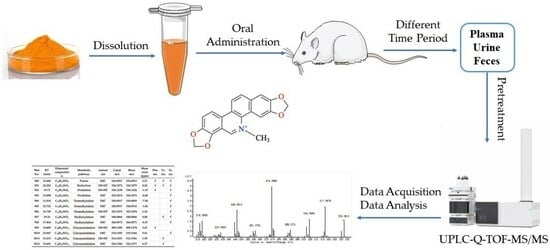
Identification of Sanguinarine Metabolites in Rats Using UPLC-Q-TOF-MS/MS
Abstract
1. Introduction
2. Results and Discussion
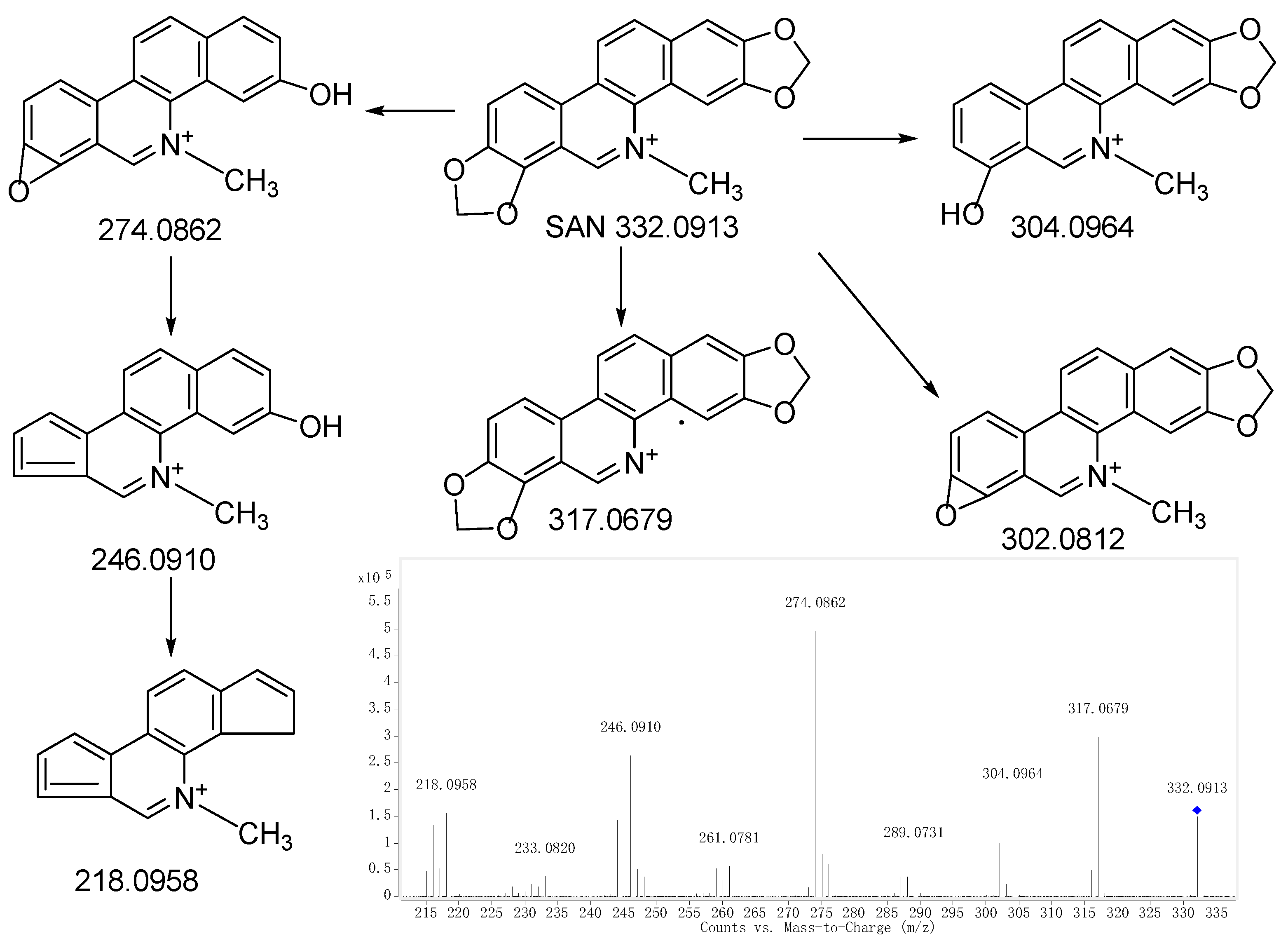
2.1. Fragmentations of Precursor Compound
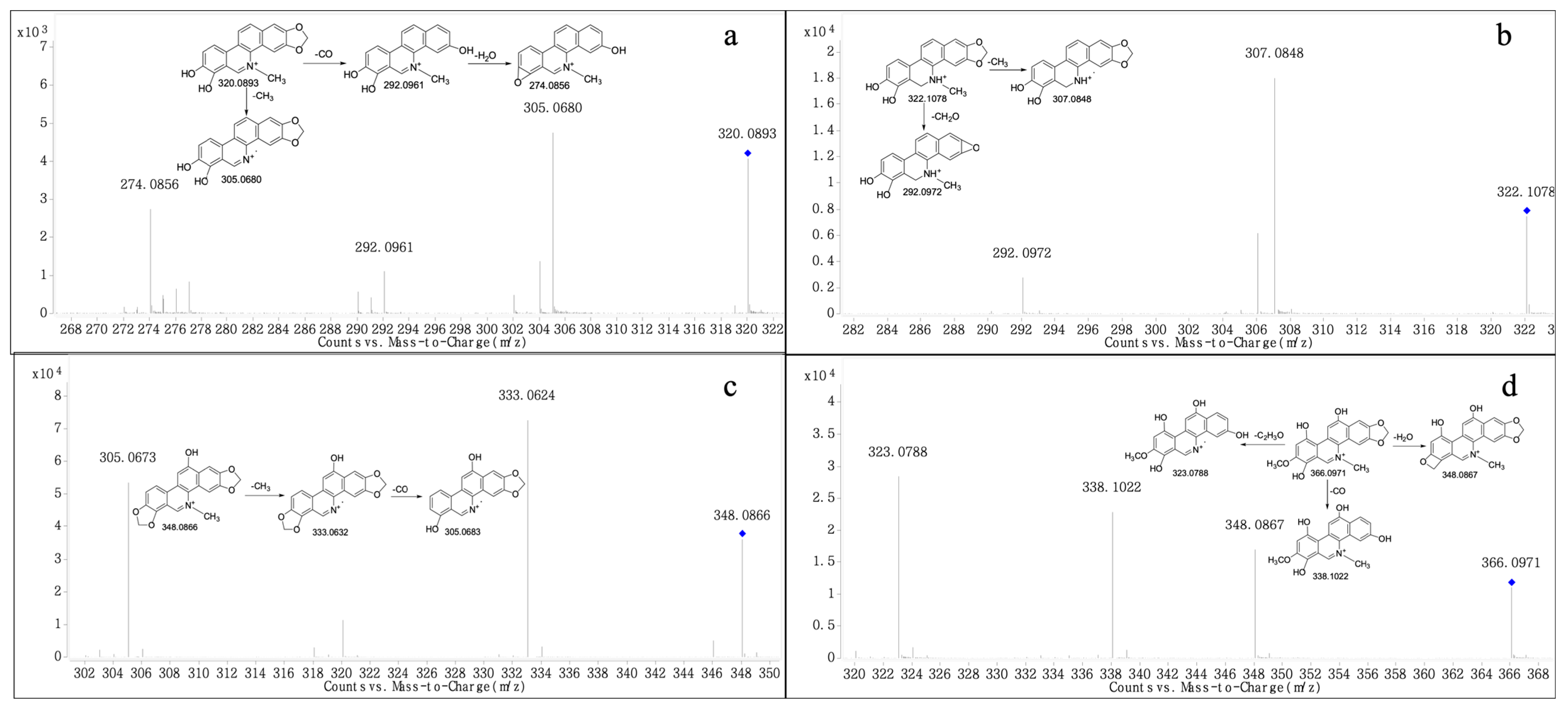
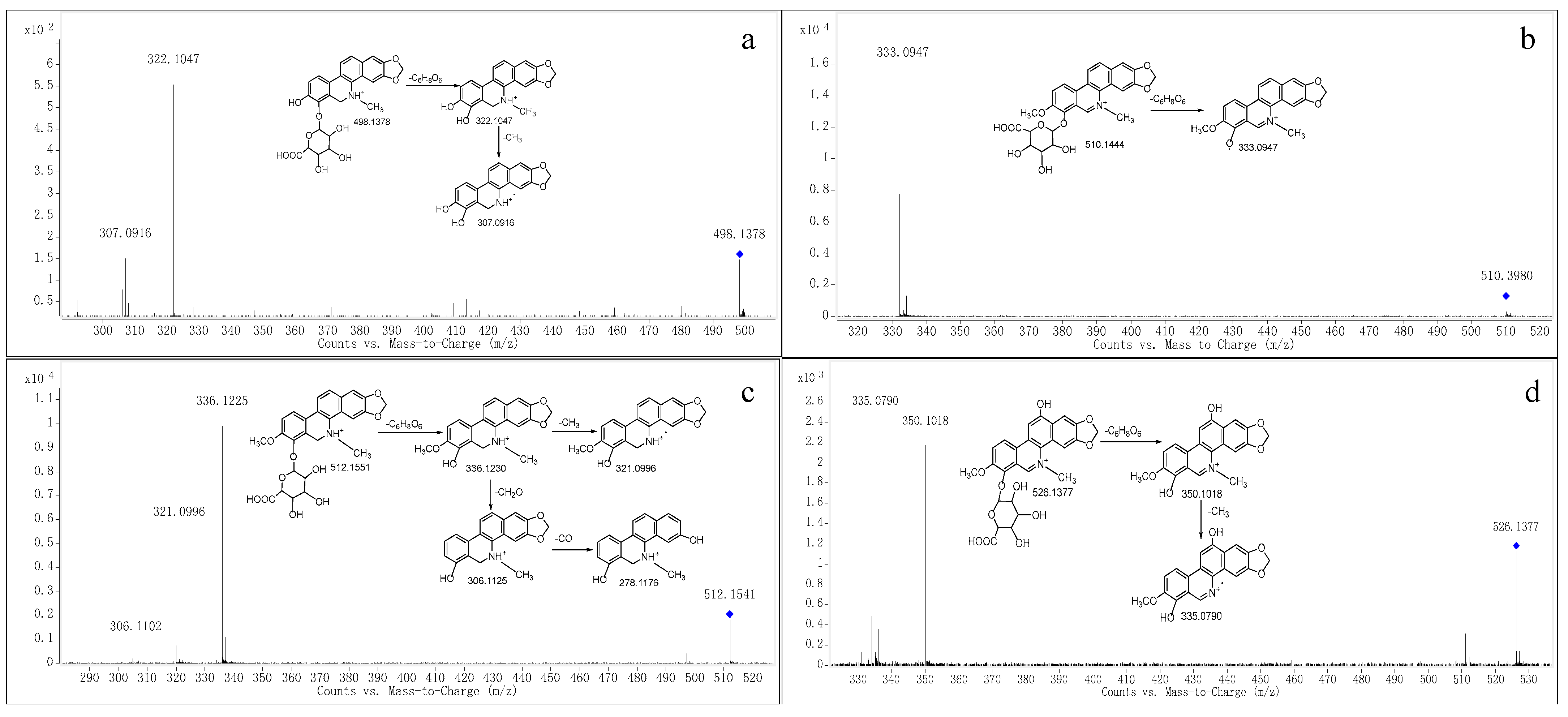
2.2. Structural Elucidation
2.3. Metabolite Profile and Metabolic Pathways of SAN
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Animals and Drug Administration
3.3. Sample Collection and Pretreatment
3.3.1. Sample Collection
3.3.2. Sample Pretreatment
3.4. Instruments and Analytical Conditions
3.5. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.; Lyu, Y.; Huang, J.; Zhang, X.; Yu, N.; Wen, Z.; Chen, S. Antibacterial Activity and Mechanism of Sanguinarine against Providencia Rettgeri in Vitro. PeerJ 2020, 8, e9543. [Google Scholar] [CrossRef] [PubMed]
- Danielewski, M.; Zielińska, S.; Matuszewska, A.; Słupski, W.; Włodarczyk, M.; Jęśkowiak, I.; Wiatrak, B.; Kowalski, K.; Jezierska-Domaradzka, A.; Ziółkowski, P.; et al. Sanguinarine-Chelerythrine Fraction of Coptis Chinensis Exerts Anti-Inflammatory Activity in Carrageenan Paw Oedema Test in Rats and Reveals Reduced Gastrotoxicity. Oxid. Med. Cell. Longev. 2022, 2022, 1504929. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; He, X.; Feng, Y.; Wang, W.; Chen, H.; Gong, M.; Liu, D.; Clarke, J.L.; van Eerde, A. Pollution by Antibiotics and Antimicrobial Resistance in LiveStock and Poultry Manure in China, and Countermeasures. Antibiotics 2021, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.D.; Wu, S.B.; Choct, M.; Pastor, A.; Steiner, T.; Swick, R.A. Impact of a Macleaya Cordata-Derived Alkaloid Extract on Necrotic Enteritis in Broilers. Poult. Sci. 2017, 96, 3581–3585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Alvensleben, S.V.; Fusconi, G.; Morlacchini, M. Safety