Characterization of Metabolite Landscape Distinguishes Medicinal Fungus Cordyceps sinensis and other Cordyceps by UHPLC-Q Exactive HF-X Untargeted Metabolomics
Abstract
1. Introduction
2. Results
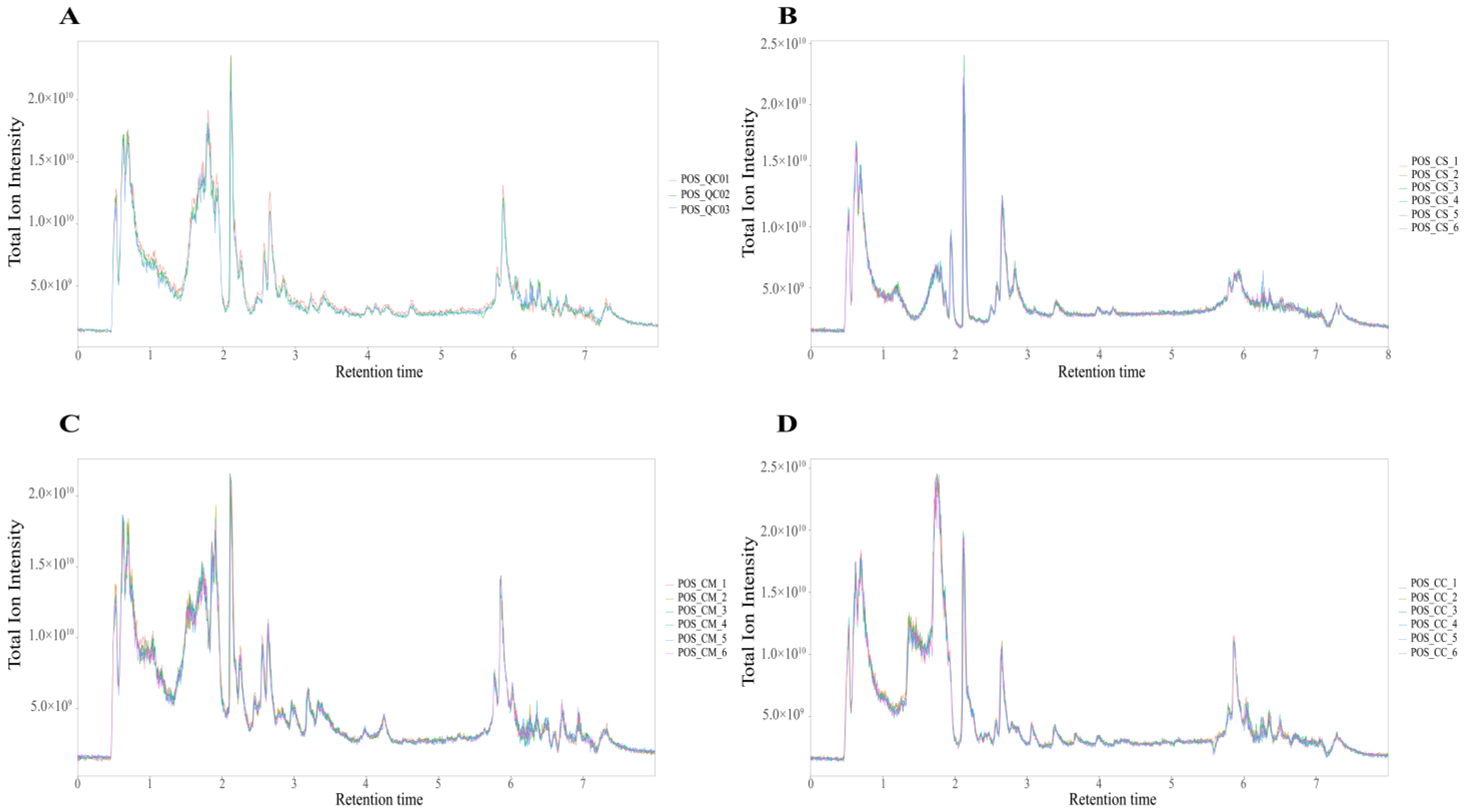
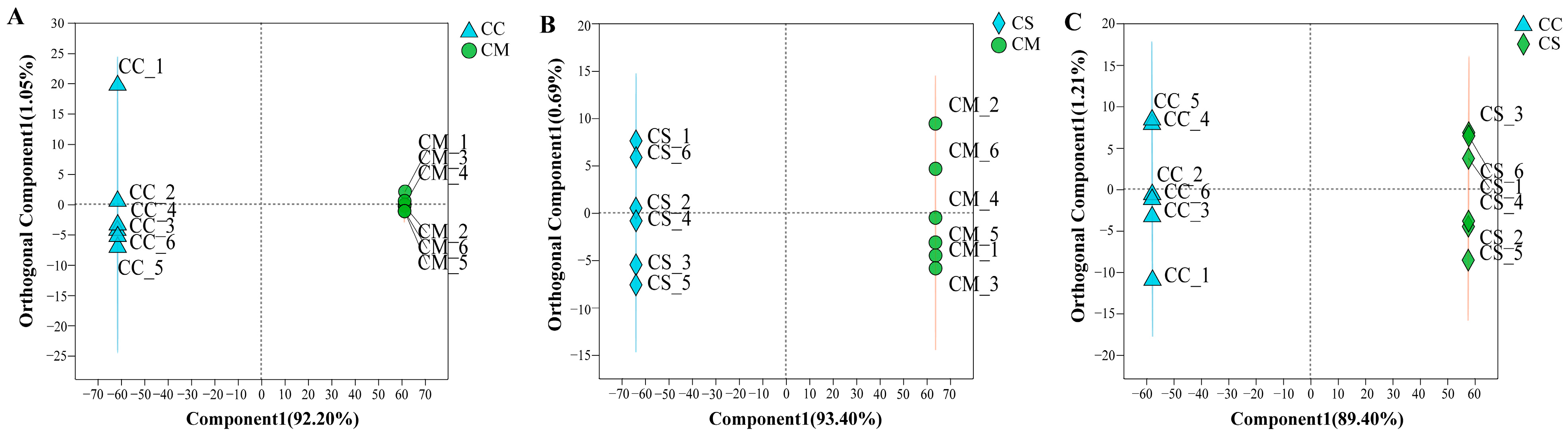
2.1. Metabolite Annotation and Comparative Analysis of Metabolite Profiles
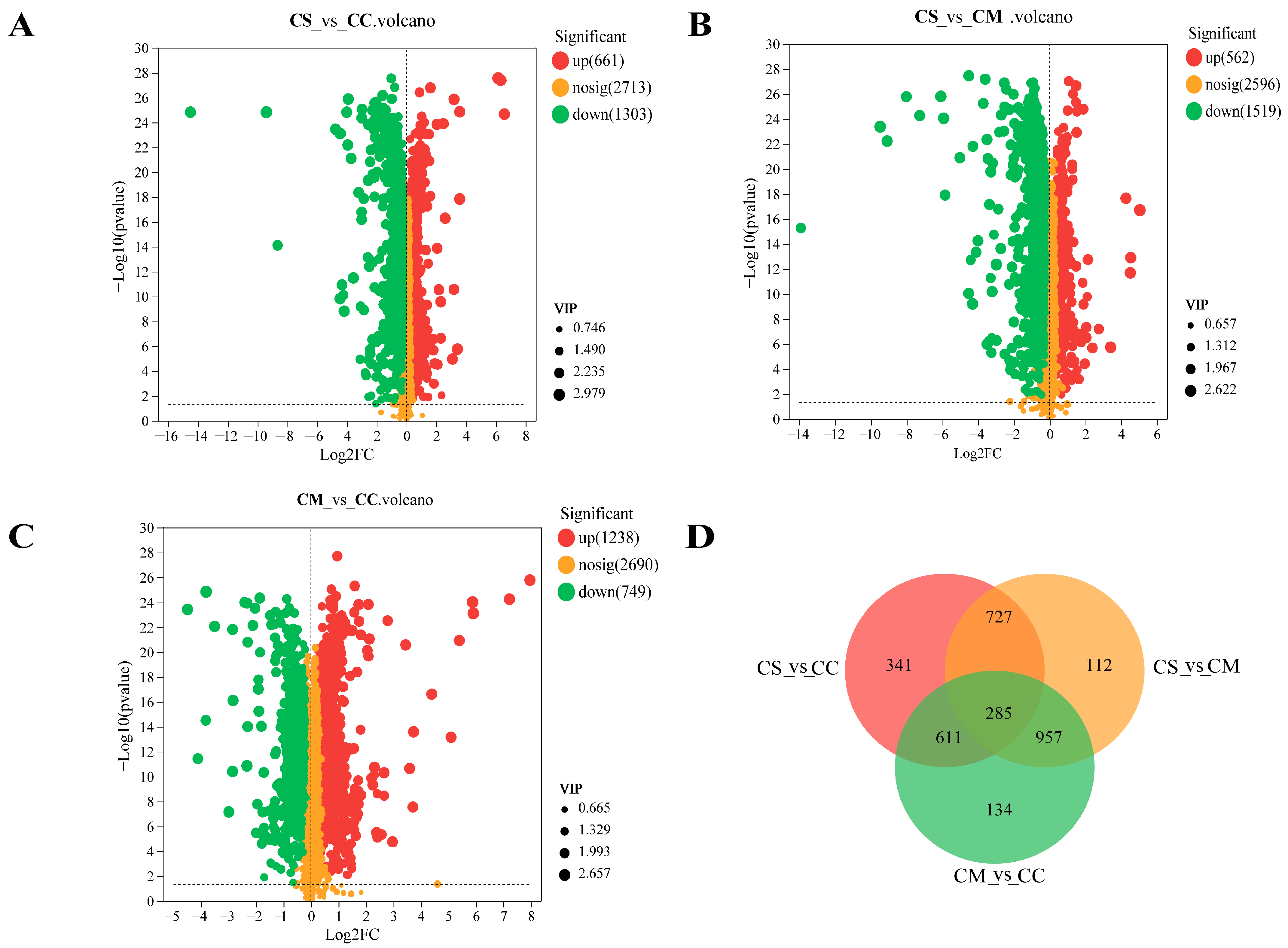
2.2. Screening of Differential Metabolites between C. sinensis and other Cordyceps
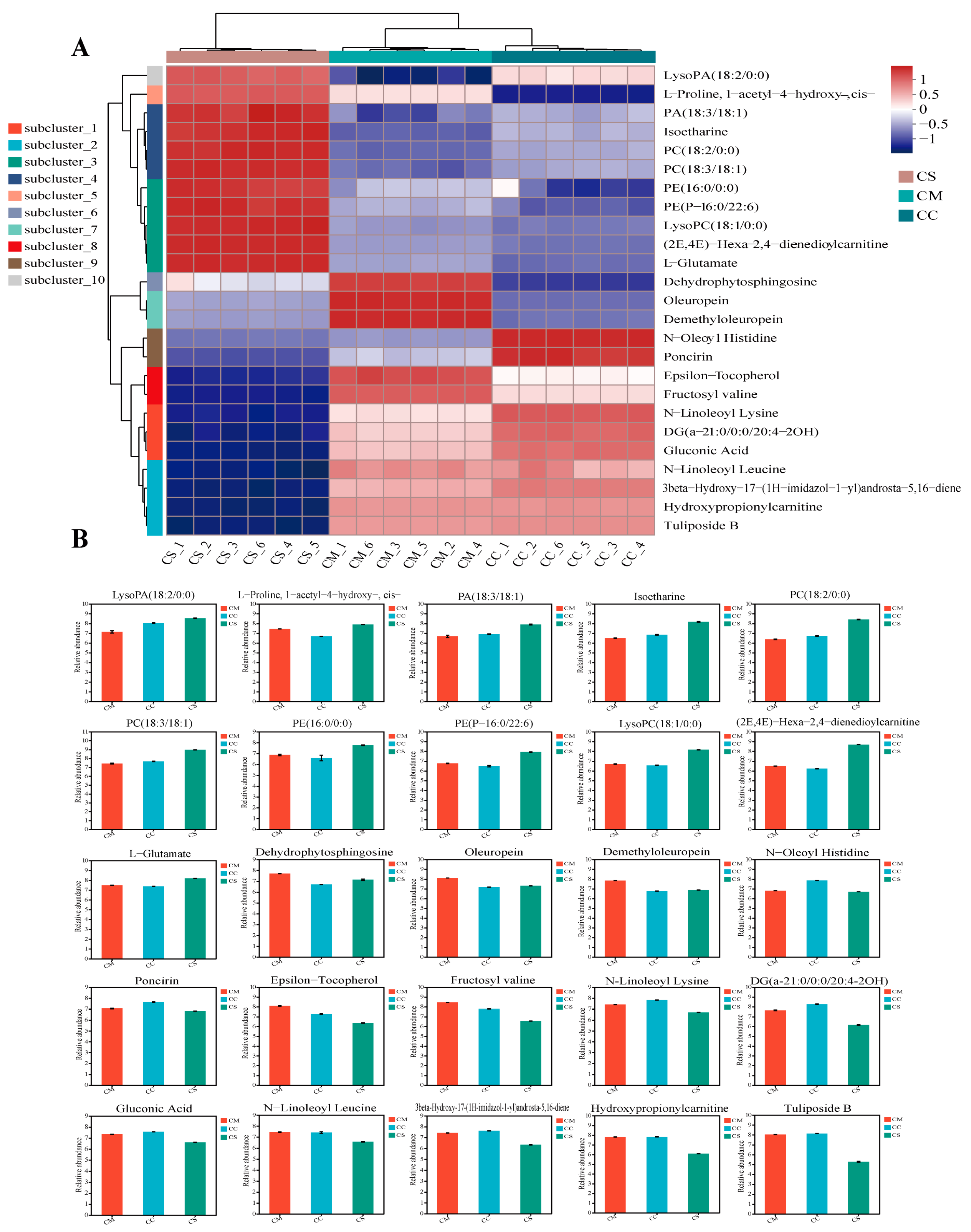
2.3. Hierarchical Cluster Analysis Showed Distinctive Metabolite Profiles
2.4. Differential Metabolite Functional Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Preparation of Sample Extracts
4.3. Quality-Control Sample
4.4. UHPLC-MS/MS Analysis
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, C.; Guo, S.; Wang, W.; Liu, X. Cordyceps industry in China. Mycology 2015, 6, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Sato, H. Situation of type specimens of Cordyceps and allies described by Dr Kobayasi. Mycology 2017, 8, 303–307. [Google Scholar] [CrossRef][Green Version]
- Olatunji, O.J.; Tang, J.; Tola, A.; Auberon, F.; Oluwaniyi, O.; Ouyang, Z. The genus Cordyceps: An extensive review of its traditional uses, phytochemistry and pharmacology. Fitoterapia 2018, 129, 293–316. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, X.; Zhang, H. Extracellular polysaccharide biosynthesis in Cordyceps. Crit. Rev. Microbiol. 2020, 46, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Lin, J.; Guo, L.; Wang, X.; Tian, S.; Liu, C.; Zhao, Y.; Zhao, R. Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 2019, 103, 7835–7841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wen, C.; Duan, Y.; Zhang, H.; Ma, H. Advance in Cordyceps militaris (Linn) Link polysaccharides: Isolation, structure, and bioactivities: A review. Int. J. Biol. Macromol. 2019, 132, 906–914. [Google Scholar] [CrossRef]
