Abstract
Based on previous results with benzoindazolequinone (BIZQ) and 3-methylnaphtho [2,3-d]isoxazole-4,9-quinone (NIQ) derivatives, a novel series of chalcone-1,4-naphthoquinone/benzohydroquinone (CNQ and CBHQ) compounds were synthesized from 2-acetyl-5,8-dihydro-6-(4-methyl-3-pentenyl)-1,4-naphthohydroquinone. Their structures were elucidated via spectroscopy. These hybrids were assessed in vivo for their antiproliferative activity on MCF-7 breast adenocarcinoma and HT-29 colorectal carcinoma cells, revealing cytotoxicity with IC50 values between 6.0 and 110.5 µM. CBHQ hybrids 5e and 5f displayed enhanced cytotoxicity against both cell lines, whereas CNQ hybrids 6a–c and 6e exhibited higher cytotoxic activity against MCF-7 cells. Docking studies showed strong binding energies (ΔGbin) of CNQs to kinase proteins involved in carcinogenic pathways. Furthermore, our in silico analysis of drug absorption, distribution, metabolism, and excretion (ADME) properties suggests their potential as candidates for cancer pre-clinical assays.
1. Introduction
The uncontrolled division and spread of abnormal cells remain a global medical challenge, with cancer being a leading cause of death [1,2]. Of the 9.9 million new cancer cases diagnosed, the most common are breast (11.7%), lung (11.4%), colorectal (10.0%), prostate (7.3%), and stomach cancer (5.6%) [3].
Quinones are widely studied compounds for their interesting biological activities [4,5,6], with daunorubicin and doxorubicin being key pillars in oncology treatments [7]. Their antitumor properties are primarily due to the inhibition of DNA topoisomerases through intercalation and the generation of reactive oxygen species (ROS), which contribute to cellular oxidative stress [8,9]. Naphthoquinones/hydroquinones regulate antiproliferative processes through p53, kinases, vascular endothelial growth factor receptor 2, COX-2, protein activators of transcription (STAT3), the inhibition of signaling via EGFR-NF-kB, and the inhibition of protein phosphatase Cdc25, among others [10,11,12,13,14].
Chalcones, 1,3-diaryl-2-propen-1-ones, are precursors to flavonoids, flavanones, and aurones [15,16,17] with diverse pharmacological benefits, including antineoplastic, antioxidant, and anti-inflammatory properties [18,19]. Studies of their antineoplastic properties have revealed interactions with various target proteins involved in proliferation [20,21,22]. These compounds disrupt the cell cycle by interfering with microtubules, which play critical roles in mitosis, motility, and transport. Their binding to tubulins prevents polymerization, altering mitotic spindle assembly and disrupting cytoskeletal function, leading to cell cycle arrest in the G2/M phase [23,24].
Chalcones also activate the p53 protein, regulating the cell cycle by binding to the human oncoprotein Mdm2 (murine double minute 2). They inhibit angiogenesis by blocking vascular endothelial growth factor (VEGF) and various enzymes, including topoisomerases, the androgen receptor, mitogen-activated protein kinases (ERK1/2), cyclooxygenase 2 (COX-2), and histone deacetylase (HDACs), ultimately inducing apoptosis [25,26,27]. The presence of a conjugated double bond with the carbonyl group is responsible for these biological properties due to its structural planarity, making this compound family a crucial pharmacophore for synthesizing new antitumor molecules [28].
In the development of new anticancer drugs, molecular hybridization is a promising strategy, involving the rational design of compounds that combine multiple pharmacophores within a single structure to enhance antiproliferative activities [29,30,31]. Chalcone hybrids have shown significant antineoplastic potential against various cancer types [32,33,34,35], while quinone-based hybrids exhibit high cytotoxicity [10,36,37]. Our research group specializes in designing and synthesizing hybrid molecules by combining 1,4-naphthoquinones with azoles, demonstrating their significant anticancer properties against breast cancer (MCF-7) and gastric cancer (KATO-III) [38,39,40]. In this work, we propose the rational design, synthesis, and in vitro anticancer evaluation of novel chalcone-naphthoquinone/hydroquinone hybrids (Figure 1). For this purpose, we use 2-acetyl-5,8-dihydro-6-(4-methyl-3-pentenyl)-1,4-naphthohydroquinone as a precursor, since it was developed in our previous studies. This compound proved to be a pharmacophore that generated structures with promising in vitro antitumor activity against neoplastic cell cultures [41]. Additionally, we conduct molecular docking studies with cancer-related proteins and in silico ADME predictions to explore potential mechanisms of anticancer action and the drug properties of these newly synthesized hybrids.
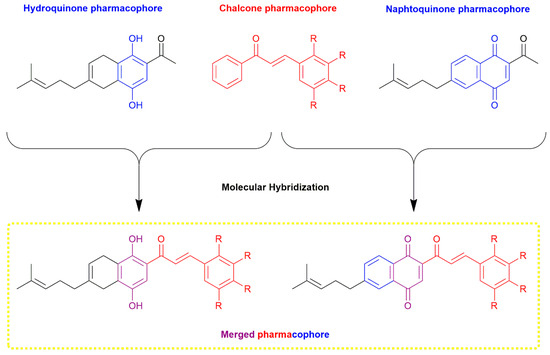
Figure 1.
Rational design of novel chalcone-naphthoquinone/hydroquinone hybrids.
2. Results and Discussion
2.1. Chemistry
As shown in Scheme 1, the synthesis of compounds 2–4a–f was performed from the precursor 2-acetyl-5,8-dihydro-6-(4-methyl-3-pentenyl)-1,4-naphthohydroquinone 1, following previous experimental methods [41,42]. Initially, it was necessary to protect the hydroxyl group attached to C-4 of the precursor to eliminate the acidic properties attributed to the phenolic 4-OH group. This protection was achieved using 3,4-dihydro-2H-pyran (DHP) as a protective agent, in the presence of p- pyridinium toluenesulfonate (PPTS), yielding 80%. The phenolic 1-OH group, on the other hand, remained unaltered due to its stabilization through intramolecular hydrogen bonding with the adjacent carbonyl group, forming a stable six-membered cyclic system [43].
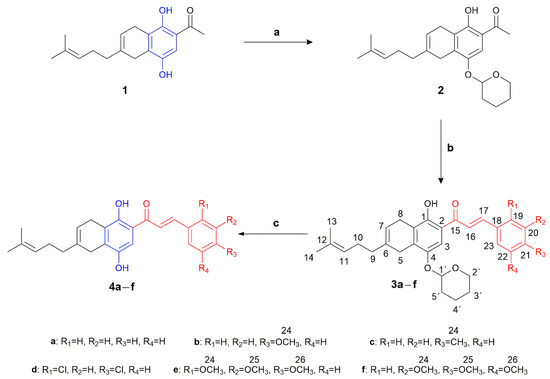
Scheme 1.
Synthesis route of compounds 4a–f. Reagents and conditions: (a) DHP, PPTS, CH2Cl2, rt, 19 h. (b) Ba(OH)2·8H2O, benzaldehydes, EtOH, 90 °C, 25 min; (c) PTSA, MeOH, rt, 3 h.
The synthesis of the new chalcone-1,4-benzohydroquinone (CBHQ) 4a–f hybrids involved Claisen–Schmidt condensation of the protected derivative 2 (as tetrahydropyranyl ether, 4-OTHP) with the respective benzaldehyde in the presence of barium hydroxide octahydrate [44]. By using an ace-pressure tube to carry out the reaction mixture, it was possible to increase the synthesis yields of these products and significantly reduce the reaction time compared to the traditional use of reflux heating. This process yielded chalcones 3a–f with moderate yields (60–70%). The subsequent deprotection of the THP group by acid hydrolysis using 4-toluenesulfonic acid monohydrate (PTSA) was carried out with good yields (93–96%).
The structures of derivatives 2–4a–f, along with other newly synthesized compounds, were elucidated spectroscopically by IR, 1H NMR, 13C NMR, and elemental analyses. In the infrared (IR) spectra, the characteristic stretching vibration (stv) absorption bands of phenolic O-H bonds were observed in the range of 3272–3492 cm−1, in addition to the stv bands of C=O bonds at 1630–1662 cm−1. The stv band of the C=C bond in the α,β-unsaturated fragment of chalcones was observed at 1558–1633 cm−1.
In the 1H NMR spectrum of derivative 2, signals corresponding to the protons in the methylene groups of the tetrahydropyranyl protecting ring (H-3′, H-4′, H-2′, and H-5′) were observed between δ 1.66 and 3.90 ppm. The signal for the methine group H-1′ was observed at δ 5.33 ppm. Additionally, the methyl group H-16 appeared at δ 2.58 ppm, while the phenolic proton 1-OH exhibited a higher chemical shift (δ 12.45 ppm). The 1H NMR and 13C NMR spectra of compound 2 are shown in Figure S1 and Figure S2, respectively.