Evaluation of a Standardized Macleaya Cordata Extract in a Ninety Day Feeding Study in Weaned Piglets. Open J. Anim. Sci. 2017, 7, 213–231. [Google Scholar] [CrossRef][Green Version]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The Chicken Gut Metagenome and the Modulatory Effects of Plant-Derived Benzylisoquinoline Alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef]
- Wang, F.; Yin, Y.; Yang, M.; Chen, J.; Fu, C.; Huang, K. Effects of Combined Supplementation of Macleaya Cordata Extract and Benzoic Acid on the Growth Performance, Immune Responses, Antioxidant Capacity, Intestinal Morphology, and Microbial Composition in Weaned Piglets. Front. Vet. Sci. 2021, 8, 708597. [Google Scholar] [CrossRef]
- Li, J.; Wu, Y.; Dong, S.; Yu, Y.; Wu, Y.; Xiang, B.; Li, Q. Research Progress on Neuroprotective Effects of Isoquinoline Alkaloids. Molecules 2023, 28, 4797. [Google Scholar] [CrossRef]
- Ke, Q.; Duan, K.; Cheng, Y.; Xu, S.; Xiao, S.; Fang, L. Sanguinarine Exhibits Antiviral Activity against Porcine Reproductive and Respiratory Syndrome Virus via Multisite Inhibition Mechanisms. Viruses 2023, 15, 688. [Google Scholar] [CrossRef]
- Sarkar, S.M. Isolation from Argemone Oil of Dihydrosanguinarine and Sanguinarine; Toxicity of Sanguinarine. Nature 1948, 162, 265. [Google Scholar] [CrossRef]
- Sharma, B.D.; Bhatia, V.; Rahtee, M.; Kumar, R.; Mukharjee, A. Epidemic Dropsy: Observations on Pathophysiology and Clinical Features during the Delhi Epidemic of 1998. Trop. Doct. 2002, 32, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Ansari, K.M.; Dhawan, A.; Khanna, S.K.; Das, M. Protective Effect of Bioantioxidants on Argemone Oil/Sanguinarine Alkaloid Induced Genotoxicity in Mice. Cancer Lett. 2006, 244, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, I.; Mukherjee, A. Argemone Oil Induces Genotoxicity in Mice. Drug Chem. Toxicol. 