- Wang, G.; Sun, C.; Xie, B.; Wang, T.; Liu, H.; Chen, X.; Huang, Q.; Zhang, C.; Li, T.; Deng, W. Cordyceps guangdongensis lipid-lowering formula alleviates fat and lipid accumulation by modulating gut microbiota and short-chain fatty acids in high-fat diet mice. Front. Nutr. 2022, 9, 1038740. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.L.; Xie, J.; Wang, J.T.; Li, G.H.; Pan, X.R.; Zhao, P.J. Activities and metabolomics of Cordyceps gunnii under different culture conditions. Front. Microbiol. 2022, 13, 1076577. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, L.; Wang, J.; Wang, W.; Niyati, N.; Guo, Y.; Wang, X. Chinese caterpillar fungus (Ophiocordyceps sinensis) in China: Current distribution, trading, and futures under climate change and overexploitation. Sci. Total Environ. 2021, 755, 142548. [Google Scholar] [CrossRef]
- Lin, M.; Guo, S.; Xie, D.; Li, S.; Hu, H. Lipidomic profiling of wild cordyceps and its substituents by liquid chromatography-electrospray ionization-tandem mass spectrometry. LWT 2022, 163, 113497. [Google Scholar] [CrossRef]
- Tang, C.-Y.; Wang, J.; Liu, X.; Chen, J.-B.; Liang, J.; Wang, T.; Simpson, W.R.; Li, Y.-L.; Li, X.-Z. Medium optimization for high mycelial soluble protein content of Ophiocordyceps sinensis using response surface methodology. Front. Microbiol. 2022, 13, 1055055. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Chen, X.X.; Zheng, S.C.; Chen, P.; Mo, M.H. The mechanisms of pharmacological activities of Ophiocordyceps sinensis Fungi. Phytother. Res. PTR 2016, 30, 1572–1583. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Huang, Q.; Zheng, Z.; Guan, H.; Liu, S. Construction of a Cordyceps sinensis exopolysaccharide-conjugated selenium nanoparticles and enhancement of their antioxidant activities. Int. J. Biol. Macromol. 2017, 99, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.K.; Wang, W.Q.; Wu, J.Y. Recent advances in Cordyceps sinensis polysaccharides: Mycelial fermentation, isolation, structure, and bioactivities: A review. J. Funct. Foods 2014, 6, 33–47. [Google Scholar] [CrossRef]
