CrossRef]
- Krohn, K.; Biele, C.; Aust, H.J.; Draeger, S.; Schulz, B. Herbarulide, a ketodivinyllactone steroid with an unprecedented homo-6-oxaergostane skeleton from the endophytic fungus Pleospora herbarum. J. Nat. Prod. 1999, 62, 629–630. [Google Scholar] [CrossRef]
- Yu, J.-H.; Yu, S.-J.; Liu, K.-L.; Wang, C.; Liu, C.; Sun, J.-y.; Zhang, H. Cytotoxic ergostane-type steroids from Ganoderma lingzhi. Steroids 2021, 165, 8767–8773. [Google Scholar] [CrossRef]
- Liu, D.; Liu, J. Peroxy natural products. Nat. Prod. Bioprospect. 2013, 3, 161–206. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.-M.; Li, C.-S.; Wang, B.-G. Nigerasterols A and B, antiproliferative sterols from the mangrove-derived endophytic fungus Aspergillus niger MA-132. Helv. Chim. Acta 2013, 96, 1055–1061. [Google Scholar] [CrossRef]
- Dericcardis, F.; Spinella, A.; Izzo, I.; Giordano, A.; Sodano, G. Synthesis of (17R)-17-methylincisterol, a highly degraded marine steroid. Tetrahedron Lett. 1995, 36, 4303–4306. [Google Scholar] [CrossRef]
- Mills, C.D.; Lenz, L.L.; Harris, R.A. A breakthrough: Macrophage-directed cancer immunotherapy. Cancer Res. 2016, 76, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Dobbs, N.; Yang, K.; Yan, N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 