Exploring the Release of Elastin Peptides Generated from Enzymatic Hydrolysis of Bovine Elastin via Peptide Mapping
Abstract
:1. Introduction
2. Results and Discussion
2.1. Hydrolysis of Elastin: A Process to Produce Elastase Inhibitory Peptides
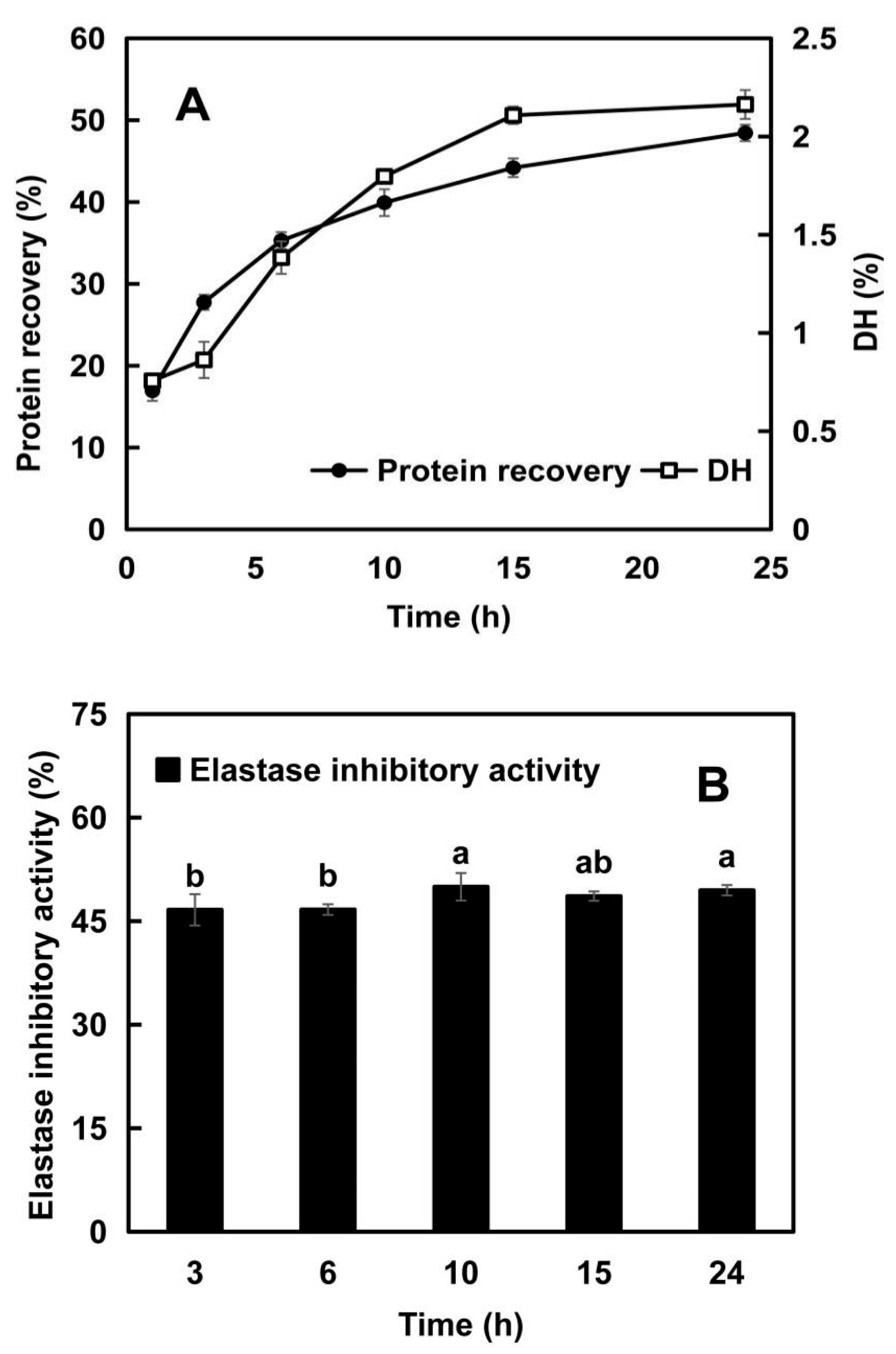
2.1.1. Changes in Protein Recovery and Degree of Hydrolysis (DH)
2.1.2. Changes in Elastase Inhibitory Activity with the Hydrolysis Time
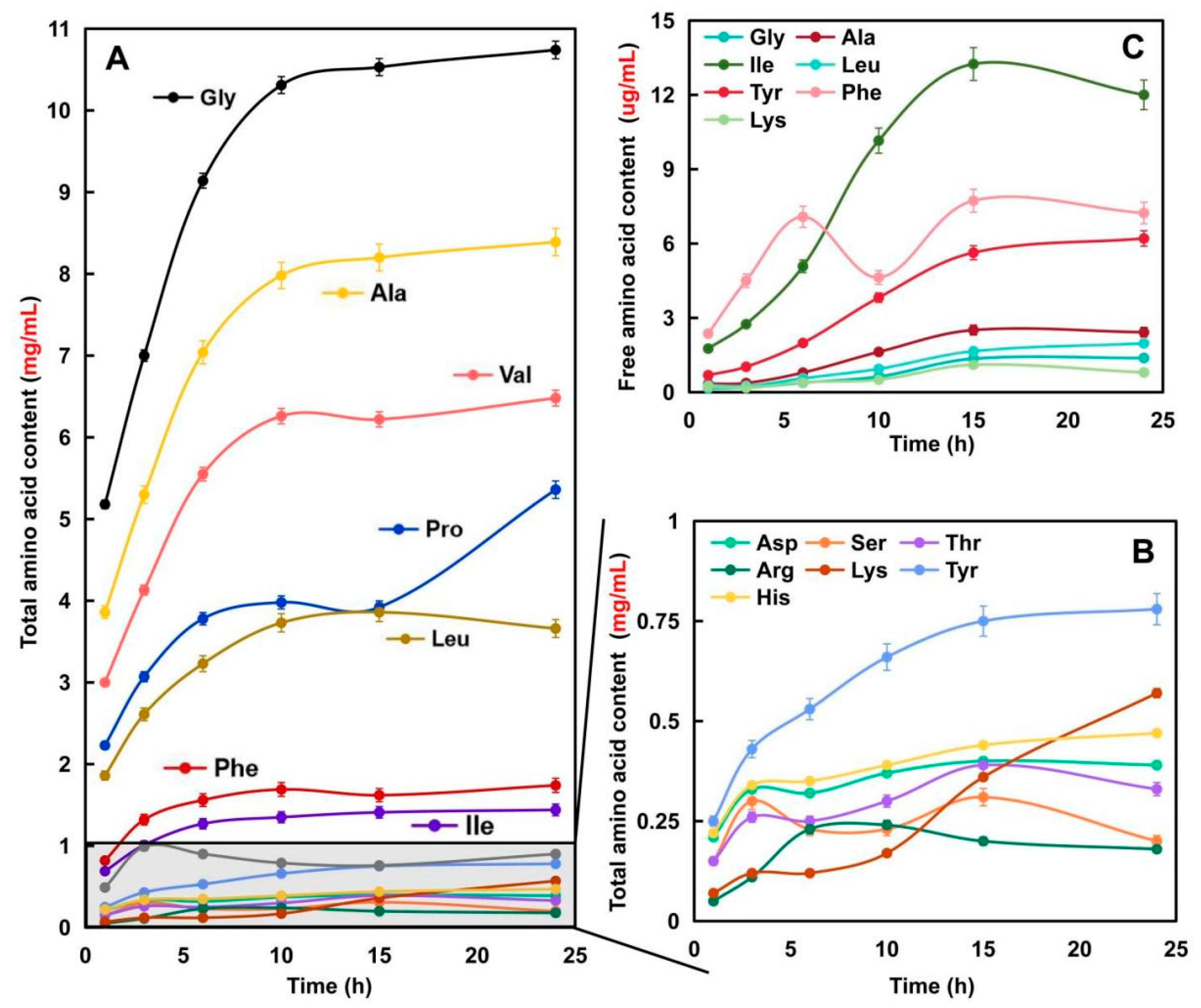
2.1.3. Amino Acid Composition of Elastin Hydrolysates Obtained after Hydrolysis for Different Times
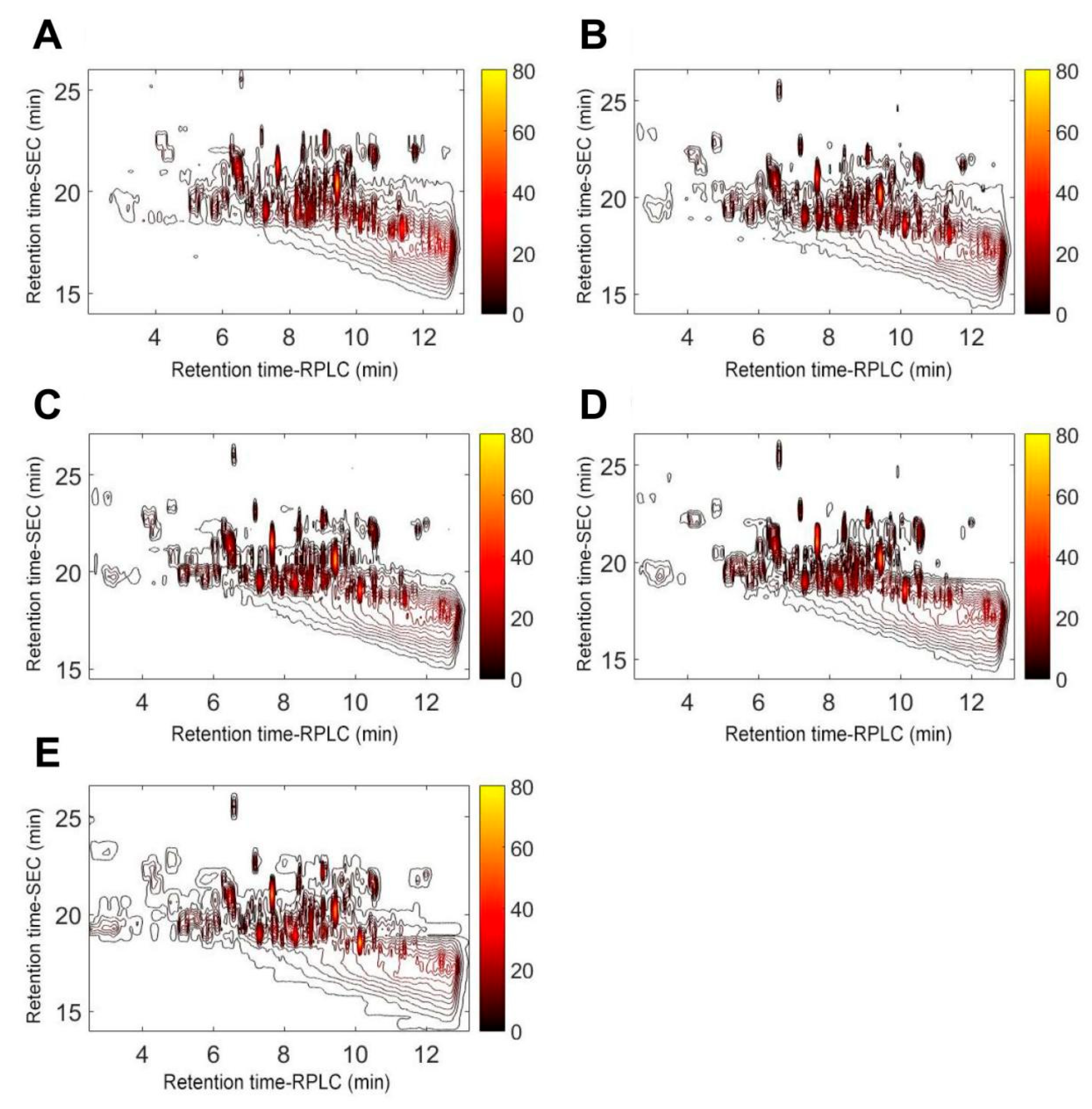
2.1.4. Peptide Fingerprints of Elastin Hydrolysates Obtained after Hydrolysis for Different Time
2.2. Peptide Mapping of Elastin Hydrolysates
2.3. Theoretical Content of the Identified Peptides from Elastin Hydrolysate
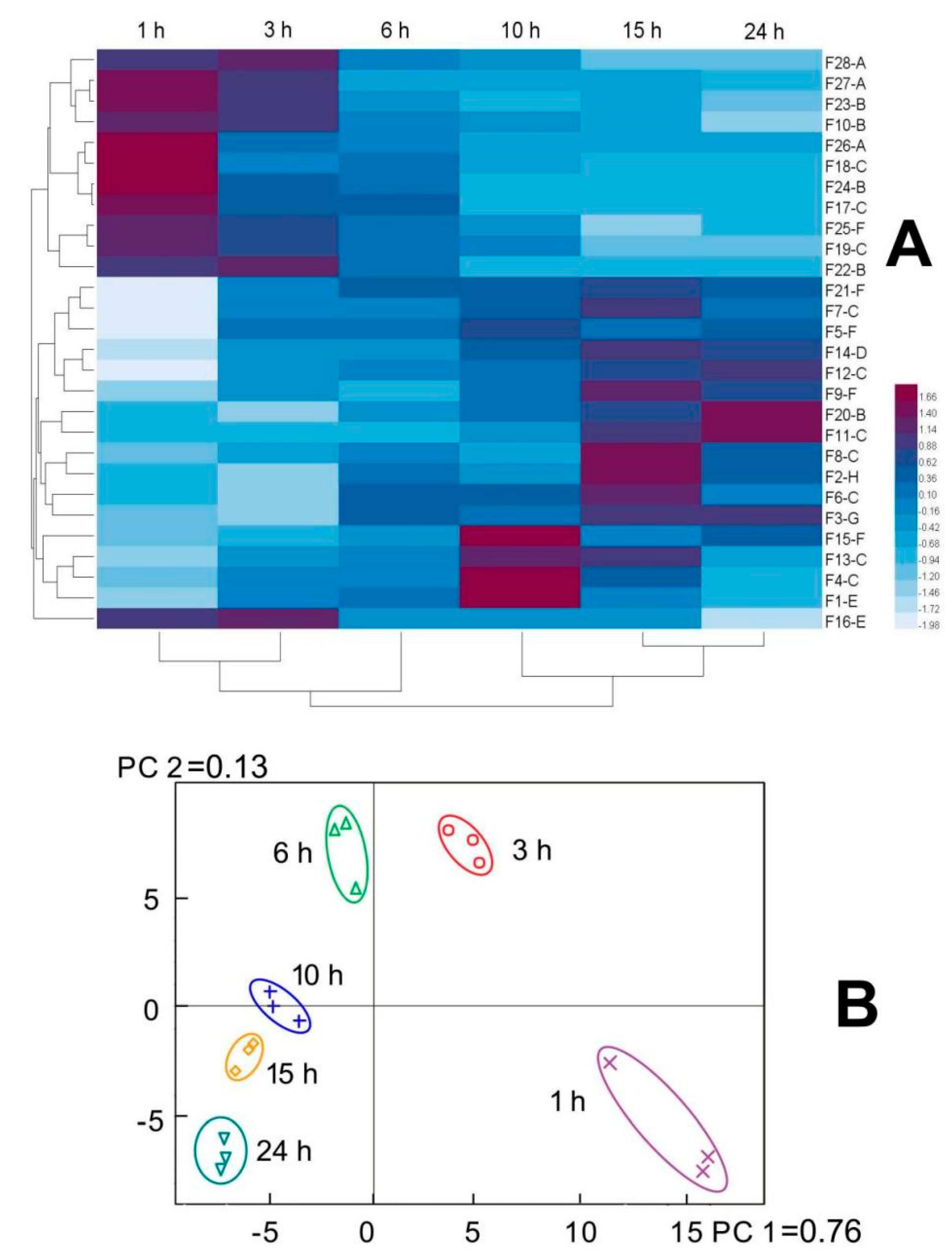
2.4. Release of the Peptide Markers during Hydrolysis of Elastin
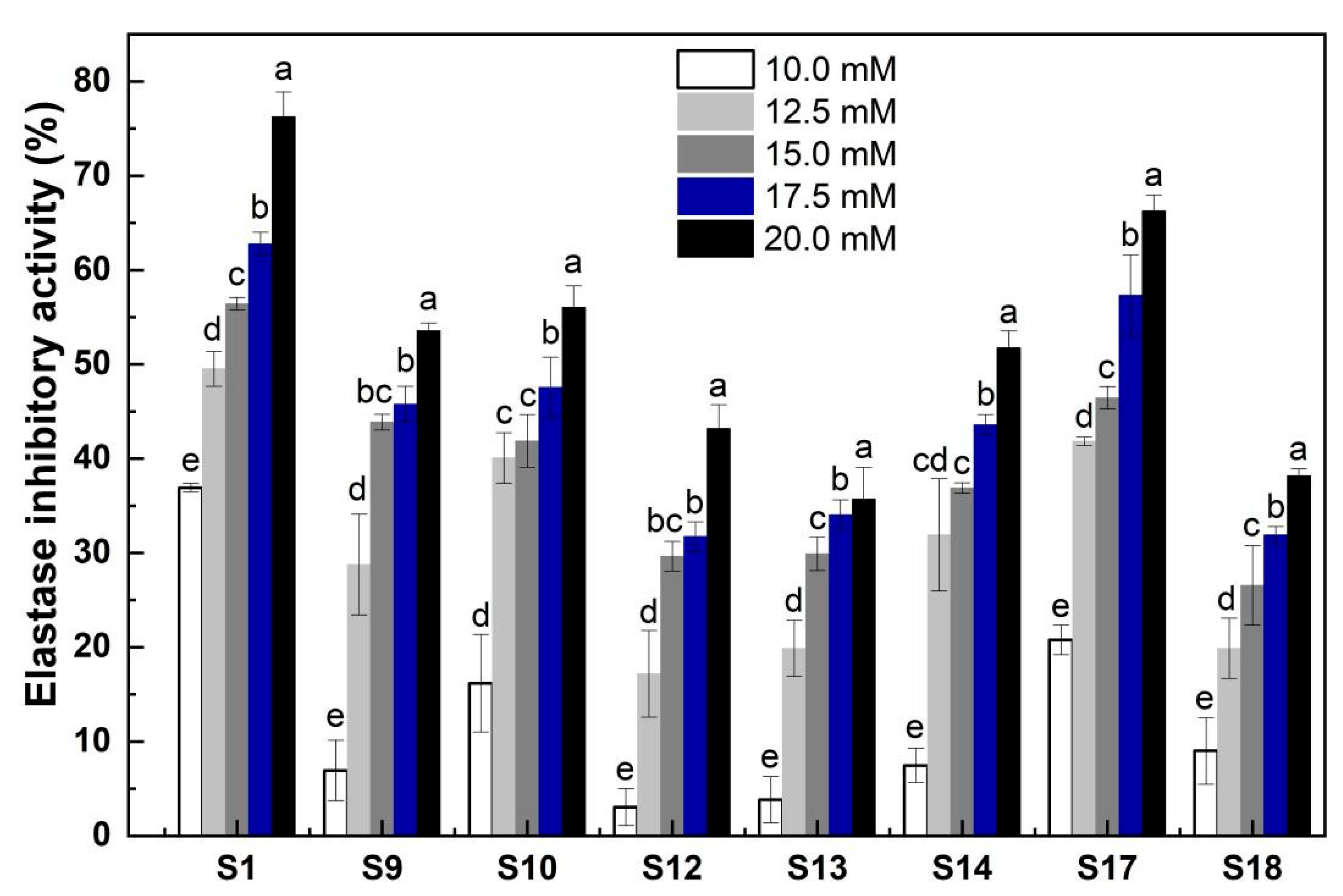
2.5. Elastase Inhibitory Activity of the Marked Peptides in the Elastin Hydrolysates
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Isolation of Elastin from Bovine Arteries
3.3. Preparation of Elastin Hydrolysates (EHs)
3.4. Protein Recovery and DH of the Elastin Hydrolysates
3.5. Amino Acid Analysis
3.6. Stop-Flow Size-Exclusion Chromatography (SEC) × Reversed Phase Liquid Chromatography (RPLC) for Peptide Separation
3.7. Peptide Analysis by UPLC-ESI-QTOF-MS/MS
3.8. Elastase Inhibitory Activity Measurements