- Lo, H.C.; Hsieh, C.; Lin, F.Y.; Hsu, T.H. A systematic review of the mysterious Caterpillar fungus Ophiocordyceps sinensis in Dong-ChongXiaCao (Dōng Chóng Xià Cǎo) and related bioactive ingredients. J. Tradit. Complement. Med. 2013, 3, 16–32. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Li, W.; Li, Q.; Qian, Z.; Liu, X.; Dong, C. A breakthrough in the artificial cultivation of Chinese cordyceps on a large-scale and its impact on science, the economy, and industry. Crit. Rev. Biotechnol. 2019, 39, 181–191. [Google Scholar] [CrossRef]
- Chatnarin, S.; Thirabunyanon, M. Potential bioactivities via anticancer, antioxidant, and immunomodulatory properties of cultured mycelial enriched β-D-glucan polysaccharides from a novel fungus Ophiocordyceps sinensis OS8. Front. Immunol. 2023, 14, 1150287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, H.; Li, S.; Zhong, X.; Wang, H.; Liu, X. Comparative metabolic profiling of Ophiocordyceps sinensis and its cultured mycelia using GC-MS. Food Res. Int. 2020, 134, 109241. [Google Scholar] [CrossRef]
- Guo, L.X.; Hong, Y.H.; Zhou, Q.Z.; Zhu, Q.; Xu, X.M.; Wang, J.H. Fungus-larva relation in the formation of Cordyceps sinensis as revealed by stable carbon isotope analysis. Sci. Rep. 2017, 7, 7789. [Google Scholar] [CrossRef]
- Liu, G.; Han, R.; Cao, L. Artificial cultivation of the Chinese cordyceps from injected ghost moth larvae. Environ. Entomol. 2019, 48, 1088–1094. [Google Scholar] [CrossRef]
- Nxumalo, W.; Elateeq, A.A.; Sun, Y. Can Cordyceps cicadae be used as an alternative to Cordyceps militaris and Cordyceps sinensis? —A review. J. Ethnopharmacol. 2020, 257, 112879. [Google Scholar] [CrossRef]
- Zhao, H.; Li, M.; Liu, L.; Li, D.; Zhao, L.; Wu, Z.; Zhou, M.; Jia, L.; Yang, F. Cordyceps militaris polysaccharide alleviates diabetic symptoms by regulating gut microbiota against TLR4/NF-κB pathway. Int. J. Biol. Macromol. 2023, 230, 123241. [Google Scholar] [CrossRef]