2020, 53, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Li, S.R.; Mirlekar, B.; Johnson, B.M.; Brickey, W.J.; Wrobel, J.A.; Yang, N.; Song, D.K.; Entwistle, S.; Tan, X.M.; Deng, M.; et al. STING-induced regulatory B cells compromise NK function in cancer immunity. Nature 2022, 610, 373–380. [Google Scholar] [CrossRef]
- Haag, S.M.; Gulen, M.F.; Reymond, L.; Gibelin, A.; Abrami, L.; Decout, A.; Heymann, M.; van der Goot, F.G.; Turcatti, G.; Behrendt, R.; et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018, 559, 269–273. [Google Scholar] [CrossRef]
- Garland, K.M.; Sheehy, T.L.; Wilson, J.T. Chemical and biomolecular strategies for STING pathway activation in cancer immunotherapy. Chem. Rev. 2022, 122, 5977–6039. [Google Scholar] [CrossRef]
- Wu, J.X.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef]
- Li, S.L.; Hong, Z.; Wang, Z.; Li, F.; Mei, J.H.; Huang, L.L.; Lou, X.W.; Zhao, S.M.; Song, L.H.; Chen, W.; et al. The cyclopeptide Astin C specifically inhibits the innate immune CDN sensor STING. Cell Rep. 2018, 25, 3405–3421. [Google Scholar] [CrossRef]
- Service, R.F. SYNTHETIC BIOLOGY Fluorine-adding bacteria may transform natural product medicines. Science 2013, 341, 1052–1053. [Google Scholar] [PubMed]
