Effects of Branched-Chain Fatty Acids Derived from Yak Ghee on Lipid Metabolism and the Gut Microbiota in Normal-Fat Diet-Fed Mice
Abstract
:1. Introduction
2. Results
2.1. Effect of Purified BCFAs from Yak Ghee on Body Weight and Food Intake
2.2. Effect of Purified BCFAs from Yak Ghee on Serum Biochemical Indices in Mice
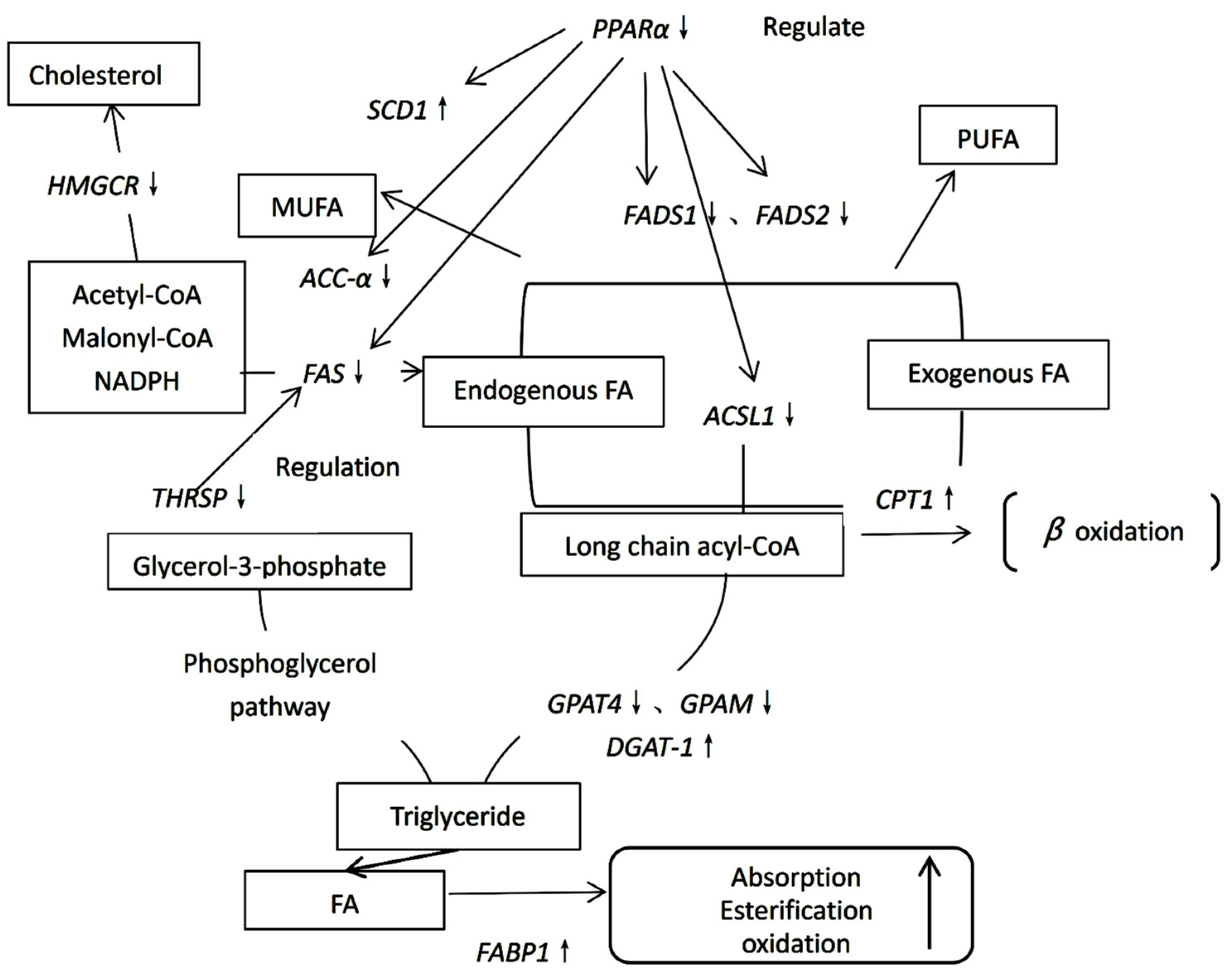
2.3. Profile of Gene Expression of Fatty Acid Synthesis-Related Genes
2.4. Gene Expression of Genes Involved in Triacylglycerol Synthesis
2.5. Expression of the Cholesterol Synthesis Gene HMGCR
2.6. Gene Expression of Lipid Metabolism-Related Genes
2.7. Analysis of the Diversity of Gut Microbiota in Each Group of Mice
2.8. Analysis of the Species Composition of the Intestinal Flora of Mice
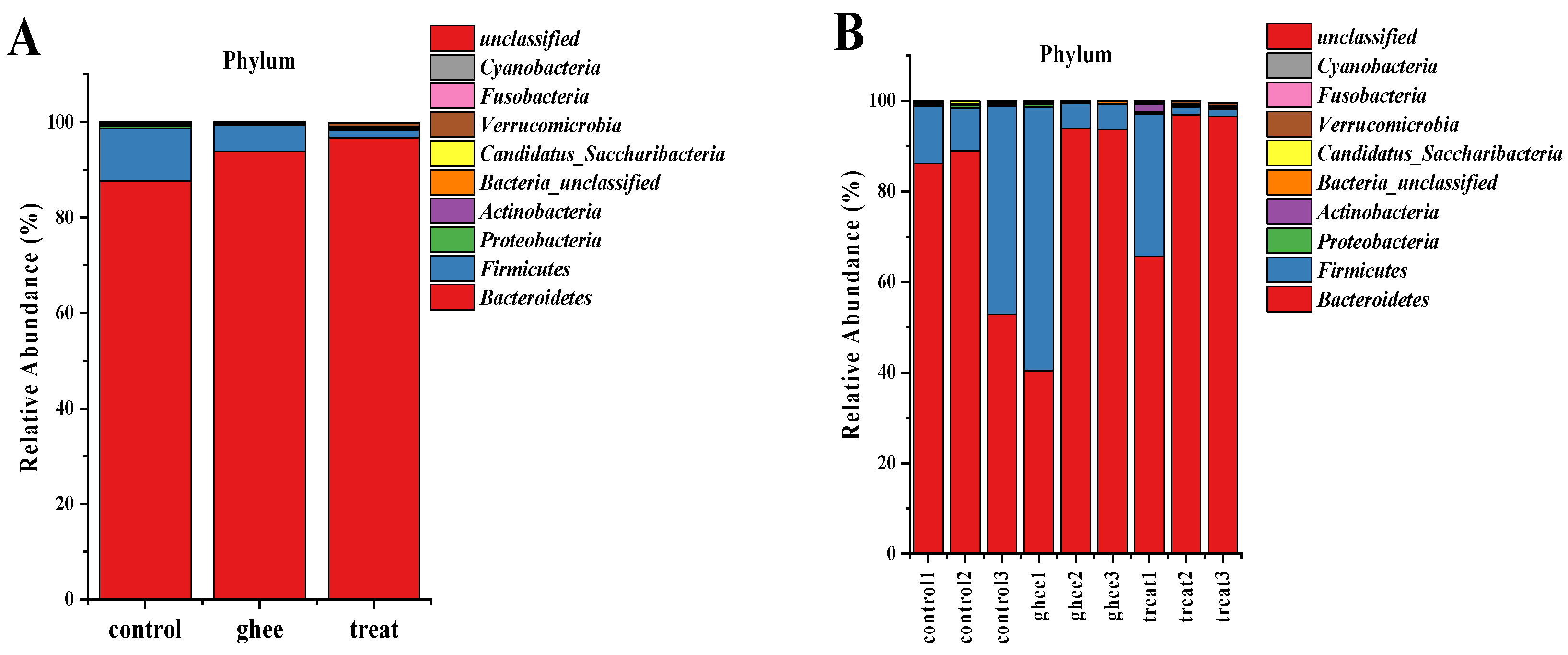
2.8.1. Relative Abundance of Major Phyla of the Intestinal Flora
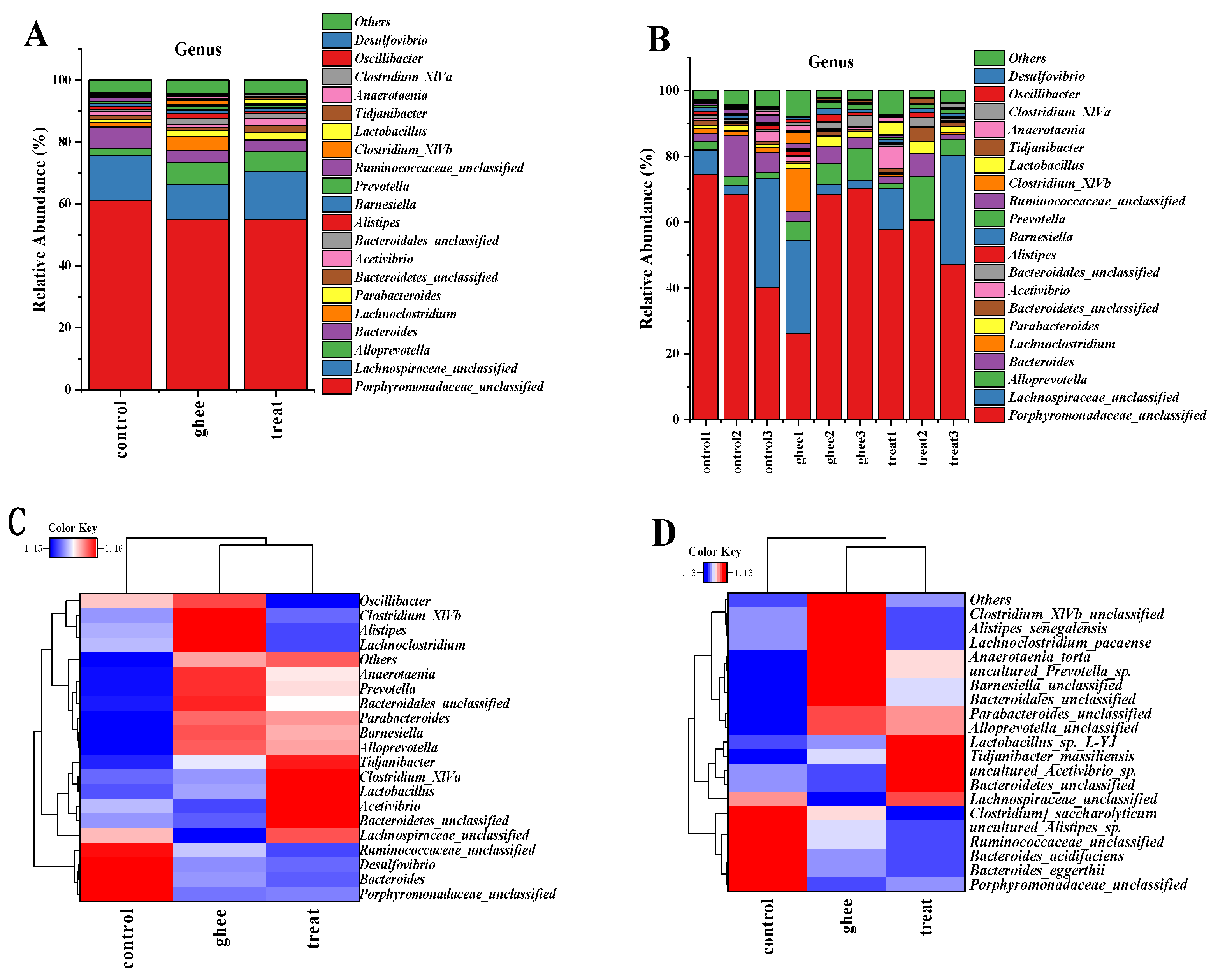
2.8.2. Relative Abundance of Major Genera and Species of Intestinal Flora
2.9. Analysis of KEGG Pathways in the SK-BR-3 Transcriptome of Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. The Experimental Animals and Materials
4.2. Animals and Tissue Sampling
4.3. Measurement of Serum Biochemical Indicators
4.4. RNA Preparation and Quantitative Real-Time PCR
4.5. 16S rDNA Gene Sequencing
4.5.1. Total DNA of the Microbe Preparation
4.5.2. 16S rDNA High-Throughput Sequencing Analysis
4.6. Cell Culture
4.7. Illumina Sequencing
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sun, M.Q.; Ma, Y.; Cheng, J.J. Study on physical and chemical properties of yak ghee. China Dairy Ind. 2016, 44, 9–11. [Google Scholar]
- Ma, L. Characterization of Milk Components of Dairy Cows, Buffaloes, Yaks, Juanshan Cows, Goats, Camels and Horses. Ph.D. Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2015. [Google Scholar]