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.; Tavano, O.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Use of alcalase in the production of bioactive peptides: A review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.; Rafiq, S.; Gat, Y.; Gat, P.; Waghmare, R.; Kumar, V. A review on bioactive peptides: Physiological functions, bioavailability and safety. Int. J. Pept. Res. Ther. 2020, 26, 139–150. [Google Scholar] [CrossRef]
- Ahmed, T.; Sun, X.; Udenigwe, C.C. Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: A systematic review. Trends Food Sci. Technol. 2022, 120, 265–273. [Google Scholar] [CrossRef]
- Miralles, B.; Amigo, L.; Recio, I. Critical review and perspectives on food-derived antihypertensive peptides. J. Agric. Food Chem. 2018, 66, 9384–9390. [Google Scholar] [CrossRef] [PubMed]
- Oliver-Cervelló, L.; Martin-Gómez, H.; Mas-Moruno, C. New trends in the development of multifunctional peptides to functionalize biomaterials. J. Pept. Sci. 2022, 28, e3335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, M.; Su, G.; Lin, L. Identification and taste characteristics of novel umami and umami-enhancing peptides separated from peanut protein isolate hydrolysate by consecutive chromatography and UOLC–ESI–QTOF–MS/MS. Food Chem. 2019, 278, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Xie, Y.; Liu, R.; Cui, G.; Zhao, M.; Zhang, J. Effect of transglutaminase on taste characteristics of pea protein hydrolysates through altering the composition of amino acids and peptides. Food Biosci. 2023, 56, 103261. [Google Scholar] [CrossRef]