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: Current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef]
- Ke, B.J.; Lee, C.L. Using submerged fermentation to fast increase N6-(2-hydroxyethyl)-adenosine, adenosine and polysaccharide productions of Cordyceps cicadae NTTU 868. AMB Express 2019, 9, 198. [Google Scholar] [CrossRef]
- Li, L.; Zhang, T.; Li, C.; Xie, L.; Li, N.; Hou, T.; Wang, Y.; Wang, B. Potential therapeutic effects of Cordyceps cicadae and Paecilomyces cicadae on adenine-induced chronic renal failure in rats and their phytochemical analysis. Drug Des. Dev. Ther. 2019, 13, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Kmonickova, E.; Han, R.L.; Zhou, W.; Yang, K.B.; Lu, H.F.; Wang, Z.Q.; Zhao, H.; Wang, H. Comprehensive evaluation of wild Cordyceps cicadae from different geographical origins by TOPSIS method based on the macroscopic infrared spectroscopy (IR) fingerprint. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 214, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Mapuranga, J.; Chang, J.; Zhang, L.; Zhang, N.; Yang, W. Fungal secondary metabolites and small RNAs enhance pathogenicity during plant-fungal pathogen interactions. J. Fungi 2022, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, G. Analysis of secondary metabolites from plant endophytic fungi. Methods Mol. Biol. 2018, 1848, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Alberti, F.; Kaleem, S.; Weaver, J.A. Recent developments of tools for genome and metabolome studies in basidiomycete fungi and their application to natural product research. Biol. Open 2020, 9, bio056010. [Google Scholar] [CrossRef]
