Comparative Analysis of Amino Acid Profiles in Patients with Glioblastoma and Meningioma Using Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)
Abstract
:1. Introduction
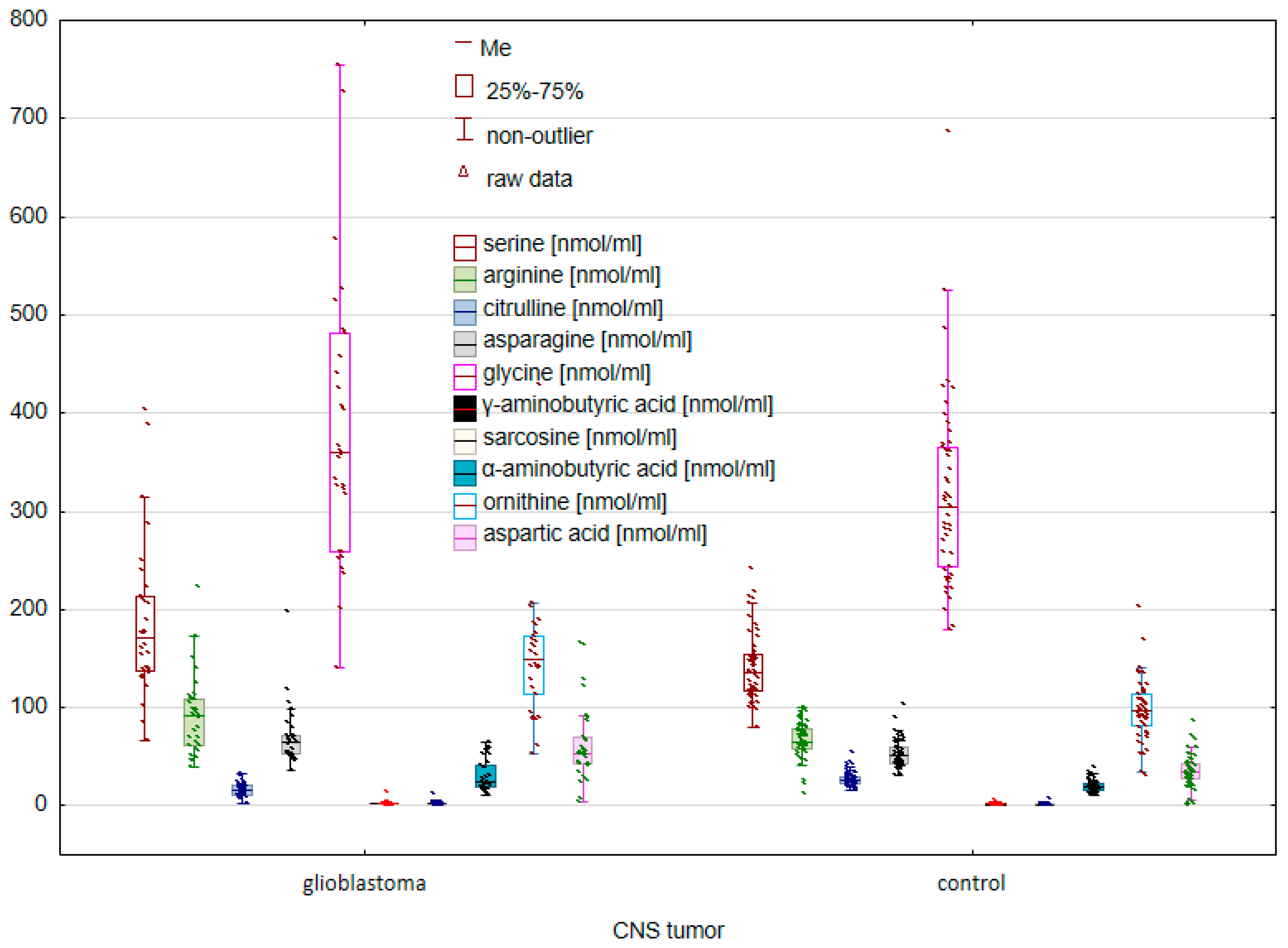
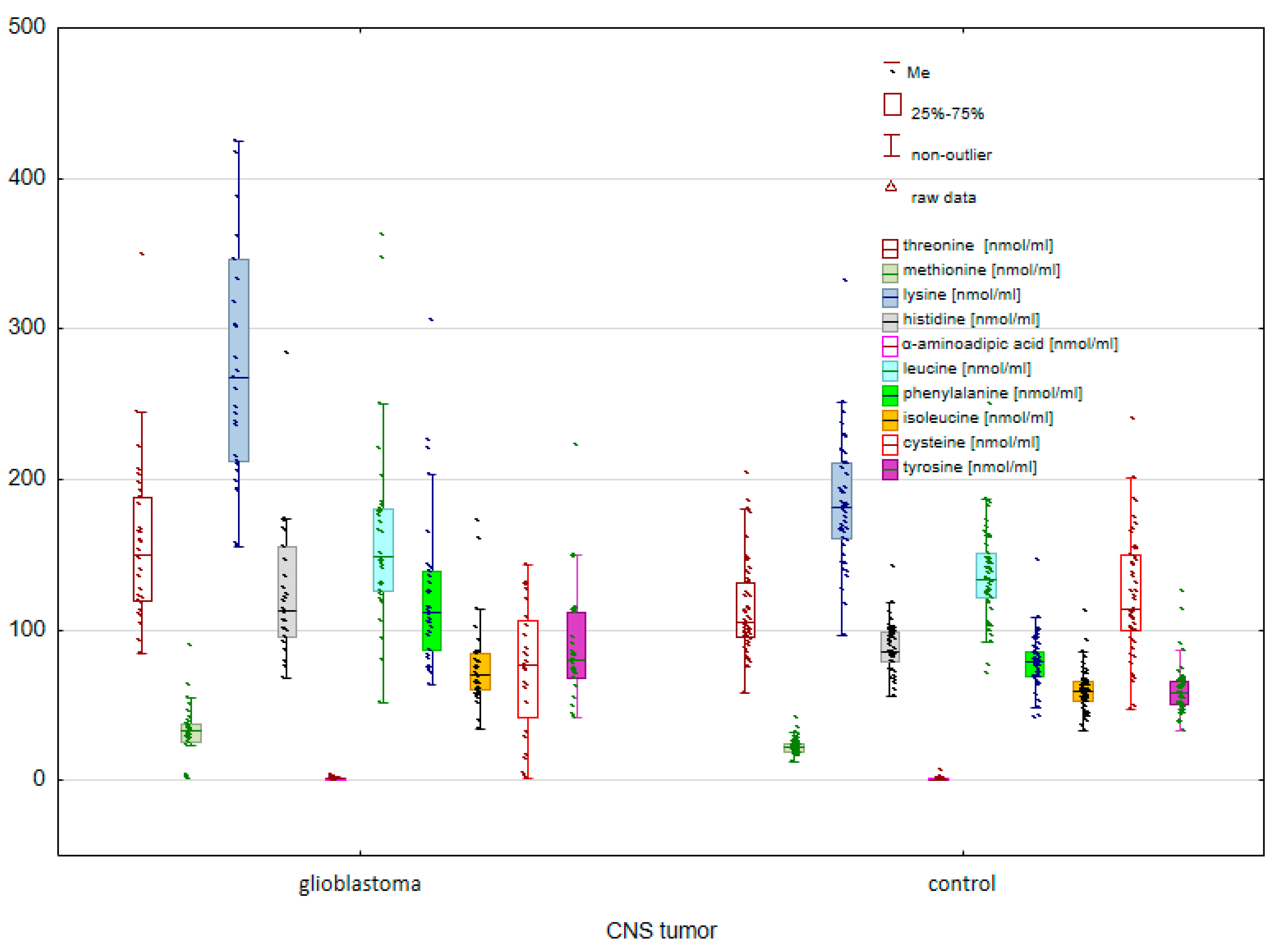
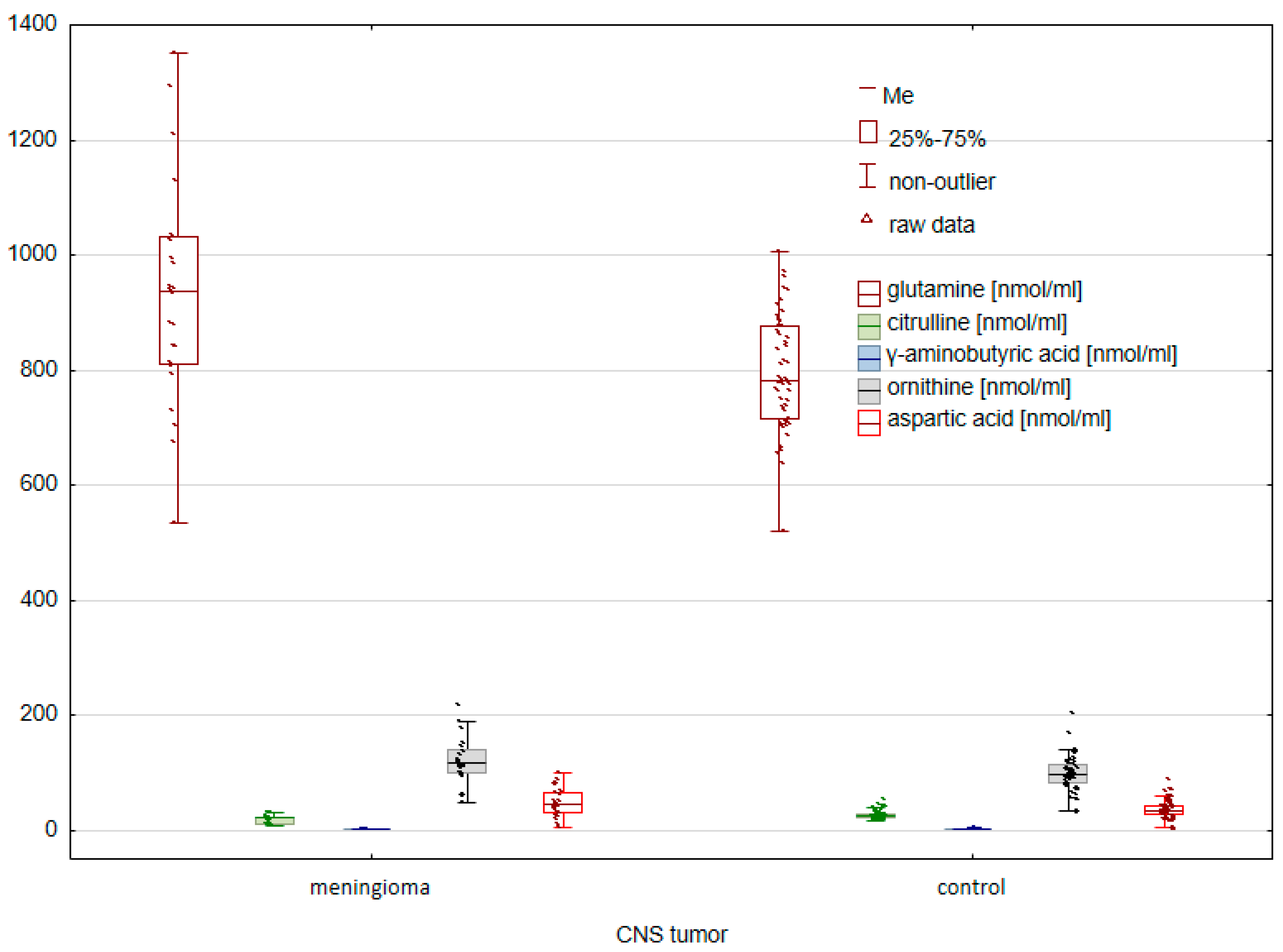
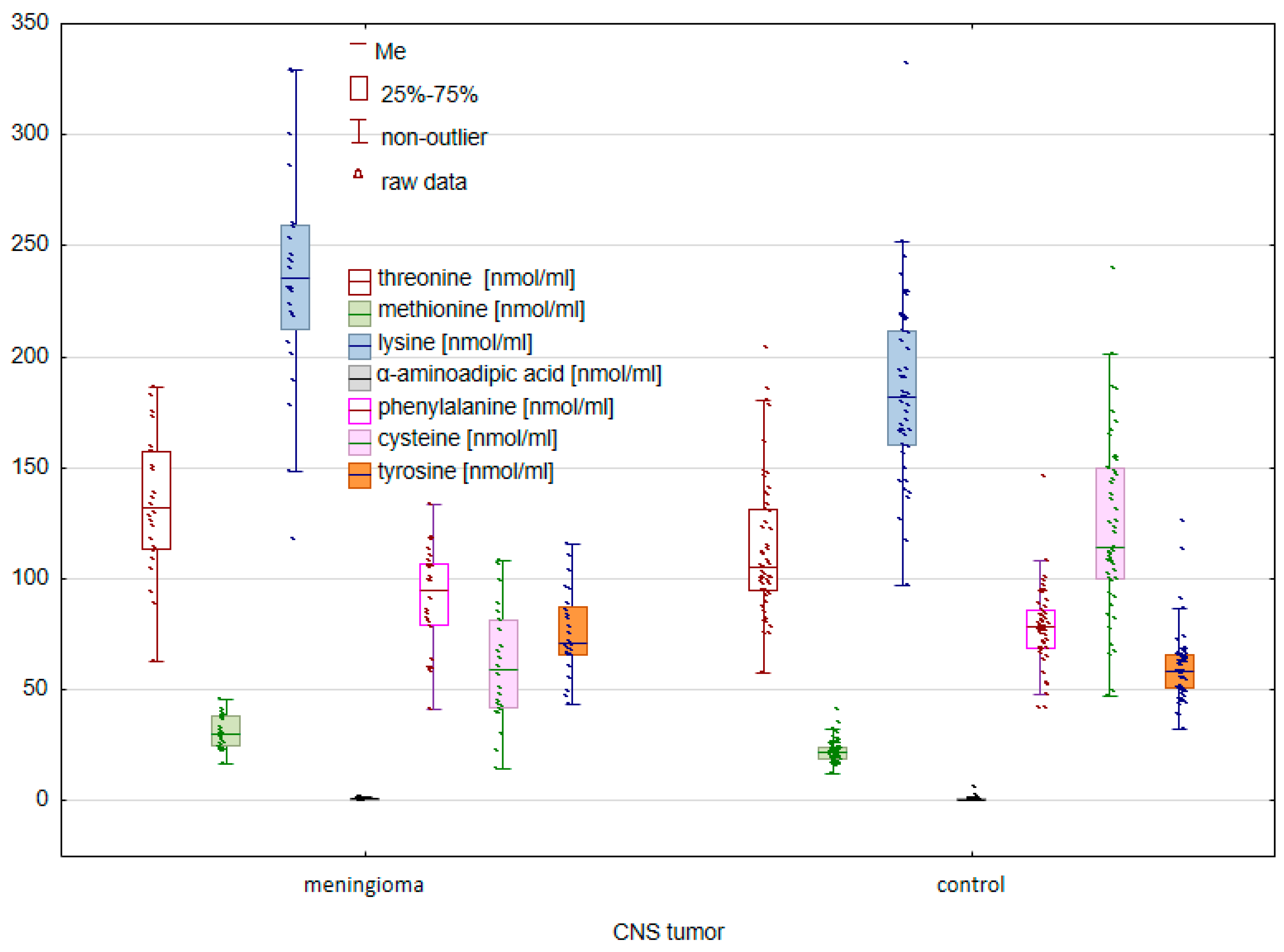
2. Results
2.1. Comparison of the Control Group and Cancer Patients
2.2. Classification Analysis
2.2.1. Receiver Operating Characteristic Curves
2.2.2. Decision Trees
3. Discussion
4. Materials and Methods
4.1. Subjects and Serum Samples
4.2. Sample Preparation
4.3. Instrumentation and Conditions
4.4. Reagents and Solvents
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, J.L.; Schwartzbaum, J.A.; Wrensch, M.; Wiemels, J.L. Epidemiology of Brain Tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef] [PubMed]
- Darlix, A.; Zouaoui, S.; Rigau, V.; Bessaoud, F.; Figarella-Branger, D.; Mathieu-Daudé, H.; Trétarre, B.; Bauchet, F.; Duffau, H.; Taillandier, L.; et al. Epidemiology for Primary Brain Tumors: A Nationwide Population-Based Study. J. Neurooncol. 2017, 131, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Polly, F.P.; Shil, S.K.; Hossain, M.A.; Ayman, A.; Jang, Y.M. Detection and Classification of HGG and LGG Brain Tumor Using Machine Learning. In Proceedings of the 2018 International Conference on Information Networking (ICOIN), Chiang Mai, Thailand, 10–12 January 2018; pp. 813–817. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Epidemiology and Etiology of Gliomas. Acta Neuropathol. 2005, 109, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-oncology 2021, 23, III1–III105. [Google Scholar] [CrossRef]
- Cao, J.; Yan, W.; Zhan, Z.; Hong, X.; Yan, H. Epidemiology and Risk Stratification of Low-Grade Gliomas in the United States, 2004-2019: A Competing-Risk Regression Model for Survival Analysis. Front. Oncol. 2023, 13, 1079597. [Google Scholar] [CrossRef] [PubMed]
- Diwanji, T.P.; Engelman, A.; Snider, J.W.; Mohindra, P. Epidemiology, Diagnosis, and Optimal Management of Glioma in Adolescents and Young Adults. Adolesc. Health Med. Ther. 2017, 8, 99–113. [Google Scholar] [CrossRef]