The 1H NMR spectra of derivatives 3a–f showed signals belonging to the protons of the α,β-unsaturated fragment characteristic of chalcones H-16 and H-17 in the ranges of δ 7.46–7.69 and δ 7.90–8.20 ppm, respectively. The signals of the analogous protons H-16 and H-17 of hybrids 4a–f were observed at δ 7.67–7.88 and δ 7.84–8.12 ppm, respectively, with both coupled protons of the unsaturated system having trans geometry (J 15, 3–16.0 Hz). Most of the phenolic protons at 4-OH and 1-OH appeared as singlets in the ranges of δ 7.94–8.12 and 12.78–13.08 ppm, respectively. Notably, in the 1H NMR spectra of hybrids 4b, 4e, and 4f, methoxyl singlets were observed between δ 3.81 and 3.98 ppm, and the signal corresponding to the methyl group H-24 of compound 4c appeared at δ 2.38 ppm. The 1H NMR spectra of compounds 3a–f and 4a–f are shown in Figures S3–S14.
In the 13C NMR spectra of compounds 3a–f and 4a–f, the characteristic carbonyl of chalcones C-15 is in the range of δ 192.7–193.6 and δ 192.7–193.4 ppm, respectively. For compounds 3a–f, the presence of the signals corresponding to C-4 and C-1 occurred between δ 144.8 and 146.4 and δ 156.8 and 157.0 ppm, respectively. The signals of the analogous carbons C-4 and C-1 of the 1,4-benzohydroquinone fragment of hybrids 4a–f were observed at δ 144.9–146.4 and δ 155.2–156.5 ppm, respectively. The 13C NMR spectrum of compounds 3a–f and 4a–f are shown in Figures S15–S26.
The acetylation of the phenolic groups in the CBHQ 4a–f hybrids was carried out using acetic anhydride in pyridine [45]. As depicted in Scheme 2, this process yielded 1,4-diacetylated derivatives 5e and 5f, as well as 4-monoacetylated derivatives 5′b, 5′c, and 5′d. The formation of these derivatives resulted from the stabilization of the proton through intramolecular hydrogen bonding with the adjacent carbonyl group. These compounds were synthesized with favorable yields ranging from 85% to 95%, except for derivative 5a, which could not be obtained in pure form.
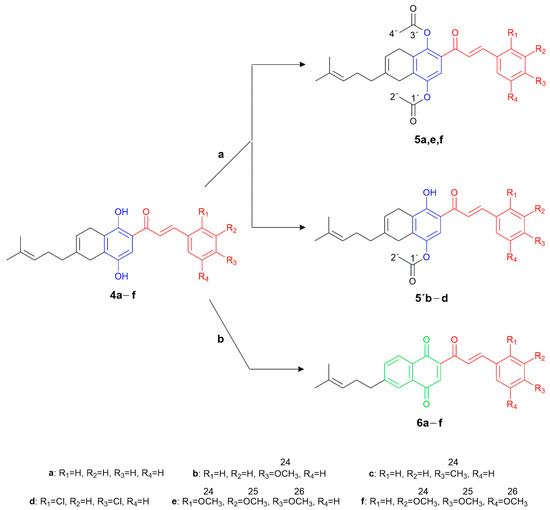
Scheme 2.
Synthesis of hybrid molecules 5a,e,f, 5′b–d, and 6a–f. Reagents and conditions: (a) acetic anhydride, pyridine, rt, 24 h. (b) DDQ, CH2CH2, rt, 30 min.
Furthermore, the obtained compounds 4a–e facilitated the synthesis of a new series of molecular hybrids, denoted as chalcones-1,4-naphthoquinones (CNQ) 6a–f (Scheme 2). These hybrids were generated through in situ reactions involving the oxidation of carbons C-1 and C-4 in the phenolic groups, along with the aromatization of the ring fused to the 1,4-benzohydroquinone moiety, using an excess of DDQ in CH2Cl2 at room temperature [39]. The new hybrids 6a–e were obtained with moderate yields ranging from 30% to 58%, while the derivative 6f could not be adequately purified.
In the infrared (IR) spectra of compounds 5e and 5f, characteristic stretching vibration (stv) absorption bands of the C=O groups in the acetyl units were observed at 1766 cm−1. On the other hand, derivatives 5′b, 5′c, and 5′d exhibited stv bands at 3379–3391 cm−1 and 1755–1758 cm−1, corresponding to the phenolic O-H bond and the C=O bond of the acetyl group, respectively. For hybrids 6a–e, the stv band related to the C=O group of the quinone moiety was observed in the range of 1654–1668 cm−1.
In the 1H NMR spectra of derivatives 5e and 5f, signals corresponding to the protons within the acetyl group (H-2′ and H-4′) were detected between δ 2.29 and 2.33 and δ 2.33 and 2.37 ppm, respectively. Additionally, the signal for the methyl group H-2′ in derivatives 5′b, 5′c, and 5′d appeared at δ 2.38–2.41 ppm, while the phenolic proton 1-OH manifested as a singlet in the range of δ 13.05–13.32 ppm. Concerning hybrids 6a–e, the signals attributed to the aromatic protons of the naphthoquinone moiety (H-7, H-5, and H-8) were observed in the ranges of δ 7.47–7.64, δ 7.66–7.94, and δ 8.00–8.07 ppm, respectively. The 1H NMR spectrum of compounds 5′b–f and 6a–e are shown in Figures S27–S36.
In the 13C NMR spectra of compounds 5e and 5f, intense signals corresponding to both carbonyl groups of the acetyl units (C-1′ and C-3′) were evident at δ 169.0 ppm. For the derivatives 5′b, 5′c, and 5′d, the signal of C-1′ appeared between δ 169.8 and 169.9 ppm. In the case of hybrids 6a–e, signals attributed to the carbonyl groups of the quinone system (C-4 and C-1) were observed in the ranges of δ 183.4–183.5 ppm and δ 185.1–185.4 ppm, respectively. The 13C NMR spectra of compounds 5′b–f and 6a–e are shown in Figures S37–S46.
2.2. In Vitro Cytotoxicity Assays
The antiproliferative activity of the new derivatives 4a–f, 5e–f, 5′b–d, and 6a–e was evaluated on MCF-7 and HT-29 cancer cell lines using a CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS). The results, presented in Table 1, are expressed as the half-maximal inhibitory concentration (IC50 ± standard deviation) for cell proliferation inhibition, along with the corresponding -log10 values (pIC50).

Table 1.
In vitro cytotoxicity data for compounds 4a–f, 5a,e,f, 5′b–d, and 6a–f on MCF-7 breast adenocarcinoma and HT-29 colon adenocarcinoma cells.
Overall, the compounds exhibiting noteworthy antineoplastic activity in this study demonstrated a higher cytotoxic effect against MCF-7 breast carcinoma, with pIC50 values ranging from 4.07 to 5.09. Similar results were observed for HT-29 colon cancer, with pIC50 values ranging from 3.74 to 5.22. Notably, the derivatives 5e, 5f, and 6b exhibited outstanding cytotoxicity against MCF-7, while the compounds 5e and 5f demonstrated significant activity against the HT-29 cell line. Conversely, the series of CBHQ hybrids (4a–f) and the 4-monoacetylated CBHQ derivatives (5′b–d) did not exhibit significant cytotoxic activity in either cancer cell cultures (pIC50 < 3.52).
Regarding the HT-29 cell line, an analysis of cytotoxicity based on the type of substituent in the aromatic ring of the chalcone fragment revealed that the methoxyl groups enhanced antiproliferative effects. An increase in the number of these substituents corresponded to a marked increase in cytotoxicity, with the derivatives 5e and 5f displaying high pIC50 values of 4.87 and 5.22, respectively.
A similar trend was observed against the MCF-7 cell line, in which the trimethoxylated derivatives 5e and 5f achieved pIC50 values of 4.96 and 5.09, respectively. In contrast, the presence of 2,4-dichloro substituents in compound 6d resulted in limited cytotoxic effects, with pIC50 values lower than 3.52 against both neoplastic cultures.
Regarding the in vitro cytotoxicity data (Table 1), the majority of the compounds in the CNQ hybrid series exhibited superior pIC50 values compared to those of CBHQs 4a–f and the monoa-cetylated 5′a–d hybrid series. This highlights the significance of the naphthoquinone and 1,4-diacetylated benzohydroquinone pharmacophores present in these structures in terms of their cytotoxic activity against both assessed carcinogenic cell lines.
In terms of the structure–activity relationship, these findings underscore the exceptional cytotoxicity of the trimethoxylated 1,4-diacetylated CBHQ compounds 5e and 5f against both cancer lines. Compound 5f, in particular, displayed the highest potency against the HT-29 line (pIC50 5.22), comparable to that of the reference drug doxorubicin (pIC50 5.39). This underscores the pivotal role played by the -OCH3 and 1,4-COCH3 substituents, which significantly influence the cytotoxicity exhibited by CBHQs 5e–f derivatives (Figure 2). These results align with those of previous reports identifying various methoxychalcones with anti-tumor properties, attributed to the electron-donating properties of the aryl ring of chalcones. The presence of the methoxy group enhances the anticancer activity of these structures [46,47,48].