- Zheng, Y.C.; Gong, W.H.; Du, X.; Yang, M.; Jing, S.Y.; Hong, J. Fatty acid composition of yak ghee and its effect on blood lipids in mice. China Dairy Ind. 2009, 37, 18–20. [Google Scholar]
- Jane, L.Y. Effects of Different Edible Fats and Oils Intake on Fatty Acid Composition and Lipid Metabolism of Liver and Blood in Mice. Ph.D. Thesis, Dalian University of Technology, Dalian, China, 2018. [Google Scholar]
- Yang, X.; Karrar, E.; Cong, F.; Lu, H.; Jin, Q.; Xu, X.; Huppertz, T.; Wei, W.; Wang, X. Identification and quantification of branched-chain fatty acids and odd-chain fatty acids of mammalian milk, dairy products, and vegetable oils using GC/TOF-MS. Int. Dairy J. 2023, 140, 105587. [Google Scholar] [CrossRef]
- Sutton, J.D.; Dhanoa, M.S.; Morant, S.V.; France, J.; Napper, D.J.; Schuller, E. Rates of production of acetate, propionate, and butyrate in the rumen of lactating dairy cows given normal and low-roughage diets. J. Dairy Sci. 2003, 86, 3620–3633. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, L.Z.; Wang, Z.S.; Xue, B.; Peng, Q.H. Relationship between rumen microorganisms and fatty acid composition of ruminant products. Feed Ind. 2018, 9, 51–55. [Google Scholar]
- Picklo, M.J.; Kalscheur, K.F.; Magnuson, A.; Bukowski, M.R.; Harnly, J.; Fukagawa, N.K.; Finley, J.W. Identification of High and Low Branched-Chain Fatty Acid-Producing Phenotypes in Holstein Cows following High-Forage and Low-Forage Diets in a Crossover Designed Trial. Curr. Dev. Nutr. 2022, 28, 154. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Roseblade, A.; Rawling, T. Expansion of the structure-activity relationship of branched chain fatty acids: Effect of unsaturation and branching group size on anticancer activity. Chem. Phys. Lipids 2020, 232, 104952. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, S.; Chen, X.; Chen, H.; Huang, M.; Zheng, J. Induction of apoptotic cell death and in vivo growth inhibition of human cancer cells by a saturated branched-chain fatty acid 13-methyltetradecanoic acid. Cancer Res. 2000, 60, 505–509. [Google Scholar]