- Chen, Y.; Bian, H.; Lv, J.; Song, W.; Xing, C.; Hui, C.; Zhang, D.; Zhang, C.; Zhao, L.; Li, Y.; et al. Gelsevirine is a novel STING-specific inhibitor and mitigates STING-related inflammation in sepsis. Front. Immunol. 2023, 14, 1190707. [Google Scholar] [CrossRef] [PubMed]
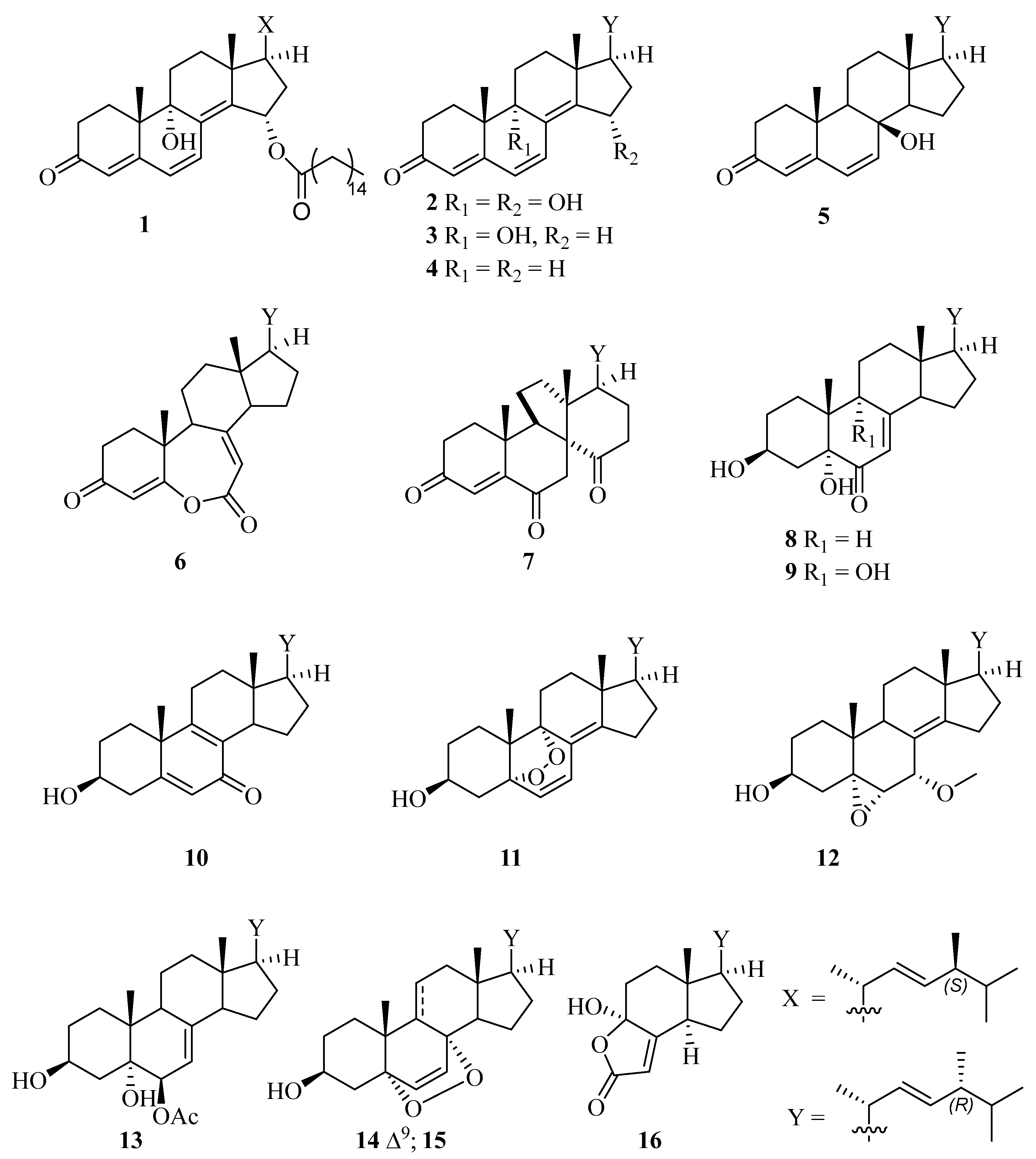
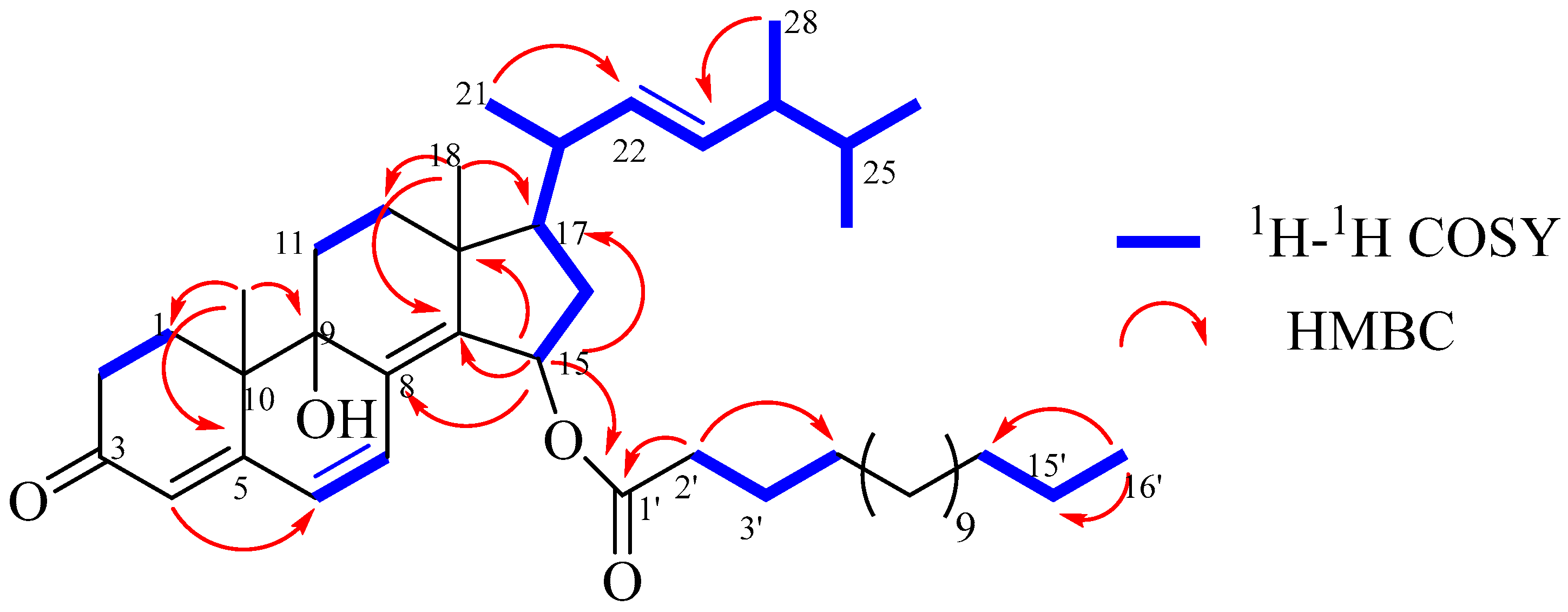
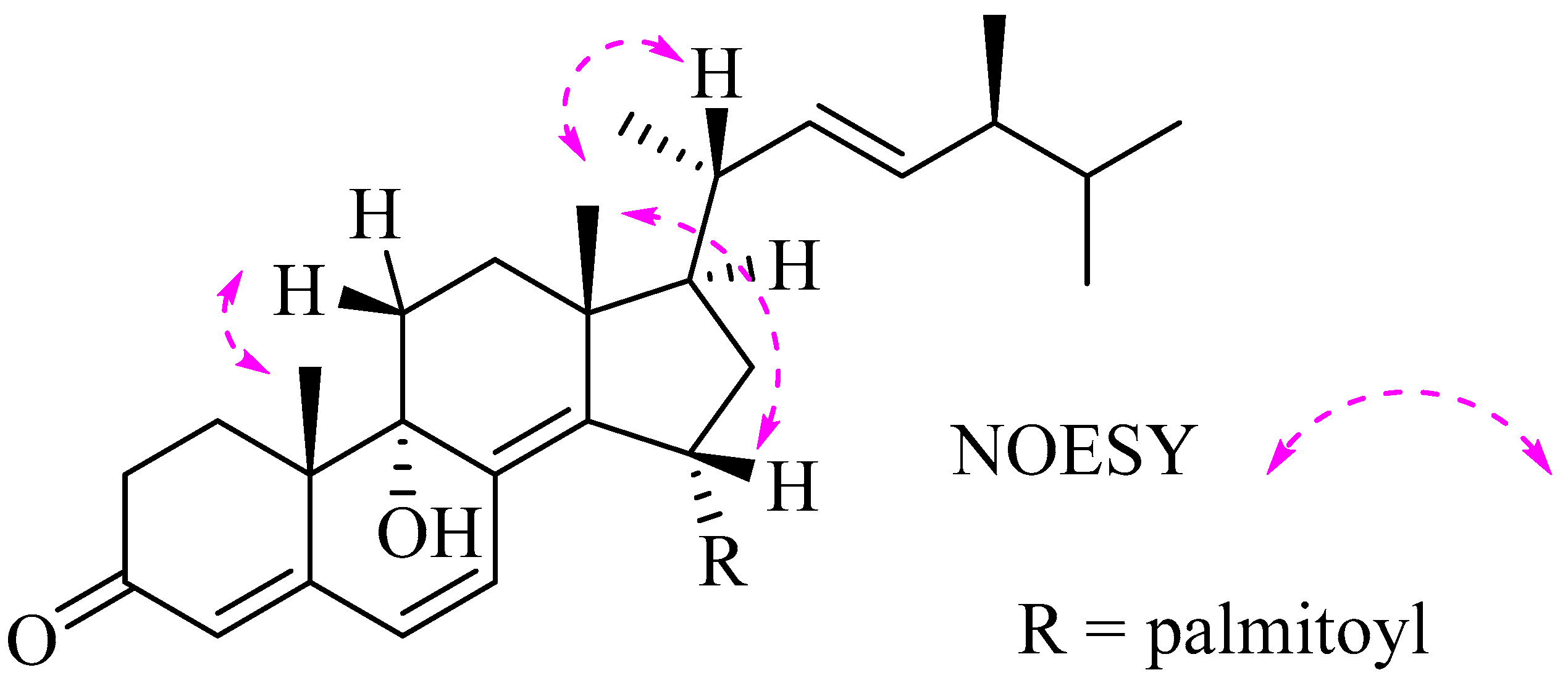

| Position | δC | δH (J in Hz) | Position | δC | δH (J in Hz) |
|---|---|---|---|---|---|
| 1 | 27.5 | 2.53 m, 1.80 m | 19 | 21.0 | 1.14 s |
| 2 | 33.9 | 2.53 m | 20 | 38.4 | 2.12 m |
| 3 | 199.3 | - | 21 | 21.3 | 1.09 d (6.6) |
| 4 | 127.3 | 5.91 s | 22 | 134.4 | 5.19 dd (15.3, 8.2) |
| 5 | 160.4 | - | 23 | 133.5 | 5.26 dd (15.3, 7.6) |
| 6 | 126.5 | 6.13 d (9.8) | 24 | 43.1 | 1.85 m |
| 7 | 129.9 | 6.41 d (9.8) | 25 | 33.1 | 1.47 m |
| 8 | 132.0 | - | 26 | 20.1 | 0.83 d (6.7) |
| 9 | 72.8 | - | 27 | 19.7 | 0.81 d (6.8) |
| 10 | 42.4 | - | 28 | 17.9 | 0.92 d (6.8) |
| 11 | 25.6 | 2.02 m, 1.75 m | 1′ | 173.6 | - |
| 12 | 32.1 | 2.00 m, 1.72 m | 2′ | 34.7 | 2.30 t (7.2) |
| 13 | 44.8 | - | 3′ | 25.2 | 1.61 m |
| 14 | 154.0 | - | 4′–13′ | 29.8–29.3 | 1.30–1.23 m |
| 15 | 71.8 | 5.81 d (7.1) | 14′ | 32.0 | 1.25 m |