- Claus, E.B.; Black, P.M. Survival Rates and Patterns of Care for Patients Diagnosed with Supratentorial Low-Grade Gliomas: Data from the SEER Program, 1973–2001. Cancer 2006, 106, 1358–1363. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH Mutation in Glioma: Molecular Mechanisms and Potential Therapeutic Targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Wiemels, J.; Wrensch, M.; Claus, E.B. Epidemiology and Etiology of Meningioma. J. Neurooncol. 2010, 99, 307–314. [Google Scholar] [CrossRef]
- Yamanaka, R.; Hayano, A.; Kanayama, T. Radiation-Induced Meningiomas: An Exhaustive Review of the Literature. World Neurosurg. 2017, 97, 635–644.e8. [Google Scholar] [CrossRef]
- Franz, G. Immunohistochemical Detection of Female Sex Hormone Receptors in Meningiomas: Correlation with Clinical and Histological Features Clinical Study. Neurosurgery 1992, 33, 212. [Google Scholar]
- Alentorn, A.; Hoang-Xuan, K.; Mikkelsen, T. Presenting Signs and Symptoms in Brain Tumors. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 134, pp. 19–26. ISBN 9780128029978. [Google Scholar]
- Snyder, H.; Robinson, K.; Shah, D.; Brennan, R.; Handrigan, M. Signs and Symptoms of Patients with Brain Tumors Presenting to the Emergency Department. J. Emerg. Med. 1993, 11, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Huse, J.T.; Holland, E.C. Targeting Brain Cancer: Advances in the Molecular Pathology of Malignant Glioma and Medulloblastoma. Nat. Rev. Cancer 2010, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Doroudchi, M.; Pishe, Z.G.; Malekzadeh, M.; Golmoghaddam, H.; Taghipour, M.; Ghaderi, A. Elevated Serum IL-17A but Not IL-6 in Glioma versus Meningioma and Schwannoma. Asian Pac. J. Cancer Prev. 2013, 14, 5225–5230. [Google Scholar] [CrossRef]
- Holst, C.B.; Christensen, I.J.; Skjøth-Rasmussen, J.; Hamerlik, P.; Poulsen, H.S.; Johansen, J.S. Systemic Immune Modulation in Gliomas: Prognostic Value of Plasma IL-6, YKL-40, and Genetic Variation in YKL-40. Front. Oncol. 2020, 10, 478. [Google Scholar] [CrossRef]
- Popescu, I.D.; Codrici, E.; Albulescu, L.; Mihai, S.; Enciu, A.M.; Albulescu, R.; Tanase, C.P. Potential Serum Biomarkers for Glioblastoma Diagnostic Assessed by Proteomic Approaches. Proteome Sci. 2014, 12, 47. [Google Scholar] [CrossRef]
- Gautam, P.; Nair, S.C.; Gupta, M.K.; Sharma, R.; Polisetty, R.V.; Uppin, M.S.; Sundaram, C.; Puligopu, A.K.; Ankathi, P.; Purohit, A.K.; et al. Proteins with Altered Levels in Plasma from Glioblastoma Patients as Revealed by ITRAQ-Based Quantitative Proteomic Analysis. PLoS ONE 2012, 7, e46153. [Google Scholar] [CrossRef]