- Yuan, L.; Chu, Q.; Yang, B.; Zhang, W.; Sun, Q.; Gao, R. Purification and identification of anti-inflammatory peptides from sturgeon (Acipenser schrenckii) cartilage. Food Sci. Hum. Wellness 2023, 12, 2175–2183. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. Biopep-uwm database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Xu, J.; Sun-Waterhouse, D.; Qiu, C.; Zhao, M.; Sun, B.; Lin, L.; Su, G. Additional band broadening of peptides in the first size-exclusion chromatographic dimension of an automated stop-flow two-dimensional high performance liquid chromatography. J. Chromatogr. A 2017, 1521, 80–89. [Google Scholar] [CrossRef]
- Salekeen, R.; Ahmed, A.; Islam, M.E.; Billah, M.M.; Rahman, H.; Islam, K.M.D. In-silico screening of bioactive phytopeptides for novel anti-ageing therapeutics. J. Biomol. Struct. Dyn. 2022, 40, 4475–4487. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Cy, E. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, L.; Lin, L.; Sun, B.; Su, G.; Zhao, M. Stop-flow reversed phase liquid chromatography × size-exclusion chromatography for separation of peptides. Anal. Chim. Acta 2018, 1018, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Gilar, M.; Fridrich, J.; Schure, M.R.; Jaworski, A. Comparison of orthogonality estimation methods for the two-dimensional separations of peptides. Anal. Chem. 2012, 84, 8722–8732. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Su, G.; Zhou, F.; Zhang, J.; Zheng, L.; Zhao, M. Protective effect of bovine elastin peptides against photoaging in mice and identification of novel antiphotoaging peptides. J. Agric. Food Chem. 2018, 66, 10760–10768. [Google Scholar] [CrossRef] [PubMed]