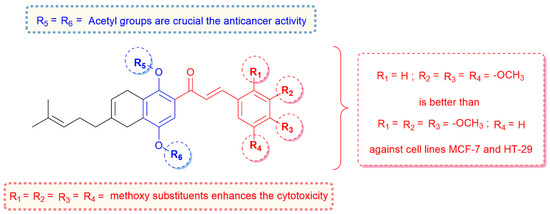
Figure 2.
SAR of chalcone–hydroquinone hybrids as anticancer agents.
Furthermore, CNQs 6a–f generally demonstrated superior pIC50 values compared to those of benzohydroquinone CBHQs 4a–f and the monoacetylated benzohydroquinone compounds 5b–d. Hence, the diacetylation of both hydroxyl groups and the presence of the quinone ring also play significant roles in their cytotoxic activity. These compounds hold promise for exploring their cytotoxic potential against other malignant cell lines. Indeed, research is evolving towards optimizing these compounds to develop lead molecules for potential antitumor drugs. This includes incorporating -OCH3 in different positions of the aryl group of diacetylated chalcones and acetylating both hydroxyl groups.
It is worth noting that the mechanism of action for doxorubicin, the reference drug in Table 1, involves the inhibition of DNA topoisomerases I and II. Doxorubicin stabilizes the tertiary adduct (DNA-drug-Topo), which prevents DNA strand rewinding, disrupts the cell cycle equilibrium, and ultimately induces apoptosis [38,49]. Since the tested compounds in this study contain a quinone system, they may potentially share a similar mechanism of action with doxorubicin. However, these compounds did not exhibit strong binding energies (ΔGbin) for topoisomerases compared to that of doxorubicin (Table S1). Therefore, in this study, we aimed to explore alternative mechanisms for the antineoplastic action of these cytotoxic hybrids through molecular-docking-based virtual screening with various cancer-related proteins, including growth factor receptors, transcription regulators, and enzymes (such as reductases, oxidases, and kinases).
2.3. In Silico Virtual Screening for Potential Antineoplastic Targets of Synthesized Cytotoxic Hybrids
Considering the higher cytotoxicity of the compounds (with pIC50 values greater than 4, as shown in Table 1), we conducted in silico molecular docking studies to identify potential biological targets for these new antiproliferative hybrids, providing insights into their possible mechanisms of action. To achieve this, we predicted the potential docking sites of these cytotoxic hybrids in several cancer-related proteins and calculated their corresponding ΔGbin values. For robust results, we focused on a subset of cancer-related proteins with known 3D structures, conducting independent searches with the compounds and utilizing their most stable conformers during interactions with these biological targets. The selected proteins are known to be overexpressed in breast cancer cells, including epidermal growth factor receptor (EGFR), epidermal growth factor receptor 2 (HER2), mesenchymal-epithelial transition factor (c-MET), tropomyosin receptor kinase A (TRKA), mitogen-activated protein kinases (MAPK1, ERK2, MEK1), tyrosine protein kinase (TPK), dihydrofolate reductase (DHFR), fibroblast growth factor receptor 2 (FGFR-2), vascular endothelial growth factor receptor 2 (VEGFR-2), estrogen receptors (ERs), cyclooxygenase (COX-2), and tubulin (TUB), among others. Additionally, COX-2 and MEK1 are overexpressed in colon adenocarcinoma cell lines [50,51,52,53,54,55,56,57].
The results of the virtual screening, encompassing all the mentioned proteins, indicated that the majority of the synthesized cytotoxic compounds exhibit a stronger binding affinity for kinase proteins. As shown in Table S1, when considering only kinase proteins (Table 2), it is evident that most of these compounds bind more strongly to c-MET, a receptor tyrosine kinase, with ΔGbin values ranging from −10.6 to −9.7 (with an average of −10.08) kcal/mol. This is followed by TRKA, with binding energies ranging from −11.0 to −8.9 (with an average of −9.97) kcal/mol. Interestingly, despite the higher cytotoxicity of CBHQ hybrids 5e and 5f, naphthoquinone derivatives 6a–c and 6e demonstrated a greater affinity for most of the studied proteins, with ΔGbin values surpassing those of the diacetylated benzohydroquinone derivative CBHQs. Among them, compound 6c displayed the most favorable ΔGbin values for several kinase proteins, including TRKA (−11.0 kcal/mol), c-MET (−10.6 kcal/mol), and TPK (−10.4 kcal/mol), as detailed in Table 2. These findings underscore the significance of the naphthoquinone planar system in CNQ derivatives 6, both in their affinity for cancer-related proteins and their cytotoxicity. In contrast, monoacetylated or diacetylated derivatives 5 exhibited lower binding energies due to their reduced potential for strong hydrogen bond interactions with amino acid residues, as they protect one or both of the hydroquinone hydroxyl groups.

Table 2.
Comparison (ΔGbin, kcal/mol) of synthesized cytotoxic hybrids and kinase inhibitors approved by the FDA for cancer.
While CBHQ derivatives 4 did not exhibit cytotoxicity against the tested cancer cell lines (Table 1), they demonstrated favorable binding energies within the active sites of kinase proteins, as presented in Table S2. This observation can be attributed to robust hydrogen bond interactions between the free hydroxyl groups of the hydroquinone system and oxygen- or nitrogen-containing groups within the proteins, as illustrated in Figures S47–S49.
Despite previous studies suggesting chalcone derivatives as tubulin polymerization inhibitors, the evaluated compounds did not consistently yield the best ΔGbin average values in comparison to those of other proteins, including c-MET and TRKA, among others (Table S1). Furthermore, it is worth noting that most of the compounds exhibited superior ΔGbin values compared to reference antiproliferative drugs such as erlotinib, larotrectinib, almonertinib, and anlotinib, all of which function as kinase inhibitors. Erlotinib, almonertinib, and larotrectininb are utilized for treating non-small cell lung cancer (NSCLC), while anlotinib is employed for various cancers, including NSCLC and different sarcoma types [58,59,60].
2.4. Binding Site and Docking of Synthesized Cytotoxic Hybrids in c-MET, TRKA, and HER2 Targets
As previously mentioned, the virtual screening results indicated that the majority of the cytotoxic hybrids exhibit a high affinity for target proteins, with an average ΔGbin of less than −8.6 kcal/mol. In general, these compounds displayed stronger binding to the c-MET receptor (with an average of −10.08 kcal/mol), followed by TRKA (with an average of −8.96 kcal/mol) and HER2 (with an average of −9.75 kcal/mol). These findings suggest that these hybrids might serve as potent inhibitors of c-MET, TRKA, and HER2, all of which are overexpressed in certain types of cancer, including human breast and colorectal cancer [55,61,62,63]. Thus, these synthesized hybrids hold promise for treating diseases driven by these enzymes and could be effective against proliferative disorders.
Detailed configurations of the binding sites, along with the amino acids involved in the docking of synthesized cytotoxic hybrids and their corresponding ΔGbin values for c-MET, TRKA, and HER2, are presented in Table 3 and depicted in 2D maps in Figure 3. Additionally, 3D docking complexes of c-MET with 6a, 6b, and 6c are illustrated in Figure 4. Complementary 2D maps for complexes involving 5e and 5f can be found in Figure S1, and binding site interactions of the synthesized cytotoxic hybrids with amino acids of MEK-1, TPK, and EGFR are outlined in Table S3.

Table 3.
Predicted binding free energy values (ΔGbin, kcal/mol) and binding site contacts of synthesized cytotoxic hybrids with amino acids of c-MET, TRKA, and HER2.
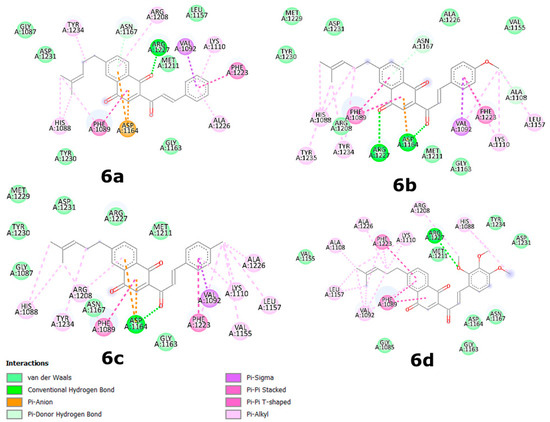
Figure 3.
Plotted 2D maps of H-bonds and hydrophobic interactions of CNQ 6a, 6b, 6c, and 6d with c-MET residues. Van der Waals, Pi–Anion, Pi–Sigma, Pi–Pi stacked, and Pi–alkyl are considered hydrophobic interactions.