| 16 | 37.6 | 1.94 m, 1.72 m | 15′ | 22.8 | 1.31 m, 1.24 m |
| 17 | 53.1 | 1.63 m | 16′ | 14.2 | 0.88, t (6.6) |
| 18 | 19.3 | 0.95 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, C.; Chen, Y.-H.; Bian, H.-H.; Zhang, J.-P.; Su, L.; Han, H.; Zhang, W. Anti-Inflammatory Ergosteroid Derivatives from the Coral-Associated Fungi Penicillium oxalicum HL-44. Molecules 2023, 28, 7784. https://doi.org/10.3390/molecules28237784
Pang C, Chen Y-H, Bian H-H, Zhang J-P, Su L, Han H, Zhang W. Anti-Inflammatory Ergosteroid Derivatives from the Coral-Associated Fungi Penicillium oxalicum HL-44. Molecules. 2023; 28(23):7784. https://doi.org/10.3390/molecules28237784
Chicago/Turabian StylePang, Cheng, Yu-Hong Chen, Hui-Hui Bian, Jie-Ping Zhang, Li Su, Hua Han, and Wen Zhang. 2023. "Anti-Inflammatory Ergosteroid Derivatives from the Coral-Associated Fungi Penicillium oxalicum HL-44" Molecules 28, no. 23: 7784. https://doi.org/10.3390/molecules28237784
APA StylePang, C., Chen, Y.-H., Bian, H.-H., Zhang, J.-P., Su, L., Han, H., & Zhang, W. (2023). Anti-Inflammatory Ergosteroid Derivatives from the Coral-Associated Fungi Penicillium oxalicum HL-44. Molecules, 28(23), 7784. https://doi.org/10.3390/molecules28237784