- Green, E.M.; Mansfield, J.C.; Bell, J.S.; Winlove, C.P. The structure and micromechanics of elastic tissue. Interface Focus 2014, 4, 20130058. [Google Scholar] [CrossRef]
- Niklason, L.E.; Abbott, W.; Gao, J.; Klagges, B.; Hirschi, K.K.; Ulubayram, K.; Conroy, N.; Jones, R.; Vasanawala, A.; Sanzgiri, S.; et al. Morphologic and mechanical characteristics of engineered bovine arteries. J. Vasc. Surg. 2001, 33, 628–638. [Google Scholar] [CrossRef]
- Taheri, A.; Abedian Kenari, A.; Motamedzadegan, A.; Habibi Rezaie, M. Optimization of goldstripe sardine (Sardinella Gibbosa) protein hydrolysate using alcalase® 2.4l by response surface methodology. CyTA J. Food 2011, 9, 114–120. [Google Scholar] [CrossRef]
- Daamen, W.F.; Hafmans, T.; Veerkamp, J.H.; van Kuppevelt, T.H. Comparison of five procedures for the purification of insoluble elastin. Biomaterials 2001, 22, 1997–2005. [Google Scholar] [CrossRef]
- Yang, B.; Yang, H.; Li, J.; Li, Z.; Jiang, Y. Amino acid composition, molecular weight distribution and antioxidant activity of protein hydrolysates of soy sauce lees. Food Chem. 2011, 124, 551–555. [Google Scholar] [CrossRef]
- Wu, Q.; Du, J.; Jia, J.; Kuang, C. Production of ace inhibitory peptides from sweet sorghum grain protein using Alcalase: Hydrolysis kinetic, purification and molecular docking study. Food Chem. 2016, 199, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, B.L.; Bruenger, E.; Sandberg, L.B. A new method for purification of mature elastin. Anal. Biochem. 