- Svoboda, P.; Sander, D.; Plachká, K.; Nováková, L. Development of matrix effect-free MISPE-UHPLC-MS/MS method for determination of lovastatin in Pu-erh tea, oyster mushroom, and red yeast rice. J. Pharm. Biomed. Anal. 2017, 140, 367–376. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, X.; Miao, Y.; Li, M.; Huang, L. Cordyceps cicadae and Cordyceps gunnii have closer species correlation with Cordyceps sinensis: From the perspective of metabonomic and MaxEnt models. Sci. Rep. 2022, 12, 20469. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Jahan, M.S.; Tang, K.; Jiang, S.; Guo, J.; Luo, S.; Luo, W.; Li, G. Comparative analysis of the medicinal and nutritional components of different varieties of Pueraria thomsonii and Pueraria lobata. Front. Plant Sci. 2023, 14, 1115782. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Geng, F.; Xu, Y.; Li, X.; Liu, D.; Liu, Z.; Luo, Z.; Wang, J. Quantitative transcriptomic and metabolomic analyses reveal the changes in Tricholoma matsutake fruiting bodies during cold storage. Food Chem. 2022, 381, 132292. [Google Scholar] [CrossRef]
- Stohs, S.J.; Hartman, M.J. A review of the receptor binding and pharmacological effects of N-methyltyramine. Phytother. Res. PTR 2015, 29, 14–16. [Google Scholar] [CrossRef]
- Ley, J.P.; Engelhart, K.; Bernhardt, J.; Bertram, H.J. 3,4-Dihydroxymandelic acid, a noradrenalin metabolite with powerful antioxidative potential. J. Agric. Food Chem. 2002, 50, 5897–5902. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, 603–610. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, 622–631. [Google Scholar] [CrossRef]
- Li, T.; Wu, F.; Zhang, A.; Dong, H.; Ullah, I.; Lin, H.; Miao, J.; Sun, H.; Han, Y.; He, Y.; et al. High-Throughput Chinmedomics Strategy Discovers the Quality Markers and Mechanisms of Wutou Decoction Therapeutic for Rheumatoid Arthritis. Front. Pharmacol. 2022, 13, 854087. [Google Scholar] [CrossRef]
- Guo, S.; Lin, M.; Xie, D.; Zhang, W.; Zhang, M.; Zhou, L.; Li, S.; Hu, H. Comparative metabolic profiling of wild Cordyceps species and their substituents by liquid chromatography-tandem mass spectrometry. Front. Pharmacol. 2022, 13, 1036589. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, J.T.; Brosnan, M.E. Glutamate: A truly functional amino acid. Amino Acids 2013, 45, 413–418. [Google Scholar] [CrossRef]