- Wongtangtintharn, S.; Oku, H.; Iwasaki, H.; Toda, T. Effect of branched-chain fatty acids on fatty acid biosynthesis of human breast cancer cells. J. Nutr. Sci. Vitaminol. 2004, 50, 137–143. [Google Scholar] [CrossRef]
- Yu, J.; Yang, L.N.; Wu, Y.Y.; Lee, B.H.; Weng, S.M.; Hu, C.L.; Han, Y.L. 13-Methyltetradecanoic acid mitigates cerebral ischemia/reperfusion injury. Neural Regen. Res. 2016, 11, 1431–1437. [Google Scholar] [CrossRef]
- Ran-Ressler, R.R.; Khailova, L.; Arganbright, K.M.; Adkins-Rieck, C.K.; Jouni, Z.E.; Koren, O.; Ley, R.E.; Brenna, J.T.; Dvorak, B. Branched Chain Fatty Acids Reduce the Incidence of Necrotizing Enterocolitis and Alter Gastrointestinal Microbial Ecology in a Neonatal Rat Model. PLoS ONE 2011, 6, e29032. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, Z.; Greenwald, J.; Kothapalli, K.S.D.; Park, H.G.; Liu, R.; Mendralla, E.; Lawrence, P.; Wang, X.; Brenna, J.T. BCFA suppresses LPS induced IL-8 mRNA expression in human intestinal epithelial cells. Prostaglandins Leukot. Essent. Fat. Acids 2017, 116, 27. [Google Scholar] [CrossRef]
- Li, J. In Vitro Hypolipidemic Effects and Mechanisms of Branched-Chain Fatty Acids iso-15:0 and iso-18:0. Ph.D. Thesis, Hunan Agricultural University, Changsha, China, 2018. [Google Scholar]
- Zhou, Z.K.; Wang, Y.Y.; Diao, Y.J.; Wang, F. Effects of excessive frying oil on glucose and lipid metabolism and related gene expression in rats. J. Chin. Cereals Oils Assoc. 2017, 32, 70–73. [Google Scholar]
- Xing, J.Y.; Zhang, Y.J.; Zhang, N.B.; Jing, W.Q.; Lee, Z.; Wang, X.Z.; Xu, Y.C. Effects of folic acid addition levels on expression and methylation of fat metabolism related genes in liver of broilers. J. Anim. Nutr. 2019, 31, 2098–2106. [Google Scholar]