1975, 64, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Chan, G.K.; Zhang, M.; Yao, P.; Lin, H.; Dong, T.T.; Li, G.; Lai, X.; Tsim, K.W. Characterization of edible bird’s nest by peptide fingerprinting with principal component analysis. Food Qual. Saf. 2017, 1, 83–92. [Google Scholar] [CrossRef]
- Geoffroy, T.R.; Thibodeau, J.; Faucher, M.; Langevin, M.E.; Lutin, F.; Bazinet, L. Relationship between feed concentration and bioactive cationic peptide recovery: Impact on ecoefficiency of eduf at semi-industrial scale. Sep. Purif. Technol. 2022, 286, 120403. [Google Scholar] [CrossRef]
- Kozel, B.A.; Mecham, R.P. Elastic fiber ultrastructure and assembly. Matrix Biol. 2019, 84, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun-Waterhouse, D.; Su, G.; Zhao, M. New insight into umami receptor, umami/umami-enhancing peptides and their derivatives: A review. Trends Food Sci. Technol. 2019, 88, 429–438. [Google Scholar] [CrossRef]
- Bounouala, F.Z.; Roudj, S.; Karam, N.; Recio, I.; Miralles, B. Casein hydrolysates by lactobacillus brevis and lactococcus lactis proteases: Peptide profile discriminates strain-dependent enzyme specificity. J. Agric. Food Chem. 2017, 65, 9324–9332. [Google Scholar] [CrossRef]
- Taddese, S.; Weiss, A.S.; Neubert, R.H.H.; Schmelzer, C.E.H. Mapping of macrophage elastase cleavage sites in insoluble human skin elastin. Matrix Biol. 2008, 27, 420–428. [Google Scholar] [CrossRef]