2016, 39, 407–411. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Wang, J.; Li, L.; Zhang, Y.; Jin, M.; Chen, X.; Sun, C.; Wang, R.; Liu, K. Cardiotoxicity of Sanguinarine via Regulating Apoptosis and MAPK Pathways in Zebrafish and HL1 Cardiomyocytes. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2022, 252, 109228. [Google Scholar] [CrossRef] [PubMed]
- Zdarilova, A.; Vrublova, E.; Vostalova, J.; Klejdus, B.; Stejskal, D.; Proskova, J.; Kosina, P.; Svobodova, A.; Vecera, R.; Hrbac, J.; et al. Natural Feed Additive of Macleaya Cordata: Safety Assessment in Rats a 90-Day Feeding Experiment. Food Chem. Toxicol. 2008, 46, 3721–3726. [Google Scholar] [CrossRef]
- Dong, Z.; Tang, S.-S.; Ma, X.-L.; Li, C.-H.; Tang, Z.-S.; Yang, Z.-H.; Zeng, J.-G. Preclinical Safety Evaluation of Macleaya Cordata Extract: A Re-Assessment of General Toxicity and Genotoxicity Properties in Rodents. Front. Pharmacol. 2022, 13, 980918. [Google Scholar] [CrossRef]
- Hakim, S.A.; Mijovic, V.; Walker, J. Experimental Transmission of Sanguinarine in Milk: Detection of a Metabolic Product. Nature 1961, 189, 201–204. [Google Scholar] [CrossRef]
- Dvorák, Z.; Simánek, V. Metabolism of Sanguinarine: The Facts and the Myths. Curr. Drug Metab. 2007, 8, 173–176. [Google Scholar] [CrossRef]
- Psotová, J.; Klejdus, B.; Vecera, R.; Kosina, P.; Kubán, V.; Vicar, J.; Simánek, V.; Ulrichová, J. A Liquid Chromatographic-Mass Spectrometric Evidence of Dihydrosanguinarine as a First Metabolite of Sanguinarine Transformation in Rat. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006, 830, 165–172. [Google Scholar] [CrossRef]
- Deroussent, A.; Ré, M.; Hoellinger, H.; Cresteil, T. Metabolism of Sanguinarine in Human and in Rat: Characterization of Oxidative Metabolites Produced by Human CYP1A1 and CYP1A2 and Rat Liver Microsomes Using Liquid Chromatography-Tandem Mass Spectrometry. J. Pharm. Biomed. Anal. 2010, 52, 391–397. [Google Scholar] [CrossRef]