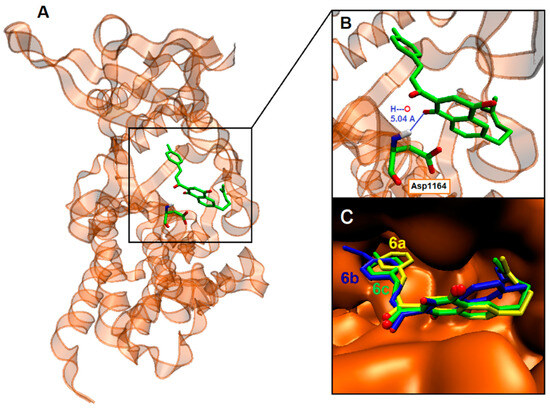
Figure 4.
(A): Visualization of the potential binding site of the CNQ hybrid 6c into c-MET; (B): Detail of its H-bonding with Asp1164; (C): Superimposition of the docking poses for CNQ hybrids 6a (yellow), 6b (blue), and 6c (green).
Overall, as demonstrated in Table 2 and Table 3, CNQ derivatives 6 exhibited superior binding affinities for kinase proteins due to the presence of the C1 and C4 carbonyl groups within the quinone ring. These groups interacted with amino acid residues through hydrogen bonding. For instance, carbonyl groups from the quinone ring in derivatives 6a and 6b interacted with Arg1127 of c-MET (Figure 3). Additionally, these interactions were favored due to the greater planarity of naphthoquinone structures compared to that of benzohydroquinone structures. Specifically, CNQs 6a, 6b, and 6c displayed excellent binding affinities for c-MET, with ΔGbin values of −10.3, −10.3, and −10.6 kcal/mol, respectively. Peptide sequences surrounding the CNQs revealed consistent docking in the same region of the enzyme, defined by the residues Arg1208 and Asp1231. All the compounds, including 5e and 5f, engaged in hydrogen bonding with c-MET residues, as well as various other interactions, including Van der Waals, Pi–Anion, Pi–Sigma, Pi–Pi stacked, and Pi–alkyl interactions.
Regarding hydrogen bonds, the residues Arg1208 and Asp1231 were most commonly involved in interactions with c-MET, serving as hydrogen bond donors toward carbonyl groups from the quinone moiety of 6a and 6b, the chalcone moiety of 6c and 6e, or the methoxy group of 5e and 5f (Figure 3 and Figure S1). In the case of TRKA, only the residues Ser672 and Arg673 interacted with carbonyl groups from the quinone and chalcone moieties of 6e, while Met592 interacted with the carbonyl group from the methoxy group of 5f through hydrogen bonds (Figure S2). HER2 exhibited interactions with residues such as Thr798, which primarily engaged in hydrogen bonds with carbonyl groups from the quinone and chalcone moieties of 6a, 6b, and 6e, as well as Ser783, which interacted with the carbonyl groups from the chalcone moiety of 6a and 6e and the methoxy group of 5e (Figure S3).
Aromatic interactions, similar to hydrogen bonds, play a crucial role in ligand–protein interfaces. Many contemporary ligand docking programs implicitly account for aromatic stacking through van der Waals and Coulombic potentials [64]. Residues Phe1089 and Phe1223 were notably involved in these interactions, engaging in π–π stacking with the aromatic rings of naphthoquinone systems in 6a–c/6d and the chalcone system in 5e and 5f. Additionally, the aromatic ring of Phe1223 interacted with the aromatic rings of the chalcone moiety in 6a–c through π–π stacking and with carbons of the hydroquinone system in 5e and 5f through π–alkyl interactions (Figure 3 and Figure S1).
In the case of TRKA, Phe669 was the primary residue involved in aromatic interactions, participating in π–π stacking with the aromatic rings of the chalcone moiety in 5e and 5f and the naphthoquinone moiety in 6a–c and 6e. Val524 interacted through π–sigma interactions with the aromatic rings of the quinone moiety in 5f, 6a, 6b, and 6c (Figure S2). Lastly, Phe864 played a prominent role in HER2 interactions, engaging in π–π stacking with the aromatic rings of the chalcone moiety in 5f and the naphthoquinone moiety in 6b, 6c, and 6e. Leu852 also contributed through π–sigma interactions with the aromatic rings of the chalcone moiety in 5e, 6b, 6c, and 6e, as well as the naphthoquinone moiety in 6a (Figure S3).
To validate the binding sites of the synthesized cytotoxic hybrids within the kinases, we conducted a comparative analysis of CNQ 6c complexes with those of known kinase ligands. The results revealed that the binding regions of CNQ 6c indeed overlap with the catalytic sites of the target enzymes, sharing a common set of contacts with the respective inhibitors (Table 4). Notably, the active site residues involved in these interactions include Phe1089, Val1092, Lys1110, Leu1157, Gly1163, Met1211, Ala1226, and Arg1227 for c-MET, Val524, Ala542, Kys544, Glu560, Val573, Phe589, Leu657, Gly667, Asp668, and Phe669 for TRKA, and Leu726, Val734, Ala751, Kys753, Leu785, Leu796, Thr798, Gln799, Leu800, Met801, Gly804, Leu852, Thr862, Asp863, and Phe864 for HER2. These residues served as common contact points for CNQ 6c and ligands 1, 2, and 3 in all three enzymes, respectively.

Table 4.
Binding site contacts of compound 6c, ligand, and drug into c-MET, TRKA, and HER2.
Of particular interest is the observation that the energetic aspects of these interactions favored CNQ 6c in comparison to erlotinib, with a favorable energy difference of 1.5 kcal/mol for c-MET and 2.2 kcal/mol for HER2. Moreover, 6c exhibited the same in silico affinity as larotrectinib for TRKA, both achieving a ΔGbin value of −11.0 kcal/mol. Importantly, the aromatic ring within the chalcone moiety of CNQs plays a pivotal role in these interactions, directly contributing to the overlap with the ligands at the catalytic sites of the enzymes (Figure 5). This crucial involvement of the chalcone moiety is consistently observed in the case of TRKA and HER2 as well (Figures S50 and S51).
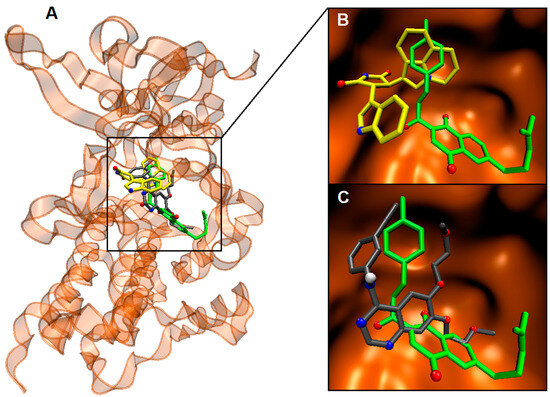
Figure 5.
(A): Overlapping of the docking poses for CNQ hybrid 6c (green), ligand 1 (yellow), and erlotinib (grey) into c-MET. Superimposition of the docking poses for (B): 6c and ligand 1 as well as (C): 6c and erlotinib.
To strengthen our research, it is important to identify and analyze the correlations between our calculated properties and experimental results. In this regard, we examined the correlation between experimental cytotoxicity (pIC50) and the hydrophobicity index (cLogP, as detailed in Table S2). Figure 6 illustrates the positive correlation between pIC50 values and the predicted cLogP values. Notably, the results indicate a stronger correlation between the pIC50 and cLogP values obtained in MCF-7 cell lines (R = 0.95) compared to those in HT-29 (R = 0.84).
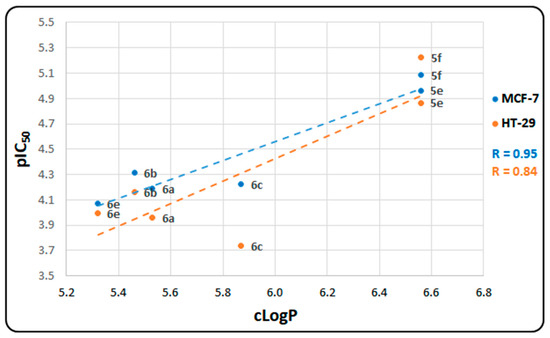
Figure 6.
Relationship between pIC50 and cLogP values for synthesized cytotoxic hybrids.
Of particular interest, CBHQ derivatives 5e and 5f exhibit higher pIC50 values for both the MCF-7 and HT-29 cell lines. This observation aligns with their greater ability to traverse the cell membrane, as evidenced by their higher cLogP values compared to those of the other synthesized cytotoxic hybrids. However, despite their superior pIC50 values in both cell lines, 5e and 5f display lower binding affinities for the evaluated proteins, including c-MET, TRKA, and HER2, compared to the rest of the cytotoxic hybrids, including 6a–c and 6e.