- Chen, J.; Yu, X.; Chen, Q.; Wu, Q.; He, Q. Screening and mechanisms of novel angiotensin-i-converting enzyme inhibitory peptides from rabbit meat proteins: A combined in silico and in vitro study. Food Chem. 2022, 370, 131070. [Google Scholar] [CrossRef]
- Duffuler, P.; Bhullar, K.S.; de Campos Zani, S.C.; Wu, J. Bioactive peptides: From basic research to clinical trials and commercialization. J. Agric. Food Chem. 2022, 70, 3585–3595. [Google Scholar] [CrossRef]
- Sáez-Plaza, P.; Navas, M.J.; Wybraniec, S.; Michałowski, T.; Asuero, A.G. An overview of the kjeldahl method of nitrogen determination. Part ii. Sample preparation, working scale, instrumental finish, and quality control. Crit. Rev. Anal. Chem. 2013, 43, 224–272. [Google Scholar] [CrossRef]
- Spellman, D.; Mcevoy, E.; Cuinn, G.O.; Fitzgerald, R.J. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the tnbs, opa and ph stat methods for quantification of degree of hydrolysis. Int. Dairy J. 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Yin, M.; Wuyun, T.; Jiang, Z.; Zeng, J. Amino acid profiles and protein quality of siberian apricot (Prunus sibirica L.) Kernels from inner mongolia. J. For. Res. 2020, 31, 1391–1397. [Google Scholar] [CrossRef]
- Yoo, J.; Yoo, Y.; Yang, H. Effects of egg shell membrane hydrolysates on anti-inflammatory, anti-wrinkle, anti-microbial activity and moisture-protection. Korean J. Food Sci. Anim. Resour. 2014, 34, 26–32. [Google Scholar] [CrossRef]

| Serial Number | Sequence | Frequency | Molecular Weight | Content (on a Dry Basis) | |
|---|---|---|---|---|---|