- Zhang, R.; Chen, S.Y. Review of fatty acid synthase (FAS) research. Rural. Econ. Technol. 2018, 29, 111–113. [Google Scholar]
- Kang, J.; Zou, M.Y.; Ma, Y.J.; Song, X.H.; Sun, C.H.; Hao, Y.Q.; Xia, W.; Wu, L.J. Association between FADS1, FADS2 gene polymorphism and Chinese Hanautism spectrum disorder. Chin. J. Child Health 2017, 25, 14–17. [Google Scholar]
- Long, S.; Zhou, Y.; Bai, D.; Hao, W.; Fang, L. Fatty Acids Regulate Porcine Reproductive and Respiratory Syndrome Virus Infection via the AMPK-ACC1 Signaling Pathway. Viruses 2019, 11, 1145. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.H.; Sun, W.C. Cloning and analysis of yak desaturated lipase 1 (SCD1) gene. Heilongjiang Anim. Husb. Vet. Med. 2017, 9, 237–240. [Google Scholar]
- Cai, L.J.; Su, R.; Lee, N.N. Discussion on the effect of acetyl-CoA synthetase 2 on tumor metastasis. J. Guangzhou Med. Univ. 2018, 46, 103–106. [Google Scholar]
- Cao, Y.; Jin, H.G.; Yu, Y.S.; Zhang, L.C. Effects of ACSL1 gene on fat metabolism in sheep. Chin. J. Vet. Med. 2018, 38, 218–221. [Google Scholar]
- Tian, Z.Y.; Li, K.J.; Men, X.L.; Wu, J. Research progress of glycerol-3-phosphate acyltransferases (GPATs). Prog. Physiol. Sci. 2015, 46, 5. [Google Scholar]
- Compe, E.; Sousa, J.D.; Franccois, K.; Roche, R.; Rahmani, R.; Torresani, J.; Raymondjean, M.; Planells, R. Spot 14 protein interacts and cooperates with chicken ovalbumin upstream promoter-transcription factor 1 in the transcription of the L-type pyruvate kinase gene through a specificity protein 1 (Sp1) binding Site. Biochem. J. 2001, 358, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.J.; Lin, Y.Q.; Yu, Y.Y.; Wang, Y.; Zhu, J.J. Clonal expression of goat CPT1A gene and correlation analysis of intramuscular fat content. J. North China Agric. 2019, 34, 231–238. [Google Scholar]