Based on these findings, the compounds with the best cytotoxicity values tend to be less polar and possess lower cLogP values (as indicated in Table S1). These characteristics correspond to the diacetylated CBHQs and CNQs, which also exhibit lower cLogP values and feature a planar bicyclic system due to the aromatization of the fused cycle with the quinone system. These factors enhance their ability to permeate the cell membrane.
Furthermore, it is possible to speculate that the CBHQs 5e and 5f may act as prodrugs. They could undergo hydrolysis through deacetylation within the cell, catalyzed by a “deacetylase” enzyme, releasing the molecules in the form of the CBHQs 4e and 4f. Subsequently, these benzohydroquinone compounds might exhibit an inhibitory effect on cancer-related kinases. This assumption is supported by their favorable binding energies in the active site of the kinase domain of EGFR, as detailed in Table S2.
2.5. In Silico Drug-Likeness, Toxicity Risks, and ADME Predictions
The drug-likeness scores for compounds 5e, 5f, and 6a–c,e were computed using the DataWarrior algorithm, and the results are presented in Table 5. Notably, derivative 5f stands out as the only compound with a positive drug-likeness value of 2.15. This significant finding suggests that compound 5f could be a promising lead candidate for further investigation. It is noteworthy that 5f incorporates essential structural elements, such as the hydroquinone and chalcone fragments, which are commonly found in approved drugs. Additionally, both 5f and 5e feature hydroquinone fragments with acetylation at positions 1 and 4. These substituents are known to contribute significantly to the enhancement of the antineoplastic cytotoxicity of potential anticancer agents.

Table 5.
Comparative toxicity risks a predicted and drug-likeness scores a for compounds 5e,f, and 6a–c,e.
In terms of toxicity risks, compounds 5e and 5f are likely to exhibit a high level of irritant risk, whereas compounds 6a, 6c, and 6e are expected to have no adverse effects, except for 6b, which may present a high irritant risk and low effects on the reproductive system (Table 5). The high irritant risk associated with 5e and 5f can be attributed to the acetylation in the hydroquinone moiety, while for 6b, it is likely due to the presence of a methoxy group in the naphthoquinone moiety.
The predicted values for several pharmacokinetic parameters of compounds 5e and 5f as well as 6a, 6b, 6c, and 6e related to oral absorption, Caco-2 cell permeability, blood–brain barrier permeability, and binding to human serum albumin, among others, are summarized in Table S4. These ADME descriptor values indicate that the percentage of predicted oral absorption for these compounds ranges from 84% to 100%, suggesting good oral bioavailability. Furthermore, all the evaluated compounds demonstrate good to excellent predicted values for Caco-2 cell permeability, with QPlogBB values falling between −1.44 and −0.98. Additionally, all the tested compounds are within the range of interaction with human serum albumin, suggesting their potential transport by plasma proteins to the target site.
However, it is worth noting that all compounds may block HERG K+-channels, which play a crucial role in cardiac repolarization, potentially increasing the risk of cardiac arrhythmias. Moreover, some compounds, including 6a, 6b, and 6e, are expected to have sufficient to excellent solubility in water, while 5e, 5f, and 6c are considered higher-lipophilicity compounds, enhancing their ability to penetrate cell membranes. In terms of compliance with Jorgensen’s rule of three, practically all the CNQs exhibit 1 or 2 violations, all of which remain within permissible limits.
Moreover, nearly all the evaluated compounds meet Lipinski’s rule of five and its Weber extension criteria, except for 5e and 5f (mol_MW > 500 amu and QPlog/Po/w > 5). However, even these two compounds have violations that fall within acceptable limits (Table S5). These results collectively suggest that, from a pharmacokinetic perspective, most of these compounds hold promise as potential candidates for preclinical assays.
3. Materials and Methods
3.1. Chemistry
All chemical reactions were carried out using commercially available solvents and reagent grade chemicals without further purification. The initial substrate, 2-acetyl-6-(4-methyl-3-pentenyl)-5,8-dihydro-1,4-naphthohydroquinone (designated as compound 1), was synthesized according to the method we previously described [41]. To record the IR and NMR spectra and carry out the elemental analyses of C, H, and N of the synthesized compounds, the experimental conditions that we previously reported were used [38].
3.1.1. Procedure for the Synthesis and Molecular Characterization of Precursor 2
Synthesis of 1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}ethanone (2)
First, 3,4-dihydro-2H-pyran (5.00 mmol) and 1.14 mmol of pyridinium p-toluenesulfonate were added to a solution of 2-acetyl-5,8-dihydro-6-(4-methyl-3-pentenyl)-1,4-naphthohydroquinone 1 (1.00 mmol) in CH2Cl2 (10 mL). The reaction mixture was stirred for 19 h at rt. Then, the reaction mixture was washed with distilled H2O (2 × 10 mL); after separating the phases, the organic phase was dried with anhydrous Na2SO4, filtered, and evaporated under reduced pressure. The crude product was purified by recrystallization using hexane as solvent. White solid (333 mg, 80%), m.p. 82–84 °C. IR υmax cm−1 (film) 3407 (O-H), 1632 (C=O), and 1033 (C-O). 1H NMR (CDCl3, TMS, ppm) δ 1.66 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 1.91 (m, 2H, CH2, H2′), 2.17 (m, 4H, 2CH2, H10, H9), 2.58 (s, 3H, CH3, H16), 3.29 (m, 4H, 2CH2, H8, H5), 3.64 (m, 1H, CH2, H5′), 3.90 (m, 1H, CH2, H5′), 5.16 (t, 1H, J = 6.7 Hz, CH, H11), 5.33 (t, 1H, J = 3.2 Hz, CH, H1′), 5.64 (s, 1H, CH, H7), 7.28 (s, 1H, CH, H3), and 12.45 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 19.0 (C3′), 24.6 (C8), 25.3 (C4′), 25.7 (C14), 26.1 (C10), 26.6 (C16), 28.4 (C5), 30.7 (C2′), 37.2 (C9), 62.0 (C5′), 97.1 (C1′), 111.6 (C3), 116.0 (C2), 117.6 (C11), 124.1 (C7), 124.8 (C8a), 131.8 (C4a), 133.4 (C12), 135.2 (C6), 146.3 (C4), 155.5 (C1), and 204.0 (C15). Elemental analysis calculated for C23H30O4: C, 74.56; H, 8.16. Found: C, 74.51; H, 8.22.
3.1.2. General Procedure for the Synthesis and Characterization of Compounds 3a–f
A solution containing the precursor 2 (1.00 mmol) and 1.00 mmol of barium hydroxide octahydrate in ethanol (8 mL) was maintained by constantly stirring it for 10 min. Then, the equivalent of 1.10 mmoles of the respective benzaldehyde was added and maintaining by stirring it for 25 min at 90 °C. After the end of the reaction time, the mixture was added to an ice/water bath and then vacuum-filtered, obtaining the respective impure products 3a–f.
Synthesis of (E)-1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}-3-phenylprop-2-en-1-one (3a)
This compound was synthesized by the general procedure, using precursor 2 and benzaldehyde, and purified by recrystallization using methanol as solvent. Yellow solid (275 mg, 60%), m.p. 139–140 °C. IR υmax cm−1 (film) 3406 (O-H), 1637 (C=O), 1574 (C=C), and 1033 (C-O). 1H NMR (CDCl3, TMS, ppm) δ 1.70 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 1.94 (m, 2H, CH2, H2′), 2.19 (m, 4H, 2CH2, H10, H9), 3.33 (m, 4H, 2CH2, H8, H5), 3.68 (m, 1H, CH2, H5′), 3.95 (m, 1H, CH2, H5′), 5.19 (t, 1H, J = 6.8 Hz, CH, H11), 5.42 (t, 1H, J = 3.2 Hz, CH, H1′), 5.68 (s, 1H, CH, H7), 7.48 (m, 4H, 4CH, H3, H20, H21, H22), 7.61 (d, 1H, J = 15.7 Hz, CH, H16), 7.69 (d, 2H, J = 8.6 Hz, 2CH, H19, H23), 7.92 (d, 1H, J = 15.7 Hz, CH, H17), and 13.15 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 19.0 (C3′), 24.8 (C8), 25.3 (C4′), 25.7 (C14), 26.2 (C10), 28.4 (C5), 30.7 (C2′), 37.2 (C9), 62.0 (C5′), 97.2 (C1′), 110.7 (C3), 116.3 (C2), 117.7 (C11), 120.6 (C16), 124.1 (C7), 125.0 (C8a), 128.6 (C19, C23), 129.0 (C20, C22), 130.7 (C21), 131.8 (C4a), 133.4 (C12), 134.8 (C6), 135.5 (C18), 144.7 (C17), 146.4 (C4), 156.9 (C1), and 193.2 (C15). Elemental analysis calculated for C30H34O4: C, 78.57; H, 7.47. Found: C, 78.61; H, 7.43.