- Arora, A.; Patil, V.; Kundu, P.; Kondaiah, P.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Pal, D.; Somasundaram, K. Serum Biomarkers Identification by ITRAQ and Verification by MRM: S100A8/S100A9 Levels Predict Tumor-Stroma Involvement and Prognosis in Glioblastoma. Sci. Rep. 2019, 9, 2749. [Google Scholar] [CrossRef]
- Gielen, P.R.; Schulte, B.M.; Kers-Rebel, E.D.; Verrijp, K.; Bossman, S.A.J.F.H.; Ter Laan, M.; Wesseling, P.; Adema, G.J. Elevated Levels of Polymorphonuclear Myeloid-Derived Suppressor Cells in Patients with Glioblastoma Highly Express S100A8/9 and Arginase and Suppress T Cell Function. Neuro-oncology 2016, 18, 1253–1264. [Google Scholar] [CrossRef]
- Lyubimova, N.V.; Toms, M.G.; Popova, E.E.; Bondarenko, Y.V.; Krat, V.B.; Kushlinskii, N.E.; Blokhin, N.N. Neurospecificity Proteins in the Serum of Patients with Brain Tumors. Bull. Exp. Biol. Med. 2010, 150, 678–681. [Google Scholar]
- Gollapalli, K.; Ray, S.; Srivastava, R.; Renu, D.; Singh, P.; Dhali, S.; Bajpai Dikshit, J.; Srikanth, R.; Moiyadi, A.; Srivastava, S. Investigation of Serum Proteome Alterations in Human Glioblastoma Multiforme. Proteomics 2012, 12, 2378–2390. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Meng, Z.; Song, H.W.; Yao, K.; Duan, Z.J.; Yu, C.J.; Li, S.W.; Yan, C.X. Preoperative Changes in Hematological Markers and Predictors of Glioma Grade and Survival. Front. Pharmacol. 2018, 9, 886. [Google Scholar] [CrossRef] [PubMed]
- Nijaguna, M.B.; Schröder, C.; Patil, V.; Shwetha, S.D.; Hegde, A.S.; Chandramouli, B.A.; Arivazhagan, A.; Santosh, V.; Hoheisel, J.D.; Somasundaram, K. Definition of a Serum Marker Panel for Glioblastoma Discrimination and Identification of Interleukin 1β in the Microglial Secretome as a Novel Mediator of Endothelial Cell Survival Induced by C-Reactive Protein. J. Proteom. 2015, 128, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.H.; Huang, J.L.; Chen, M.; Wang, B.L.; Ou, Q.S.; Huang, S.Y. Diagnostic Value of Preoperative Inflammatory Markers in Patients with Glioma: A Multicenter Cohort Study. J. Neurosurg. 2018, 129, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.; Persson, O.; Ingvarsson, J.; Widegren, B.; Salford, L.; Borrebaeck, C.A.K.; Wingren, C. Plasma Proteome Profiling Reveals Biomarker Patterns Associated with Prognosis and Therapy Selection in Glioblastoma Multiforme Patients. Proteom. Clin. Appl. 2010, 4, 591–602. [Google Scholar] [CrossRef]