| (mg/g) | (μmol/g) | ||||
| S1 | VGVPGVGVPGVGVPGV | 8 | 1344.7765 | 148.76 | 110.62 |
| S2 | VGVPGVGVPGVGVPG | 9 | 1245.7081 | 155.03 | 124.45 |
| S3 | GVPGVGVPGVGVPGV | 8 | 1245.7081 | 137.81 | 110.62 |
| S4 | GVPGVGVPGVGVPG | 9 | 1146.6397 | 142.70 | 124.45 |
| S5 | VPGVGVPGVGVPG | 9 | 1090.6212 | 135.73 | 124.45 |
| S6 | PGVGVPGVGVPG | 10 | 990.5498 | 136.97 | 138.28 |
| S7 | VGVPGVGVPGV | 9 | 935.5440 | 116.43 | 124.45 |
| S8 | GVPGVGVPG | 10 | 737.4072 | 101.97 | 138.28 |
| S9 | VPGVGVPG | 10 | 680.3857 | 94.08 | 138.28 |
| S10 | VPGVG | 15 | 427.2431 | 88.62 | 207.42 |
| S11 | GVGV | 21 | 330.1903 | 95.88 | 290.39 |
| S12 | VPG | 26 | 271.1532 | 97.49 | 359.53 |
| S13 | GV | 80 | 174.1000 | 192.60 | 1106.24 |
| S14 | PG | 60 | 172.0848 | 142.78 | 829.68 |
| S15 | AAAAA | 12 | 373.1961 | 61.93 | 165.94 |
| S16 | AAAA | 20 | 302.1590 | 83.57 | 276.56 |
| S17 | AAA | 37 | 231.1219 | 118.25 | 511.64 |
| S18 | AA | 64 | 160.0847 | 141.67 | 884.99 |
| Serial Number | Sequence | Content (μmol/g, on a Dry Basis) as a Function of Hydrolysis Time | ||||
|---|---|---|---|---|---|---|
| 3 h | 6 h | 10 h | 15 h | 24 h | ||
| S1 | VGVPGVGVPGVGVPGV | 6.95 ± 0.03 a | 8.26 ± 0.14 d | 8.65 ± 0.19 e | 8.02 ± 0.03 c | 7.33 ± 0.04 b |
| S9 | VPGVGVPG | 3.67 ± 0.04 a | 5.07 ± 0.04 b | 6.06 ± 0.03 d | 5.99 ± 0.03 c | 6.23 ± 0.03 e |
| S10 | VPGVG | 0.23 ± 0.02 a | 0.38 ± 0.02 b | 0.66 ± 0.01 c | 0.72 ± 0.03 d | 0.95 ± 0.03 e |
| S12 | VPG | 0.27 ± 0.02 a | 0.36 ± 0.01 b | 0.52 ± 0.02 d | 0.45 ± 0.01 c | 0.50 ± 0.02 d |