- Jin, J.; Peng, D.Q. Advances in research of adipocyte-type fatty acid binding protein and atherosclerosis. Chin. J. Arterioscler. 2009, 17, 82–85. [Google Scholar]
- Liu, Q.; Mo, Z.C.; Guo, B.B.; Qu, L.H.; Liao, H.Q.; Xie, Y.J.; Zhang, M.X. Effect of palmitate on the expression of SR-BI and lipid contenzt in KGN cells via oxidase proliferator-activated receptor alpha. J. Pract. Med. 2019, 35, 49–54. [Google Scholar]
- Wang, J.T.; Cao, W.N.; Huo, Z.M.; Yan, X.W. Characterization of PPAR gene family in C. philippinarum and expression changes under dry dew stress. J. Dalian Ocean Univ. 2019, 34, 19–29. [Google Scholar]
- Storch, J.; McDermott, L. Structural and functional analysis of fatty acid-binding proteins. J. Lipid. Res. 2009, 50, 126–131. [Google Scholar] [CrossRef]
- Newberry, E.P.; Xie, Y.; Kennedy, S.; Han, X.L.; Buhman, K.K.; Luo, J.Y.; Gross, R.W.; Davidson, N. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein-encoding gene. J. Biol. Chem. 2003, 278, 51664–51672. [Google Scholar] [CrossRef]
- Atshaves, B.P.; Story, S.M.; Huang, H.; Schroeder, F. Liver fatty acid binding protein expression enhances branched-chain fatty acid metabolism. Mol. Cell. Biochem. 2004, 259, 115–129. [Google Scholar] [CrossRef]
- Cogburn, L.A.; Wang, X.; Carre, W.; Rejto, L.; Porter, T.E.; Aggray, S.E.; Simon, J. Systems-wide chicken DNA microarrays, gene expression profiling and discovery of functional genes. Poultry Sci. 2003, 82, 6378–6383. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Ding, Z.X.; Zhang, M.; Chen, H.Q.; Qin, H.L.; Jiang, Y.Q. Effects of probiotics on gut flora disturbance and bacterial translocation in mice with inflammatory bowel disease. J. Shanghai Jiaotong. Univ. 2010, 30, 74–78. [Google Scholar]