- Linhares, P.; Carvalho, B.; Vaz, R.; Costa, B.M. Glioblastoma: Is There Any Blood Biomarker with True Clinical Relevance? Int. J. Mol. Sci. 2020, 21, 5809. [Google Scholar] [CrossRef]
- Ali, H.; Harting, R.; de Vries, R.; Ali, M.; Wurdinger, T.; Best, M.G. Blood-Based Biomarkers for Glioma in the Context of Gliomagenesis: A Systematic Review. Front. Oncol. 2021, 11, 665235. [Google Scholar] [CrossRef]
- Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. [Google Scholar] [CrossRef]
- Maher, E.A.; Marin-Valencia, I.; Bachoo, R.M.; Mashimo, T.; Raisanen, J.; Hatanpaa, K.J.; Jindal, A.; Jeffrey, F.M.; Choi, C.; Madden, C.; et al. Metabolism of [U-13C]Glucose in Human Brain Tumors in Vivo. NMR Biomed. 2012, 25, 1234–1244. [Google Scholar] [CrossRef]
- Mashimo, T.; Pichumani, K.; Vemireddy, V.; Hatanpaa, K.J.; Singh, D.K.; Sirasanagandla, S.; Nannepaga, S.; Piccirillo, S.G.; Kovacs, Z.; Foong, C.; et al. Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell 2014, 159, 1603–1614. [Google Scholar] [CrossRef]
- Baranovičová, E.; Galanda, T.; Galanda, M.; Hatok, J.; Kolarovszki, B.; Richterová, R.; Račay, P. Metabolomic Profiling of Blood Plasma in Patients with Primary Brain Tumours: Basal Plasma Metabolites Correlated with Tumour Grade and Plasma Biomarker Analysis Predicts Feasibility of the Successful Statistical Discrimination from Healthy Subjects—A Preliminary Study. IUBMB Life 2019, 71, 1994–2002. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Caflisch, L.; Lodi, A.; Brenner, A.J.; Tiziani, S. Metabolomic Signature of Brain Cancer. Mol. Carcinog. 2017, 56, 2355–2371. [Google Scholar] [CrossRef]
- Masalha, W.; Daka, K.; Woerner, J.; Pompe, N.; Weber, S.; Delev, D.; Krüger, M.T.; Schnell, O.; Beck, J.; Heiland, D.H.; et al. Metabolic Alterations in Meningioma Reflect the Clinical Course. BMC Cancer 2021, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- Ferrasi, A.C.; Puttini, R.; Galvani, A.F.; Hamamoto Filho, P.T.; Delafiori, J.; Argente, V.D.; de Oliveira, A.N.; Dias-Audibert, F.L.; Catharino, R.R.; Silva, O.C.; et al. Metabolomics Approach Reveals Important Glioblastoma Plasma Biomarkers for Tumor Biology. Int. J. Mol. Sci. 2023, 24, 8813. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, P.; Kensicki, E.; Bloom, G.; Prabhu, A.; Sarcar, B.; Kahali, S.; Eschrich, S.; Qu, X.; Forsyth, P.; Gillies, R. The Metabolomic Signature of Malignant Glioma Reflects Accelerated Anabolic Metabolism. Cancer Res. 2012, 72, 5878–5888. [Google Scholar] [CrossRef]