- Kosina, P.; Vacek, J.; Papoušková, B.; Stiborová, M.; Stýskala, J.; Cankař, P.; Vrublová, E.; Vostálová, J.; Simánek, V.; Ulrichová, J. Identification of Benzo[c]Phenanthridine Metabolites in Human Hepatocytes by Liquid Chromatography with Electrospray Ion-Trap and Quadrupole Time-of-Flight Mass Spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-H.; Wu, Y.; Sun, Z.-L.; Liu, Z.-Y. Identification of Sanguinarine Metabolites in Pig Liver Preparations by Accurate Mass Measurements Using Electrospray Ionization Hybrid Ion Trap/Time-of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Ma, C.; Guan, H.; Chen, L.; Xie, Q.; Cheng, X.; Wang, C. Metabolites Identification and Reversible Interconversion of Chelerythrine and Dihydrochelerythrine in Vitro/in Vivo in Rats Using Ultra-Performance Liquid Chromatography Combined with Electrospray Ionization Quadrupole Time-of-Flight Tandem Mass Spectrometry. J. Pharm. Biomed. Anal. 2020, 189, 113462. [Google Scholar] [CrossRef]
- Qi, X.-Y.; Liang, S.-C.; Ge, G.-B.; Liu, Y.; Dong, P.-P.; Zhang, J.-W.; Wang, A.-X.; Hou, J.; Zhu, L.-L.; Yang, L.; et al. Inhibitory Effects of Sanguinarine on Human Liver Cytochrome P450 Enzymes. Food Chem. Toxicol. 2013, 56, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-S.; Liu, Z.-Y.; Li, Y.; Sun, Z.-L. NQO1 Involves in the Imine Bond Reduction of Sanguinarine and Recombinant Adeno-Associated Virus Mediated NQO1 Overexpression Decreases Sanguinarine-Induced Cytotoxicity in Rat BRL Cells. Toxicol. Lett. 2014, 225, 119–129. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, M.; Zhong, X.; Ou, X.; Yun, X.; Wang, M.; Ren, S.; Qing, Z.; Zeng, J. Identification of the Impurities in Bopu Powder® and Sangrovit® by LC-MS Combined with a Screening Method. Molecules 2021, 26, 3851. [Google Scholar] [CrossRef]