Synthesis of (E)-1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}-3-(4-methoxyphenyl)prop-2-en-1-one (3b)
This compound was synthesized by the general procedure, using precursor 2 and 4-methoxybenzaldehyde, and purified by recrystallization using ethanol as solvent. White solid (322 mg, 66%), m.p. 134–136 °C. IR υmax cm−1 (film) 3428 (O-H), 1632 (C=O), 1605 (C=C), and 1172 (C-O). 1H NMR (CDCl3, TMS, ppm) δ 1.70 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 1.94 (m, 2H, CH2, H2′), 2.20 (m, 4H, 2CH2, H10, H9), 3.34 (m, 4H, 2CH2, H8, H5), 3.68 (m, 1H, CH2, H5′), 3.88 (s, 3H, CH3, H24), 3.95 (m, 1H, CH2, H5′), 5.19 (t, 1H, J = 6.7 Hz, CH, H11), 5.42 (t, 1H, J = 3.1 Hz, CH, H1′), 5.68 (s, 1H, CH, H7), 6.97 (d, 2H, J = 8.6 Hz, 2CH, H20, H22), 7.48 (m, 2H, 2CH, H3, H16), 7.64 (d, 2H, J = 8.6 Hz, 2CH, H19, H23), 7.90 (d, 1H, J = 15.5 Hz, CH, H17), and 13.26 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.8 (C13), 19.0 (C3′), 24.8 (C8), 25.3 (C4′), 25.7 (C14), 26.2 (C10), 28.4 (C5), 30.7 (C2′), 37.2 (C9), 55.5 (C24), 62.0 (C5′), 97.1 (C1′), 110.7 (C3), 114.5 (C20, C22), 116.4 (C2), 117.7 (C11), 118.1 (C16), 124.1 (C7), 124.9 (C8a), 127.6 (C18), 130.5 (C19, C23), 131.8 (C4a), 133.4 (C12), 135.1 (C6), 144.6 (C17), 146.3 (C4), 156.9 (C1), 161.8 (C21), and 193.2 (C15). Elemental analysis calculated for C31H36O5: C, 76.20; H, 7.43. Found: C, 76.26; H, 7.39.
Synthesis of (E)-1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}-3-(4-methylphenyl)prop-2-en-1-one (3c)
This compound was synthesized by the general procedure, using precursor 2 and 4-methybenzaldehyde, and purified by recrystallization using ethanol as solvent. Orange solid (307 mg, 65%), m.p. 132–133 °C. IR υmax cm−1 (film) 3492 (O-H), 1634 (C=O), and 1584 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.69 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 1.94 (m, 2H, CH2, H2′), 2.19 (m, 4H, 2CH2, H10, H9), 2.42 (s, 3H, CH3, H24), 3.35 (m, 4H, 2CH2, H8, H5), 3.69 (m, 1H, CH2, H5′), 3.96 (td, 1H, J = 9.6 Hz, J = 3.3 Hz, CH2, H5′), 5.19 (t, 1H, J = 6.4 Hz, CH, H11), 5.41 (t, 1H, J = 3.1 Hz, CH, H1′), 5.67 (s, 1H, CH, H7), 7.25 (d, 2H, J = 8.1 Hz, 2CH, H20, H22), 7.49 (s, 1H, 1CH, H3), 7.55 (m, 3H, 3CH, H16, H19, H23), 7.90 (d, 1H, J = 15.7 Hz, CH, H17), and 13.16 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 19.0 (C24), 21.6 (C3′), 24.8 (C8), 25.3 (C4′), 25.7 (C14), 26.2 (C10), 28.4 (C5), 30.7 (C2′), 37.2 (C9), 62.0 (C5′), 97.2 (C1′), 110.7 (C3), 116.3 (C2), 117.7 (C11), 119.5 (C16), 124.2 (C7), 125.0 (C8a), 128.7 (C19, C23), 129.7 (C20, C22), 131.7 (C4a), 132.1 (C18), 133.5 (C12), 135.3 (C6), 141.3 (C21), 144.8 (C17), 146.3 (C4), 156.9 (C1), and 193.3 (C15). Elemental analysis calculated for C31H36O4: C, 78.78; H, 7.68. Found: C, 78.74; H, 7.72.
Synthesis of (E)-3-(2,4-Dichlorophenyl)1-{1-hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}prop-2-en-1-one (3d)
This compound was synthesized by the general procedure, using precursor 2 and 2,4-dichlorobenzaldehyde, and purified by recrystallization using methanol as solvent. Yellow solid (369 mg, 70%), m.p. 134–136 °C. IR υmax cm−1 (film) 3442 (O-H), 1634 (C=O), and 1581 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.70 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 1.94 (m, 2H, CH2, H2′), 2.20 (m, 4H, 2CH2, H10, H9), 3.34 (m, 4H, 2CH2, H8, H5), 3.66 (m, 1H, CH2, H5′), 3.92 (m, 1H, CH2, H5′), 5.19 (t, 1H, J = 6.6 Hz, CH, H11), 5.41 (t, 1H, J = 3.1 Hz, CH, H1′), 5.67 (s, 1H, CH, H7), 7.32 (dd, 1H, J = 8.4 Hz, J = 2.1 Hz, CH, H22), 7.44 (s, 1H, 1CH, H3), 7.49 (d, 1H, J = 2.1 Hz, CH, H20), 7.55 (d, 1H, J = 15.7 Hz, CH, H16), 7.69 (d, 1H, J = 8.4 Hz, CH, H23), 8.20 (d, 1H, J = 15.7 Hz, CH, H17), and 12.97 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.8 (C13), 18.9 (C3′), 24.8 (C8), 25.3 (C4′), 25.8 (C14), 26.2 (C10), 28.5 (C5), 30.7 (C2′), 37.2 (C9), 62.0 (C5′), 97.0 (C1′), 110.5 (C3), 116.1 (C2), 117.6 (C11), 123.6 (C16), 124.1 (C7), 125.1 (C8a), 127.6 (C22), 128.7 (C20), 130.2 (C23), 131.8 (C4a), 131.8 (C18), 133.4 (C12), 135.8 (C19), 136.2 (C21), 136.6 (C6), 139.1 (C17), 146.4 (C4), 157.0 (C1), and 192.7 (C15). Elemental analysis calculated for C30H32Cl2O4: C, 68.31; H, 6.11. Found: C, 68.36; H, 6.08.
Synthesis of (E)-1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}-3-(2,3,4-trimethoxyphenyl)prop-2-en-1-one (3e)
This compound was synthesized by the general procedure, using precursor 2 and 2,3,4-trimethoxybenzaldehyde, and purified by recrystallization using ethanol as solvent. Red solid (357 mg, 65%), m.p. 97–99 °C. IR υmax cm−1 (film) 3415 (O-H), 1631 (C=O), 1564 (C=C), and 1107 (C-O). 1H NMR (CDCl3, TMS, ppm) δ 1.70 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 2.09 (m, 6H, 3CH2, H2′, H10, H9), 3.33 (m, 4H, 2CH2, H8, H5), 3.69 (m, 1H, CH2, H5′), 3.92 (m, 7H, 2CH3, CH2, H25, H26, H5′), 4.00 (s, 3H, CH3, H24), 5.19 (t, 1H, J = 6.7 Hz, CH, H11), 5.39 (t, 1H, J = 3.1 Hz, CH, H1′), 5.67 (s, 1H, CH, H7), 6.74 (d, 1H, J = 8.3 Hz, CH, H22), 7.39 (d, 1H, J = 8.3 Hz, CH, H23), 7.52 (s, 1H, CH, H3), 7.69 (d, 1H, J = 15.8 Hz, CH, H16), 8.07 (d, 1H, J = 15.8 Hz, CH, H17), and 13.23 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 19.0 (C3′), 24.8 (C8), 25.3 (C4′), 25.7 (C14), 26.2 (C10), 28.4 (C5), 30.7 (C2′), 37.2 (C9), 56.1 (C24), 61.0 (C25), 61.3 (C26), 62.0 (C5′), 97.3 (C1′), 107.6 (C22), 110.8 (C3), 116.5 (C2), 117.7 (C11), 119.9 (C16), 121.9 (C18), 124.2 (C8a), 124.7 (C7), 124.8 (C23), 131.7 (C4a), 133.5 (C12), 135.0 (C6), 140.2 (C17), 142.5 (C20), 146.4 (C4), 154.0 (C19), 156.0 (C21), 156.8 (C1), and 193.6 (C15). Elemental analysis calculated for C33H40O7: C, 72.24; H, 7.35. Found: C, 72.20; H, 7.37.