| S13 | GV | 14.02 ± 2.03 a | 22.75 ± 1.92 b | 35.35 ± 2.74 c | 34.21 ± 3.08 c | 43.32 ± 0.17 d |
| S14 | PG | 5.62 ± 0.31 a | 7.04 ± 0.45 b | 10.60 ± 0.48 c | 10.26 ± 0.69 c | 12.36 ± 0.19 d |
| S17 | AAA | 35.76 ± 0.49 a | 37.74 ± 0.84 b | 36.76 ± 0.39 ab | 37.51 ± 1.40 b | 39.83 ± 0.68 c |
| S18 | AA | 50.35 ± 2.28 a | 56.31 ± 1.36 b | 60.91 ± 1.95 c | 63.91 ±2.16 c | 70.90 ± 2.92 d |
| SUM | 116.85 ± 4.45 a | 137.90 ± 3.72 b | 159.51 ± 5.11 c | 161.07 ± 6.30 d | 181.42 ± 2.95 e | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Y.; Jiang, L.; Zhao, T.; Su, G.; Zhao, M. Exploring the Release of Elastin Peptides Generated from Enzymatic Hydrolysis of Bovine Elastin via Peptide Mapping. Molecules 2023, 28, 7534. https://doi.org/10.3390/molecules28227534
Zhang J, Liu Y, Jiang L, Zhao T, Su G, Zhao M. Exploring the Release of Elastin Peptides Generated from Enzymatic Hydrolysis of Bovine Elastin via Peptide Mapping. Molecules. 2023; 28(22):7534. https://doi.org/10.3390/molecules28227534
Chicago/Turabian StyleZhang, Jianan, Yang Liu, Liwen Jiang, Tiantian Zhao, Guowan Su, and Mouming Zhao. 2023. "Exploring the Release of Elastin Peptides Generated from Enzymatic Hydrolysis of Bovine Elastin via Peptide Mapping" Molecules 28, no. 22: 7534. https://doi.org/10.3390/molecules28227534
APA StyleZhang, J., Liu, Y., Jiang, L., Zhao, T., Su, G., & Zhao, M. (2023). Exploring the Release of Elastin Peptides Generated from Enzymatic Hydrolysis of Bovine Elastin via Peptide Mapping. Molecules, 28(22), 7534. https://doi.org/10.3390/molecules28227534