- Blachier, F.; Boutry, C.; Bos, C.; Tomé, D. Metabolism and functions of L-glutamate in the epithelial cells of the small and large intestines. Am. J. Clin. Nutr. 2009, 90, 814s–821s. [Google Scholar] [CrossRef] [PubMed]
- Albarracin, S.L.; Baldeon, M.E.; Sangronis, E.; Petruschina, A.C.; Reyes, F.G.R. L-glutamate: A key amino acid for senory and metabolic functions. Arch. Latinoam. Nutr. 2016, 66, 101–112. [Google Scholar]
- Li, B.; Chen, T.; Hu, W.; Wang, Z.; Wu, J.; Zhou, Q.; Li, P. Poncirin ameliorates cardiac ischemia-reperfusion injury by activating PI3K/AKT/PGC-1α signaling. Eur. J. Pharmacol. 2022, 917, 174759. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, L.; Wang, B. Poncirin ameliorates oxygen glucose deprivation/reperfusion injury in cortical neurons via inhibiting NOX4-mediated NLRP3 inflammasome activation. Int. Immunopharmacol. 2022, 102, 107210. [Google Scholar] [CrossRef]
- Igarashi, Y.; Takagi, K.; Kajiura, T.; Furumai, T.; Oki, T. Glucosylquestiomycin, a novel antibiotic from Microbispora sp. TP-A0184: Fermentation, isolation, structure determination, synthesis and biological activities. J. Antibiot. 1998, 51, 915–920. [Google Scholar] [CrossRef]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zhao, Z. Moderate warming will expand the suitable habitat of Ophiocordyceps sinensis and expand the area of O. sinensis with high adenosine content. Sci. Total Environ. 2021, 787, 147605. [Google Scholar] [CrossRef]
- Guo, L.-X.; Xu, X.-M.; Wu, C.-F.; Lin, L.; Zou, S.-C.; Luan, T.; Yuan, J.-P.; Wang, J.-H. Fatty acid composition of lipids in wild Cordyceps sinensis from major habitats in China. Biomed. Prev. Nutr. 2012, 2, 42–50. [Google Scholar] [CrossRef]