- Lee, J.Y.; Sun, Z.Y.; Yuan, Y. Correlation between gut flora and coronary heart disease based on high-channel sequencing. Chin. Gen. Pract. 2019, 22, 3554–3560. [Google Scholar]
- Huang, W.X.; Wang, H.P.; Wang, Y.L.; Dai, W.K.; Zheng, Y.J. High-throughput sequencing reveals the change of gut microbiota in infants with pneumonia following antibiotic treatment. Chin. J. Microecol. 2016, 28, 497–500. [Google Scholar]
- Zhang, H.H.; Jiang, J.X. Overview of bacterial infection and pathogenesis of Helicobacter. China Natl. Poult. 2017, 39, 44–48. [Google Scholar]
- Wang, S.M.; Zhang, S.; Zhang, L.L.; Zhang, G.S. Research progress on the role of lactic acid bacteria in regulating organism immunity. Cereals Oils 2022, 35, 16–27. [Google Scholar]




| Group | Initial Weight/g | Weight after Four Weeks/g | Difference/g | Total Food Intake/g | Food Utilization Rate/% |
|---|---|---|---|---|---|
| control | 16.278 ± 0.373 bB | 19.969 ± 0.949 aA | 3.691 ± 0.942 aA | 814.507 ± 26.328 aA | 0.453 ± 0.361 aA |
| ghee | 16.175 ± 0.369 bB | 20.070 ± 0.940 aA | 3.895 ± 0.977 aA | 801.244 ± 30.614 aA | 0.486 ± 0.215 aA |
| treat | 16.411 ± 0.643 aA | 20.209 ± 1.168 aA | 3.798 ± 1.349 aA | 794.825 ± 27.145 aA | 0.478 ± 0.241 aA |
| Group | TC (mmol/L) | TG (mmol/L) | LDL-C (mmol/L) | HDL-C (mmol/L) |
|---|---|---|---|---|
| control | 6.646 ± 0.345 aA | 0.943 ± 0.009 bB | 1.059 ± 0.012 aA | 1.093 ± 0.018 bB |
| ghee | 6.844 ± 0.565 aA | 1.126 ± 0.078 aA | 1.104 ± 0.016 aA | 1.007 ± 0.039 cC |
| treat | 5.543 ± 0.661 bB | 0.837 ± 0.033 bB | 0.856 ± 0.044 bB | 1.214 ± 0.008 aA |
| Group | Observed_Species | Chao1 | Shannon | Simpson |
|---|---|---|---|---|
| control | 2132.00 ± 109.42 | 2734.02 ± 109.52 bB | 7.02 ± 0.27 | 0.97 ± 0.01 |
| ghee | 1980.33 ± 56.57 | 2359.89 ± 36.29 aA | 6.94 ± 0.01 | 0.97 ± 0.01 |
| treat | 2083.67 ± 133.83 | 2689.39 ± 60.43 bB | 6.93 ± 0.26 | 0.96 ± 0.02 |