- Huang, J.; Weinstein, S.J.; Kitahara, C.M.; Karoly, E.D.; Sampson, J.N.; Albanes, D. A Prospective Study of Serum Metabolites and Glioma Risk. Oncotarget 2017, 8, 70366–70377. [Google Scholar] [CrossRef]
- Locasale, J.W.; Melman, T.; Song, S.; Yang, X.; Swanson, K.D.; Cantley, L.C.; Wong, E.T.; Asara, J.M. Metabolomics of Human Cerebrospinal Fluid Identifies Signatures of Malignant Glioma. Mol. Cell. Proteom. 2012, 11, M111.014688. [Google Scholar] [CrossRef]
- Nakamizo, S.; Sasayama, T.; Shinohara, M.; Irino, Y.; Nishiumi, S.; Nishihara, M.; Tanaka, H.; Tanaka, K.; Mizukawa, K.; Itoh, T.; et al. GC/MS-Based Metabolomic Analysis of Cerebrospinal Fluid (CSF) from Glioma Patients. J. Neurooncol. 2013, 113, 65–74. [Google Scholar] [CrossRef]
- Kurokawa, G.A.; Hamamoto Filho, P.T.; Delafiori, J.; Galvani, A.F.; de Oliveira, A.N.; Dias-Audibert, F.L.; Catharino, R.R.; Pardini, M.I.M.C.; Zanini, M.A.; Lima, E.d.O.; et al. Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases. Int. J. Mol. Sci. 2023, 24, 394. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The Apogee of the Omics Trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Griffin, J.L.; Kauppinen, R.A. A Metabolomics Perspective of Human Brain Tumours. FEBS J. 2007, 274, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Xiong, N.; Gao, X.; Zhao, H.; Cai, F.; Zhang, F.-C.; Yuan, Y.; Liu, W.; He, F.; Zacharias, L.G.; Lin, H.; et al. Using Arterial–Venous Analysis to Characterize Cancer Metabolic Consumption in Patients. Nat. Commun. 2020, 11, 3169. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, 603837. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, J.; Shen, A.; Miao, Y.; Cao, Y.; Zhang, Y.; Cong, P.; Gao, P. Rewired Metabolism of Amino Acids and Its Roles in Glioma Pathology. Metabolites 2022, 12, 918. [Google Scholar] [CrossRef]
- Bernhard, C.; Reita, D.; Martin, S.; Entz-Werle, N.; Dontenwill, M. Glioblastoma Metabolism: Insights and Therapeutic Strategies. Int. J. Mol. Sci. 2023, 24, 9137. [Google Scholar] [CrossRef] [PubMed]
- Danzi, F.; Pacchiana, R.; Mafficini, A.; Scupoli, M.T.; Scarpa, A.; Donadelli, M.; Fiore, A. To Metabolomics and beyond: A Technological Portfolio to Investigate Cancer Metabolism. Signal Transduct. Target. Ther. 2023, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, K.; Xie, Q.; Wu, Q.; Mack, S.C.; Shi, Y.; Kim, L.J.Y.; Prager, B.C.; Flavahan, W.A.; Liu, X.; et al. Purine Synthesis Promotes Maintenance of Brain Tumor Initiating Cells in Glioma. Nat. Neurosci. 2017, 20, 661–673. [Google Scholar] [CrossRef]