- Vecera, R.; Klejdus, B.; Kosina, P.; Orolin, J.; Stiborová, M.; Smrcek, S.; Vicar, J.; Dvorák, Z.; Ulrichová, J.; Kubán, V.; et al. Disposition of Sanguinarine in the Rat. Xenobiotica 2007, 37, 549–558. [Google Scholar] [CrossRef]
- Xie, H.; Yang, J.; Feng, S.; Cheng, P.; Zeng, J.; Xiong, X. Simultaneous Quantitative Determination of Sanguinarine, Chelerythrine, Dihydrosanguinarine and Dihydrochelerythrine in Chicken by HPLC-MS/MS Method and Its Applications to Drug Residue and Pharmacokinetic Study. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015, 985, 124–130. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, N.-J.; Cao, Y.; Sun, Z.; Wang, Q.; Liu, Z.-Y.; Sun, Z.-L. Sanguinarine Metabolism and Pharmacokinetics Study in Vitro and in Vivo. J. Vet. Pharmacol. Ther. 2020, 43, 208–214. [Google Scholar] [CrossRef]
- Shen, L.-X.; Liu, G.-F.; Song, J.-S.; Cao, Y.-H.; Peng, X.; Wu, R.-R.; Cao, Y.; Chen, X.-J.; Liu, Z.; Sun, Z.-L.; et al. Sex Differences in the Pharmacokinetics and Tissue Residues of Macleaya Cordata Extracts in Rats. Xenobiotica 2022, 52, 46–53. [Google Scholar] [CrossRef]
- Kosina, P.; Walterová, D.; Ulrichová, J.; Lichnovský, V.; Stiborová, M.; Rýdlová, H.; Vicar, J.; Krecman, V.; Brabec, M.J.; Simánek, V. Sanguinarine and Chelerythrine: Assessment of Safety on Pigs in Ninety Days Feeding Experiment. Food Chem. Toxicol. 2004, 42, 85–91. [Google Scholar] [CrossRef] [PubMed]


| Met No. | RT (min) | Elemental Composition | Metabolic Pathway | Ionization | Calcd m/z | Meas m/z | Mass Error (ppm) | Plasma | Urine | Feces |
|---|---|---|---|---|---|---|---|---|---|---|
| M0 | 13.496 | C20H14NO4 | Parent | [M]+ | 332.0917 | 332.0913 | 3.31 | √ | √ | √ |
| M1 | 24.281 | C20H15NO4 | Reduction | [M+H]+ | 334.1074 | 334.1075 | 0.30 | √ | √ | √ |