| Group | Bacteroidetes | Firmicutes | F/B | Proteobacteria | Actinomycetes | Candidatus_Saccharibacteria | Verrucomicrobia |
|---|---|---|---|---|---|---|---|
| control | 87.601 ± 2.047 cC | 11.052 ± 2.286 aA | 0.126 | 0.533 ± 0.000 aA | 0.218 ± 0.010 bB | 0.387 ± 0.237 aA | 0.006 ± 0.005 bB |
| ghee | 93.837 ± 0.228 bB | 5.530 ± 0.026 bB | 0.059 | 0.091 ± 0.011 cC | 0.139 ± 0.067 bB | 0.058 ± 0.022 bB | 0.001 ± 0.001 bB |
| treat | 97.015 ± 3.690 aA | 1.600 ± 4.777 cC | 0.016 | 0.366 ± 0.024 bB | 1.108 ± 1.020 aA | 0.086 ± 0.098 bB | 0.189 ± 0.182 aA |
| Group | Helicobacter | Bilophila | Akkermansia | Lactobacillus |
|---|---|---|---|---|
| control | 0.069 ± 0.109 aA | NA | 0.005 ± 0.004 bB | 0.083 ± 0.054 aA |
| ghee | NA | NA | 0.001 ± 0.001 bB | 0.201 ± 0.093 aA |
| treat | NA | 0.051 ± 0.071 aA | 0.344 ± 0.100 aA | 1.381 ± 1.964 aA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, T.; Luo, Y.; Sun, W.; Li, X. Effects of Branched-Chain Fatty Acids Derived from Yak Ghee on Lipid Metabolism and the Gut Microbiota in Normal-Fat Diet-Fed Mice. Molecules 2023, 28, 7222. https://doi.org/10.3390/molecules28207222
Tan T, Luo Y, Sun W, Li X. Effects of Branched-Chain Fatty Acids Derived from Yak Ghee on Lipid Metabolism and the Gut Microbiota in Normal-Fat Diet-Fed Mice. Molecules. 2023; 28(20):7222. https://doi.org/10.3390/molecules28207222
Chicago/Turabian StyleTan, Ting, Yihao Luo, Wancheng Sun, and Xiaoxiao Li. 2023. "Effects of Branched-Chain Fatty Acids Derived from Yak Ghee on Lipid Metabolism and the Gut Microbiota in Normal-Fat Diet-Fed Mice" Molecules 28, no. 20: 7222. https://doi.org/10.3390/molecules28207222
APA StyleTan, T., Luo, Y., Sun, W., & Li, X. (2023). Effects of Branched-Chain Fatty Acids Derived from Yak Ghee on Lipid Metabolism and the Gut Microbiota in Normal-Fat Diet-Fed Mice. Molecules, 28(20), 7222. https://doi.org/10.3390/molecules28207222