- Palanichamy, K.; Thirumoorthy, K.; Kanji, S.; Gordon, N.; Singh, R.; Jacob, J.R.; Sebastian, N.; Litzenberg, K.T.; Patel, D.; Bassett, E.; et al. Methionine and Kynurenine Activate Oncogenic Kinases in Glioblastoma, and Methionine Deprivation Compromises Proliferation. Clin. Cancer Res. 2016, 22, 3513–3523. [Google Scholar] [CrossRef]
- Prabhu, A.; Sarcar, B.; Kahali, S.; Yuan, Z.; Johnson, J.J.; Adam, K.P.; Kensicki, E.; Chinnaiyan, P. Cysteine Catabolism: A Novel Metabolic Pathway Contributing to Glioblastoma Growth. Cancer Res. 2014, 74, 787–796. [Google Scholar] [CrossRef]
- Dols, M.C.; Domínguez López, M.; Ramírez Plaza, C.; Pérez Miranda, E.; Gil Calle, S.; Chamorro, E.V.; Alés Díaz, I.; Pino, A.M.; García, J.A.; Calderón, V.G. Specific Alterations in the Serum Amino Acid Profile of Patients with Lung Cancer and Head and Neck Cancer. Oncologica 2006, 29, 283–290. [Google Scholar]
- Jia, B.; Wang, W.; Lin, S.; Shi, L.; Li, Y.; Gu, Y.; Gao, F.; Qin, Y. The Free Amino Acid Profiles and Metabolic Biomarkers of Predicting the Chemotherapeutic Response in Advanced Sarcoma Patients. Clin. Transl. Oncol. 2020, 22, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Cadoni, G.; Giraldi, L.; Chiarla, C.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Settimi, S.; Galli, J.; Paludetti, G.; Arzani, D.; et al. Prognostic Role of Serum Amino Acids in Head and Neck Cancer. Dis. Markers 2020, 2020, 2291759. [Google Scholar] [CrossRef] [PubMed]
- Toklu, S.; Kemerdere, R.; Kacira, T.; Gurses, M.S.; Benli Aksungar, F.; Tanriverdi, T. Tissue and Plasma Free Amino Acid Detection by LC-MS/MS Method in High Grade Glioma Patients. J. Neurooncol 2023, 163, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Mörén, L.; Wibom, C.; Bergström, P.; Johansson, M.; Antti, H.; Bergenheim, A.T. Characterization of the Serum Metabolome Following Radiation Treatment in Patients with High-Grade Gliomas. Radiat. Oncol. 2016, 11, 51. [Google Scholar] [CrossRef]
- Wang, T.J.; Ngo, D.; Psychogios, N.; Dejam, A.; Larson, M.G.; Vasan, R.S.; Ghorbani, A.; O’sullivan, J.; Cheng, S.; Rhee, E.P.; et al. 2-Aminoadipic Acid Is a Biomarker for Diabetes Risk. J. Clin. Investig. 2013, 123, 4309. [Google Scholar] [CrossRef]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New Aspects of Amino Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Watanabe, A.; Higashi, T.; Sakata, T.; Nagashima, H. Serum Amino Acid Levels in Patients With Hepatocellular Carcinoma. Cancer 1984, 54, 1875–1882. [Google Scholar] [CrossRef]