- He, Y.; Zhang, W.; Peng, F.; Lu, R.; Zhou, H.; Bao, G.; Wang, B.; Huang, B.; Li, Z.; Hu, F. Metabolomic variation in wild and cultured cordyceps and mycelia of Isaria cicadae. Biomed. Chromatogr. BMC 2019, 33, e4478. [Google Scholar] [CrossRef]
- Mi, J.; Han, Y.; Xu, Y.; Kou, J.; Li, W.J.; Wang, J.R.; Jiang, Z.H. Deep profiling of immunosuppressive glycosphingolipids and sphingomyelins in Wild Cordyceps. J. Agric. Food Chem. 2018, 66, 8991–8998. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, Z.; Chen, J.; Duan, T.; He, H.; Tang, S. Characterization of metabolite landscape distinguishes wild from cultivated Polygonati Rhizomes by UHPLC-Q-TOF-MS untargeted metabolomics. Food Biosci. 2023, 53, 102574. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.; Li, X.; Wang, T.; Wang, J.; Xiao, M.; He, M.; Chang, X.; Fan, Y.; Li, Y. Characterization of Metabolite Landscape Distinguishes Medicinal Fungus Cordyceps sinensis and other Cordyceps by UHPLC-Q Exactive HF-X Untargeted Metabolomics. Molecules 2023, 28, 7745. https://doi.org/10.3390/molecules28237745
Tang C, Li X, Wang T, Wang J, Xiao M, He M, Chang X, Fan Y, Li Y. Characterization of Metabolite Landscape Distinguishes Medicinal Fungus Cordyceps sinensis and other Cordyceps by UHPLC-Q Exactive HF-X Untargeted Metabolomics. Molecules. 2023; 28(23):7745. https://doi.org/10.3390/molecules28237745
Chicago/Turabian StyleTang, Chuyu, Xiuzhang Li, Tao Wang, Jie Wang, Mengjun Xiao, Min He, Xiyun Chang, Yuejun Fan, and Yuling Li. 2023. "Characterization of Metabolite Landscape Distinguishes Medicinal Fungus Cordyceps sinensis and other Cordyceps by UHPLC-Q Exactive HF-X Untargeted Metabolomics" Molecules 28, no. 23: 7745. https://doi.org/10.3390/molecules28237745
APA StyleTang, C., Li, X., Wang, T., Wang, J., Xiao, M., He, M., Chang, X., Fan, Y., & Li, Y. (2023). Characterization of Metabolite Landscape Distinguishes Medicinal Fungus Cordyceps sinensis and other Cordyceps by UHPLC-Q Exactive HF-X Untargeted Metabolomics. Molecules, 28(23), 7745. https://doi.org/10.3390/molecules28237745