Synthesis of (E)-1-{1-Hydroxy-6-(4-methylpent-3-en-1-yl)-4-[(tetrahydro-2H-pyran-2-yl)oxy]-5,8-dihydronaphthalen-2-yl}-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one (3f)
This compound was synthesized by the general procedure, using precursor 2 and 3,4,5-trimethoxybenzaldehyde, and purified by recrystallization using ethanol as solvent. Orange solid (340 mg, 62%), m.p. 99–100 °C. IR υmax cm−1 (film) 3394 (O-H), 1631 (C=O), and 1563 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.68 (m, 10H, 2CH3, 2CH2, H13, H14, H3′, H4′), 2.05 (m, 6H, 3CH2, H2′, H10, H9), 3.34 (m, 4H, 2CH2, H8, H5), 3.66 (m, 1H, CH2, H5′), 3.82 (m, 1H, CH2, H5′), 3.94 (m, 9H, 3CH3, H24, H25, H26), 5.19 (t, 1H, J = 5.8 Hz, CH, H11), 5.34 (t, 1H, J = 3.3 Hz, CH, H1′), 5.67 (s, 1H, CH, H7), 6.88 (s, 2H, 2CH, H19, H23), 7.46 (d, 2H, J = 15.8 Hz, 2CH, H16, H3), 7.82 (d, 1H, J = 15.8 Hz, CH, H17), and 13.09 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 19.2 (C3′), 24.8 (C8), 25.3 (C4′), 25.7 (C14), 26.2 (C10), 28.5 (C5), 30.8 (C2′), 37.2 (C9), 56.3 (C24, C26), 61.0 (C25), 62.3 (C5′), 97.7 (C1′), 105.9 (C19, C23), 111.2 (C3), 116.3 (C2), 117.7 (C11), 120.1 (C16), 124.1 (C7), 125.0 (C8a), 130.4 (C18), 131.7 (C4a), 133.4 (C12), 135.5 (C6), 140.7 (C21), 144.8 (C4), 146.4 (C17), 153.5 (C20, C22), 156.9 (C1), and 193.1 (C15). Elemental analysis calculated for C33H40O7: C, 72.24; H, 7.35. Found: C, 72.21; H, 7.36.
General Procedure for the Synthesis and Characterization of Compounds 4a–f
Acid monohydrate 4-toluenesulfonic (0.80 mmol) was added to a solution of the respective compound 3a–f (1.00 mmol) in methanol (10 mL). The reaction mixture was stirred for 3 h at rt. After the end of the reaction time, the mixture was added to an ice/water bath and then vacuum-filtered, obtaining the respective impure products 4a–f.
Synthesis of (E)-1-[1,4-Dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]-3-phenylprop-2-en-1-one (4a)
This compound was synthesized by the general procedure using precursor 3a and purified by recrystallization using methanol as solvent. Orange solid (348 mg, 93%), m.p. 141–143 °C. IR υmax cm−1 (film) 3403 (O-H), 1662 (C=O), and 1633 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.63 (s, 3H, CH3, H13), 1.68 (s, 3H, CH3, H14), 2.20 (m, 4H, 2CH2, H10, H9), 3.30 (s, 4H, 2CH2, H8, H5), 5.19 (t, 1H, J = 6.7 Hz, CH, H11), 5.68 (s, 1H, CH, H7), 7.48 (m, 4H, 4CH, H3, H20, H21, H22), 7.85 (m, 4H, 4CH, H16, H17, H19, H23), 8.01 (s, 1H, OH, H4), and 12.95 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 16.9 (C13), 24.5 (C8), 25.7 (C14), 26.0 (C10), 28.0 (C5), 37.0 (C9), 110.6 (C3), 116.4 (C2), 117.6 (C11), 120.9 (C16), 124.0 (C7), 124.2 (C8a), 128.7 (C19, C23), 129.0 (C20, C22), 130.8 (C21), 131.2 (C4a), 133.2 (C12), 133.5 (C6), 135.0 (C18), 144.3 (C17), 146.3 (C4), 155.3 (C1), and 193.3 (C15). Elemental analysis calculated for C25H26O3: C, 80.18; H, 7.00. Found: C, 80.25; H, 6.96.
Synthesis of (E)-1-[1,4-Dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]-3-(4-methoxyphenyl)prop-2-en-1-one (4b)
This compound was synthesized by the general procedure using precursor 3b and purified by recrystallization using methanol as solvent. Orange solid (388 mg, 96%), m.p. 156–158 °C. IR υmax cm−1 (film) 3353 (O-H), 1631 (C=O), and 1605 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.63 (s, 3H, CH3, H13), 1.68 (s, 3H, CH3, H14), 2.19 (m, 4H, 2CH2, H10, H9), 3.29 (m, 4H, 2CH2, H8, H5), 3.88 (s, 3H, CH3, H24), 5.18 (t, 1H, J = 6.8 Hz, CH, H11), 5.67 (s, 1H, CH, H7), 7.05 (d, 2H, J = 8.6 Hz, 2CH, H20, H22), 7.44 (s, 1H, CH, H3), 7.67 (d, 1H, J = 15.7 Hz, CH, H16), 7.80 (d, 2H, J = 8.6 Hz, 2CH, H19, H23), 7.87 (d, 1H, J = 15.7 Hz, CH, H17), and 7.98 (s, 1H, OH, H4), 13.07 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 16.9 (C13), 24.5 (C8), 25.0 (C14), 26.0 (C10), 28.0 (C5), 37.0 (C9), 55.0 (C24), 110.5 (C3), 114.5 (C20, C22), 116.4 (C2), 117.6 (C11), 118.2 (C16), 124.1 (C7), 124.1 (C8a), 127.5 (C18), 130.6 (C19, C23), 131.2 (C4a), 132.8 (C12), 133.5 (C6), 144.4 (C17), 146.2 (C4), 155.2 (C1), 162.1 (C21), and 193.2 (C15). Elemental analysis calculated for C26H28O4: C, 77.20; H, 6.98. Found: C, 77.18; H, 7.03.
Synthesis of (E)-1-[1,4-Dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]-3-(4-methylphenyl)prop-2-en-1-one (4c)
This compound was synthesized by the general procedure using precursor 3c and purified by recrystallization using methanol as solvent. Orange solid (361 mg, 93%), m.p. 185–186 °C. IR υmax cm−1 (film) 3272 (O-H), 1630 (C=O), and 1558 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.62 (s, 3H, CH3, H13), 1.67 (s, 3H, CH3, H14), 2.18 (m, 4H, 2CH2, H10, H9), 2.38 (s, 3H, CH3, H24), 3.28 (s, 4H, 2CH2, H8, H5), 5.17 (t, 1H, J = 6.2 Hz, CH, H11), 5.66 (s, 1H, CH, H7), 7.28 (d, 2H, J = 7.7 Hz, 2CH, H20, H22), 7.41 (s, 1H, CH, H3), 7.68 (m, 3H, 3CH, H16, H19, H23), 7.85 (d, 1H, J = 15.3 Hz, CH, H17), 7.96 (s, 1H, OH, H4), and 12.96 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 17.1 (C13), 20.8 (C24), 24.5 (C8), 25.1 (C14), 26.0 (C10), 28.1 (C5), 37.1 (C9), 110.4 (C3), 116.4 (C2), 117.6 (C11), 119.7 (C16), 124.1 (C7), 124.2 (C8a), 128.7 (C19, C23), 129.7 (C20, C22), 131.2 (C4a), 132.2 (C18), 133.0 (C12), 133.5 (C6), 141.2 (C21), 144.4 (C17), 146.3 (C4), 155.3 (C1), and 193.1 (C15). Elemental analysis calculated for C26H28O3: C, 80.38; H, 7.26. Found: C, 80.43; H, 7.20.
Synthesis of (E)-3-(2,4-Dichlorophenyl)-1-[1,4-dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]prop-2-en-1-one (4d)
This compound was synthesized by the general procedure using precursor 3d and purified by recrystallization using methanol as solvent. Yellow solid (421 mg, 95%), m.p. 155–157 °C. IR υmax cm−1 (film) 3390 (O-H), 1634 (C=O), and 1581 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.63 (s, 3H, CH3, H13), 1.68 (s, 3H, CH3, H14), 2.19 (m, 4H, 2CH2, H10, H9), 2.89 (s, 4H, 2CH2, H8, H5), 5.19 (t, 1H, J = 6.3 Hz, CH, H11), 5.67 (s, 1H, CH, H7), 7.49 (m, 2H, 2CH, H22, H3), 7.63 (d, 1H, J = 2.1 Hz, CH, H20), 7.88 (d, 1H, J = 15.8 Hz, CH, H16), 8.12 (m, 3H, 2CH, H23, H17, OH, H4), and 12.78 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 16.9 (C13), 24.5 (C8), 24.9 (C14), 26.0 (C10), 28.1 (C5), 37.0 (C9), 110.7 (C3), 116.3 (C2), 117.6 (C11), 124.1 (C16), 124.3 (C8a), 124.3 (C7), 127.9 (C22), 129.5 (C20), 129.7 (C23), 131.2 (C4a), 131.8 (C18), 133.5 (C12), 133.8 (C19), 135.6 (C21), 136.2 (C6), 137.9 (C17), 146.4 (C4), 155.4 (C1), and 192.7 (C15). Elemental analysis calculated for C25H24Cl2O3: C, 67.73; H, 5.46. Found: C, 67.79; H, 5.51.