- Diez-Ruiz, A.; Tilz, G.P.; Zangerle, R.; Baier-Bitterlich, G.; Wachter, H.; Fuchs, D. Soluble Receptors for Tumour Necrosis Factor in Clinical Laboratory Diagnosis. Eur. J. Haematol. 1995, 54, 1–8. [Google Scholar] [CrossRef]
- Neurauter, G.; Grahmann, A.V.; Klieber, M.; Zeimet, A.; Ledochowski, M.; Sperner-Unterweger, B.; Fuchs, D. Serum Phenylalanine Concentrations in Patients with Ovarian Carcinoma Correlate with Concentrations of Immune Activation Markers and of Isoprostane-8. Cancer Lett. 2008, 272, 141–147. [Google Scholar] [CrossRef]
- Ploder, M.; Neurauter, G.; Spittler, A.; Schroecksnadel, K.; Roth, E.; Fuchs, D. Serum Phenylalanine in Patients Post Trauma and with Sepsis Correlate to Neopterin Concentrations. Amino Acids 2008, 35, 303–307. [Google Scholar] [CrossRef]
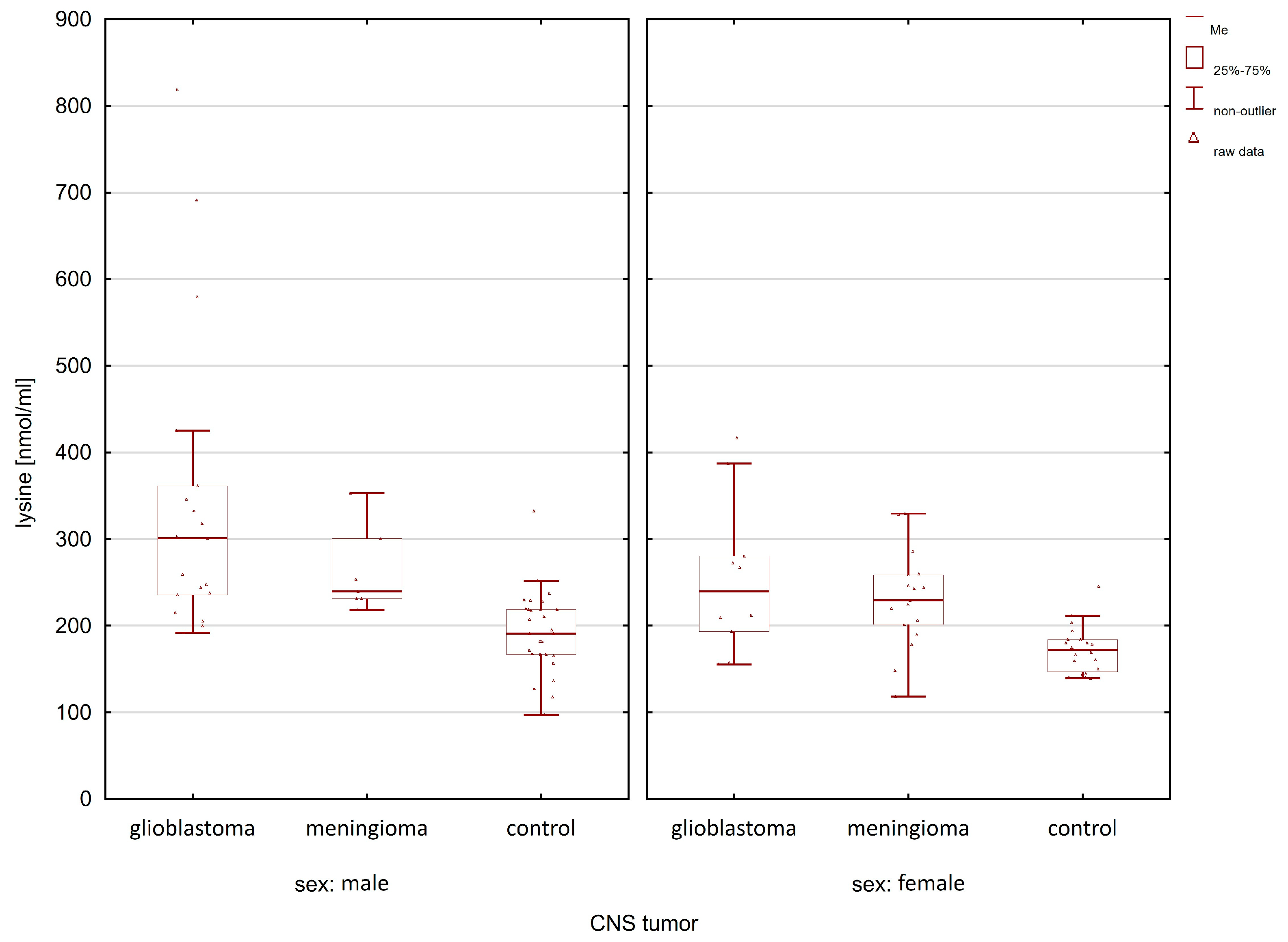
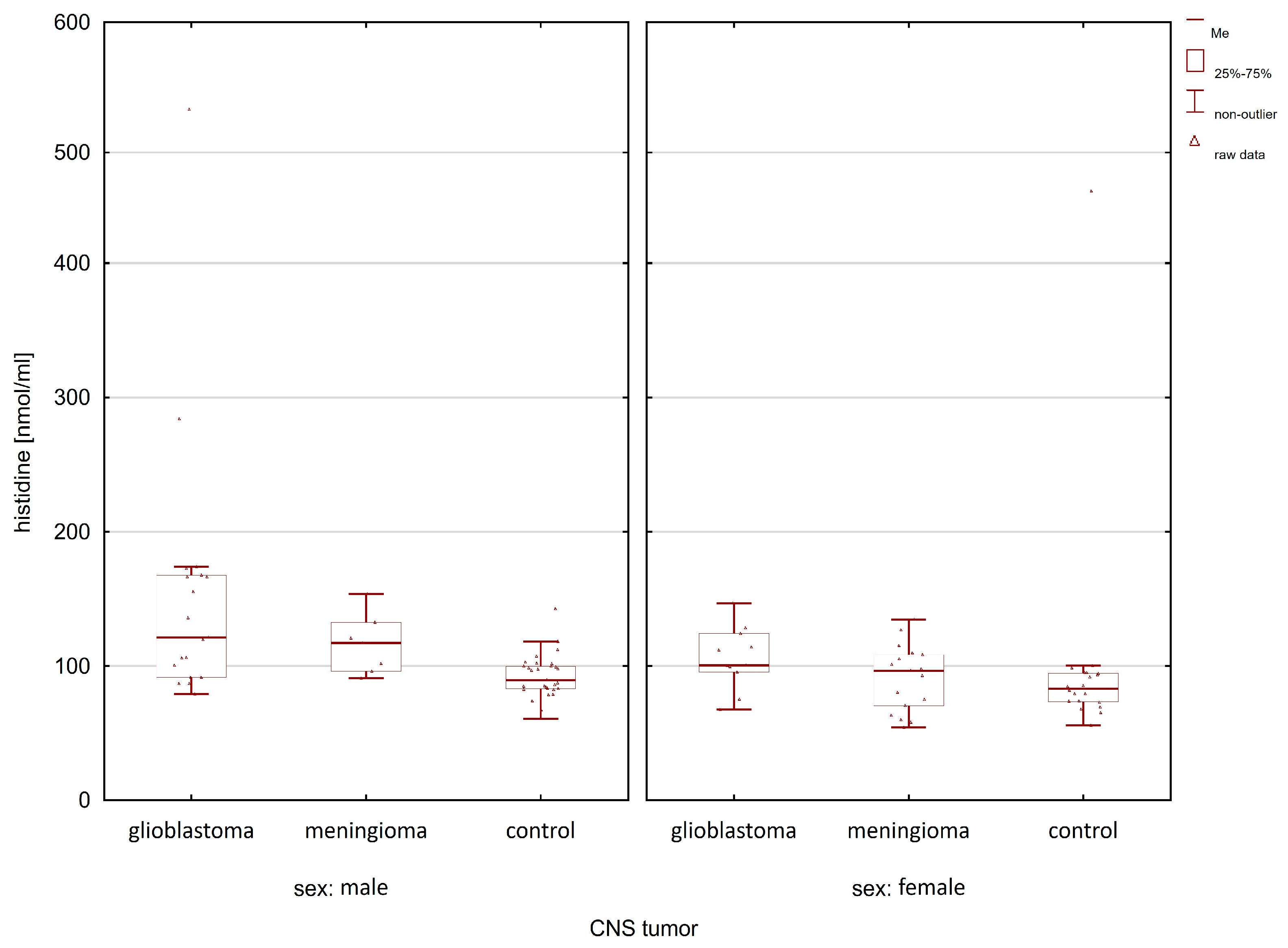



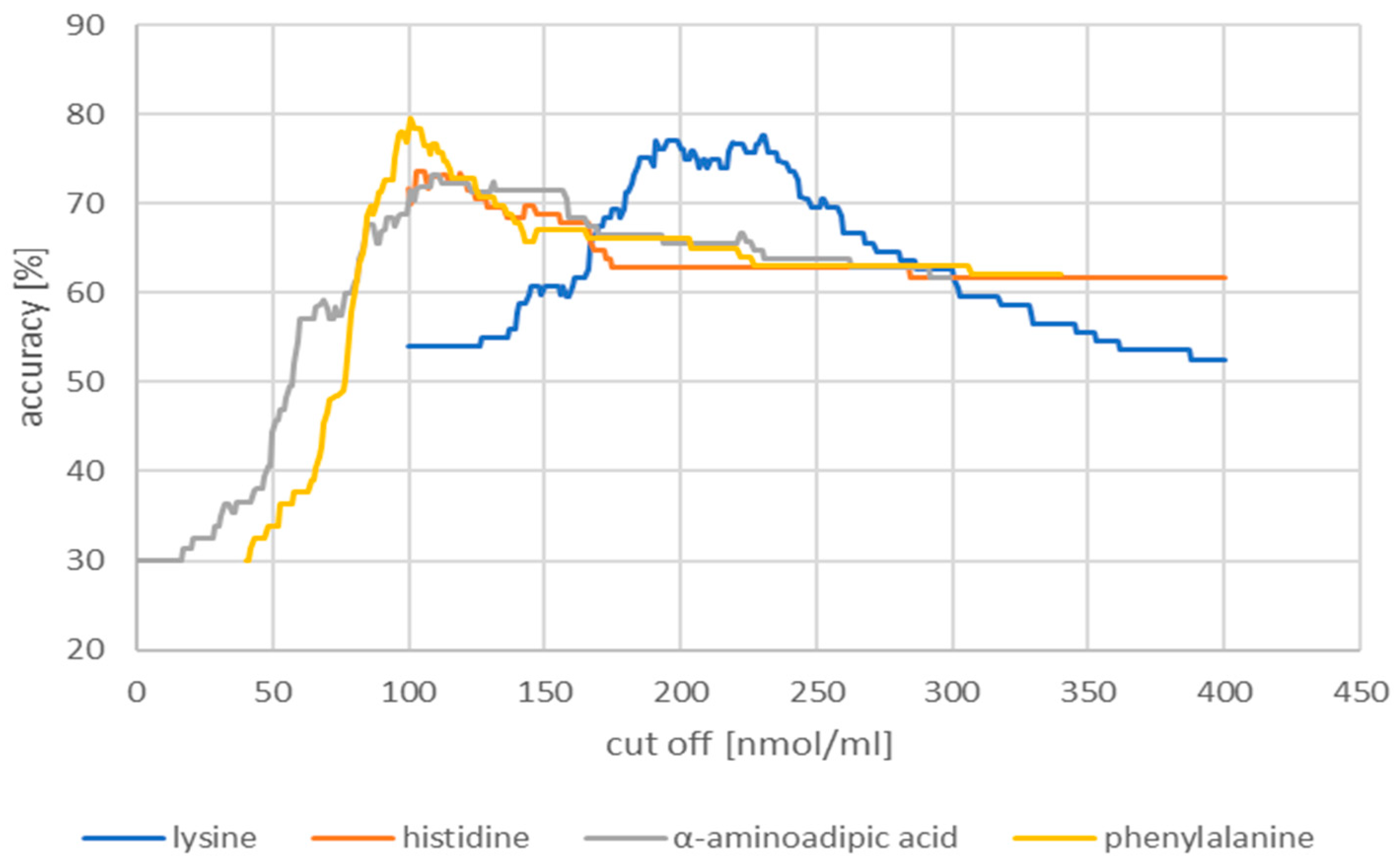


| Compounds | Women p-Value | Men p-Value |
|---|---|---|
| lysine | 0.0059 | 0.0000 |
| histidine | 0.0250 | 0.0004 |
| α-aminoadipic acid | 0.0026 | 0.0060 |
| phenylalanine | 0.0088 | 0.0001 |
| Cut-off (nmol/mL) | Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|
| Lysine | 230.00 | 77.7 | 63.0 | 91.8 |
| Histidine | 103.00 | 73.7 | 60.0 | 89.8 |
| α-aminoadipic acid | 1.09 | 73.2 | 50.0 | 91.8 |
| Phenylalanine | 101.00 | 79.5 | 66.7 | 95.9 |
| CNS Tumor | Glioma (n = 30) | Meningioma (n = 24) |
|---|---|---|
| Age | 62.5 | 59.6 |
| Sex | ||
| 11 (36.67%) | 17 (70.83%) |
| 19 (63.33%) | 7 (29.17%) |
| WHO Classification | ||
| 30 | |
| 13 | |
| 3 | |
| 3 | |
| 5 |
| Compound Name | Abbreviated Name | tR (min) | Quantification Transition |
|---|---|---|---|
| Serine | SER | 3.885 | 233.9 > 146.00 |
| Glutamine | GLN | 3.468 | 275.1 > 172.00 |
| Arginine | ARG | 3.557 | 303.1 > 69.00 |
| Citrulline | CIT | 3.549 | 304.1 > 156.00 |
| Homoarginine | HARG (IS) | 3.753 | 317.10 > 84.00 |
| Asparagine | ASN | 3.985 | 243.10 > 157.00 |
| 1-Methyl-l-histidine | 1MHIS | 4.201 | 298.1 > 96.00 |
| 3-Methyl-l-histidine | 3MHIS | 4.235 | 298.1 > 210.00 |
| 4-Hydroxyproline | HYP | 4.183 | 260.1 > 172.1 |