| M2 | 19.73 | C20H17NO4 | Oxidation | [M+H]+ | 336.1230 | 336.1226 | −1.19 | √ | √ | |
| M3 | 12.098 | C20H16NO4 | Oxidation | [M]+ | 334.1074 | 334.1072 | −0.60 | √ | ||
| M4 | 11.014 | C19H14NO4 | Demethylation | [M]+ | 320.0917 | 320.0893 | −7.50 | √ | ||
| M5 | 12.721 | C19H14NO4 | Demethylation | [M]+ | 320.0917 | 320.0912 | −1.56 | √ | ||
| M6 | 16.749 | C19H15NO4 | Demethylation | [M+H]+ | 322.1074 | 322.1078 | 1.24 | √ | ||
| M7 | 19.32 | C20H14NO5 | Hydroxylation | [M]+ | 348.0866 | 348.0866 | 0.00 | √ | √ | |
| M8 | 17.864 | C20H16NO6 | Hydroxylation | [M]+ | 366.0972 | 366.0971 | −0.27 | √ | ||
| M9 | 14.885 | C25H23NO10 | Glucuronidation | [M+H]+ | 498.1395 | 498.1378 | 3.41 | √ | ||
| M10 | 23.847 | C26H24NO10 | Glucuronidation | [M]+ | 510.1395 | 510.1444 | 9.61 | √ | ||
| M11 | 15.213 | C26H25NO10 | Glucuronidation | [M+H]+ | 512.1551 | 512.1541 | 1.95 | √ | ||
| M12 | 15.691 | C26H24NO11 | Glucuronidation | [M]+ | 526.1344 | 526.1377 | 6.27 | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Liu, Z.; Dong, Z.; Zou, X.; Zeng, J.; Yang, Z. Identification of Sanguinarine Metabolites in Rats Using UPLC-Q-TOF-MS/MS. Molecules 2023, 28, 7641. https://doi.org/10.3390/molecules28227641
Liu M, Liu Z, Dong Z, Zou X, Zeng J, Yang Z. Identification of Sanguinarine Metabolites in Rats Using UPLC-Q-TOF-MS/MS. Molecules. 2023; 28(22):7641. https://doi.org/10.3390/molecules28227641
Chicago/Turabian StyleLiu, Mengting, Zhiqin Liu, Zhuang Dong, Xianglin Zou, Jianguo Zeng, and Zihui Yang. 2023. "Identification of Sanguinarine Metabolites in Rats Using UPLC-Q-TOF-MS/MS" Molecules 28, no. 22: 7641. https://doi.org/10.3390/molecules28227641
APA StyleLiu, M., Liu, Z., Dong, Z., Zou, X., Zeng, J., & Yang, Z. (2023). Identification of Sanguinarine Metabolites in Rats Using UPLC-Q-TOF-MS/MS. Molecules, 28(22), 7641. https://doi.org/10.3390/molecules28227641