| Glycine | GLY | 4.423 | 203.9 > 144.00 |
| Threonine | THR | 4.482 | 248.10 > 160.00 |
| Alanine | ALA | 5.347 | 218.00 > 130.00 |
| Gamma-aminobutyric acid | GABA | 5.739 | 232.00 > 172.00 |
| Sarcosine | SAR | 5.948 | 217.90 > 88.00 |
| Beta-aminoisobutyric acid | BAIB | 6.202 | 232.00 > 172.00 |
| α-Aminobutyric acid | ABA | 6.567 | 232.00 > 172.00 |
| Ornithine | ORN | 6.795 | 347.00 > 287.00 |
| Methionine | MET | 7.085 | 227.9 > 190.10 |
| Methionine-d3 | Met-d3 (IS) | 7.030 | 281.1 > 193.00 |
| Proline | PRO | 7.130 | 244.00 > 156.00 |
| Lysine | LYS | 7.743 | 361.00 > 301.10 |
| Aspartic acid | ASP | 7.753 | 304.00 > 216.10 |
| Histidine | HIS | 7.826 | 370.10 > 196.00 |
| Valine | VAL | 8.116 | 246.00 > 158.00 |
| Glutamic acid | GLU | 8.223 | 317.50 > 172.10 |
| Tryptophan | TRP | 8.468 | 333.10 > 245.10 |
| α-Aminoadipic acid | AAA | 9.202 | 332.00 > 244.10 |
| Leucine | LEU | 9.593 | 260.00 > 172.10 |
| Phenylalanine | PHE | 9.679 | 294.10 > 206.10 |
| Isoleucine | ILE | 9.997 | 260.00 > 172.10 |
| Homophenylalanine | HPHE (IS) | 11.224 | 308.00 > 220.00 |
| Cystine | C-C | 11.402 | 497.00 > 248.00 |
| Tyrosine | TYR | 11.999 | 395.90 > 136.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kośliński, P.; Pluskota, R.; Koba, M.; Siedlecki, Z.; Śniegocki, M. Comparative Analysis of Amino Acid Profiles in Patients with Glioblastoma and Meningioma Using Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS). Molecules 2023, 28, 7699. https://doi.org/10.3390/molecules28237699
Kośliński P, Pluskota R, Koba M, Siedlecki Z, Śniegocki M. Comparative Analysis of Amino Acid Profiles in Patients with Glioblastoma and Meningioma Using Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS). Molecules. 2023; 28(23):7699. https://doi.org/10.3390/molecules28237699
Chicago/Turabian StyleKośliński, Piotr, Robert Pluskota, Marcin Koba, Zygmunt Siedlecki, and Maciej Śniegocki. 2023. "Comparative Analysis of Amino Acid Profiles in Patients with Glioblastoma and Meningioma Using Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)" Molecules 28, no. 23: 7699. https://doi.org/10.3390/molecules28237699
APA StyleKośliński, P., Pluskota, R., Koba, M., Siedlecki, Z., & Śniegocki, M. (2023). Comparative Analysis of Amino Acid Profiles in Patients with Glioblastoma and Meningioma Using Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS). Molecules, 28(23), 7699. https://doi.org/10.3390/molecules28237699