Synthesis of (E)-1-[1,4-Dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]-3-(2,3,4-trimethoxyphenyl)prop-2-en-1-one (4e)
This compound was synthesized by the general procedure using precursor 3e and purified by recrystallization using methanol as solvent. Red solid (441 mg, 95%), m.p. 149–151 °C. IR υmax cm−1 (film) 3434 (O-H), 1633 (C=O), and 1579 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.63 (s, 3H, CH3, H13), 1.68 (s, 3H, CH3, H14), 2.20 (m, 4H, 2CH2, H10, H9), 3.29 (s, 4H, 2CH2, H8, H5), 3.85 (s, 3H, CH3, H25), 3.93 (s, 3H, CH3, H26), 3.98 (s, 3H, CH3, H24), 5.19 (t, 1H, J = 6.8 Hz, CH, H11), 5.68 (s, 1H, CH, H7), 6.92 (d, 1H, J = 8.9 Hz, CH, H22), 7.40 (s, 1H, CH, H3), 7.59 (d, 1H, J = 8.9 Hz, CH, H23), 7.79 (d, 1H, J = 16.0 Hz, CH, H16), 8.02 (s, 1H, OH, H4), 8.07 (d, 1H, J = 16.0 Hz, CH, H17), and 13.08 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 16.9 (C13), 24.5 (C8), 24.9 (C14), 26.0 (C10), 28.0 (C5), 37.0 (C9), 55.6 (C24), 60.1 (C25), 60.9 (C26), 108.2 (C22), 110.2 (C3), 116.5 (C2), 117.7 (C11), 119.4 (C16), 121.4 (C18), 124.1 (C23), 124.2 (C8a), 124.4 (C7), 131.1 (C4a), 132.8 (C12), 133.6 (C6), 139.6 (C17), 142.6 (C20), 146.3 (C4), 153.9 (C19), 155.2 (C21), 156.5 (C1), and 193.4 (C15). Elemental analysis calculated for C28H32O6: C, 72.39; H, 6.94. Found: C, 72.43; H, 6.90.
Synthesis of (E)-1-[1,4-Dihydroxy-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-2-yl]-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one (4f)
This compound was synthesized by the general procedure using precursor 3f and purified by recrystallization using methanol as solvent. Orange solid (432 mg, 93%), m.p. 168–169 °C. IR υmax cm−1 (film) 3433 (O-H), 1634 (C=O), and 1582 (C=C). 1H NMR (Acetone-d6, TMS, ppm) δ 1.66 (m, 6H, 2CH3, H13, H14), 2.22 (m, 4H, 2CH2, H10, H9), 3.30 (s, 4H, 2CH2, H8, H5), 3.81 (s, 3H, CH3, H25), 3.93 (s, 6H, 2CH3, H24, H26), 5.19 (t, 1H, J = 6.5 Hz, CH, H11), 5.68 (s, 1H, CH, H7), 7.19 (s, 2H, 2CH, H19, H23), 7.40 (s, 1H, CH, H3), 7.74 (d, 1H, J = 15.3 Hz, CH, H16), 7.84 (d, 1H, J = 15.3 Hz, CH, H17), 7.94 (s, 1H, OH, H4), and 13.03 (s, 1H, OH, H1). 13C NMR (Acetone-d6, TMS, ppm) δ 16.9 (C13), 24.5 (C8), 25.0 (C14), 26.0 (C10), 28.0 (C5), 37.0 (C9), 55.7 (C24, C26), 69.8 (C25), 106.5 (C19, C23), 110.7 (C3), 116.4 (C2), 117.6 (C11), 119.9 (C16), 124.1 (C7), 124.2 (C8a), 130.3 (C18), 131.2 (C4a), 133.1 (C12), 133.5 (C6), 141.0 (C21), 144.9 (C4), 146.2 (C17), 153.8 (C20, C22), 155.3 (C1), and 193.2 (C15). Elemental analysis calculated for C28H32O6: C, 72.39; H, 6.94. Found: C, 72.44; H, 6.92.
General Procedure for the Synthesis and Characterization of Compounds 5′b–d and 5e,f
A total of 0.50 mL (5,3 mmol) of acetic anhydride was added to a solution of the respective compound 4b–f (0.25 mmol) in pyridine (0.50 mL), and the reaction mixture was maintained in the dark with occasional stirring for 24 h at room temperature. After the end of the reaction time, the mixture was added to an ice/water bath. Subsequently, the mixture was dissolved with CH2Cl2 (40 mL), and successive extractions were performed with 10% HCl solution (2 × 20 mL) and with H2O (2 × 10 mL) until a neutral pH of the aqueous phase was attained. The organic phase was dried with anhydrous Na2SO4, filtered, and evaporated under reduced pressure. The crude product was purified by column chromatography with hexane/ethyl acetate as eluent in varying proportions.
Synthesis of (E)-4-Hydroxy-3-[3-(4-methoxyphenyl)prop-2-enoyl]-7-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-1-yl ethanoate (5′b)
This compound was synthesized by the general procedure using precursor 4b and purified by CC with hexane/ethyl acetate 2:1. Yellow solid (94 mg, 85%), m.p. 176–177 °C. IR υmax cm−1 (film) 3391 (O-H), 1758 (C=O), and 1572 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.68 (m, 6H, 2CH3, H13, H14), 2.17 (m, 4H, 2CH2, H10, H9), 2.39 (s, 3H, CH3, H2′), 3.15 (s, 2H, CH2, H8), 3.38 (s, 2H, CH2, H5), 3.88 (s, 3H, CH3, H24), 5.16 (t, 1H, J = 6.6 Hz, CH, H11), 5.67 (s, 1H, CH, H7), 6.96 (d, 2H, J = 8.6 Hz, 2CH, H20, H22), 7.43 (d, 2H, J = 15.7 Hz, 2CH, H16, H3), 7.64 (d, 2H, J = 8.6 Hz, 2CH, H19, H23), 7.90 (d, 1H, J = 15.6 Hz, CH, H17), and 13.32 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.8 (C13), 20.9 (C2′), 24.7 (C8), 25.7 (C14), 26.1 (C10), 28.2 (C5), 37.0 (C9), 55.4 (C24), 114.5 (C20, C22), 116.8 (C2), 117.6 (C3), 117.9 (C11), 118.9 (C16), 123.9 (C7), 125.7 (C8a), 127.4 (C18), 130.6 (C19, C23), 131.9 (C4a), 132.6 (C12), 136.4 (C6), 139.9 (C4), 145.3 (C17), 159.3 (C1), 162.0 (C21), 169.9 (C1′), and 192.8 (C15). Elemental analysis calculated for C28H30O5: C, 75.31; H, 6.77. Found: C, 75.26; H, 6.80.
Synthesis of (E)-4-Hydroxy-7-(4-methylpent-3-en-1-yl)-3-[3-(4-methylphenyl)prop-2-enoyl]-5,8-dihydronaphthalen-1-yl ethanoate (5′c)
This compound was synthesized by the general procedure using precursor 4c and purified by CC with hexane/ethyl acetate 2:1. Yellow solid (99 mg, 90%), m.p. 168–169 °C. IR υmax cm−1 (film) 3389 (O-H), 1757 (C=O), and 1583 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.64 (s, 3H, CH3, H13), 1.72 (s, 3H, CH3, H14), 2.19 (m, 4H, 2CH2, H10, H9), 2.41 (m, 6H, 2CH3, H24, H2′), 3.14 (s, 2H, CH2, H8), 3.39 (s, 2H, CH2, H5), 5.16 (t, 1H, J = 6.8 Hz, CH, H11), 5.68 (s, 1H, CH, H7), 7.26 (m, 2H, 2CH, H20, H22), 7.53 (m, 4H, 4CH, H3, H16, H19, H23), 7.92 (d, 1H, J = 15.5 Hz, CH, H17), and 13.26 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.8 (C13), 20.8 (C24), 21.6 (C2′), 24.7 (C8), 25.7 (C14), 26.1 (C10), 28.3 (C5), 37.0 (C9), 116.7 (C2), 117.9 (C3), 118.9 (C11), 119.0 (C16), 123.9 (C7), 125.8 (C8a), 128.8 (C19, C23), 129.8 (C20, C22), 131.9 (C4a), 131.9 (C18), 132.6 (C12), 136.6 (C6), 140.0 (C4), 141.6 (C21), 145.5 (C17), 159.3 (C1), 169.8 (C1′), and 193.0 (C15). Elemental analysis calculated for C28H30O4: C, 78.11; H, 7.02. Found: C, 78.17; H, 6.98.
Synthesis of (E)-3-[3-(2,4-Dichlorophenyl)prop-2-enoyl]-4-hydroxy-7-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-1-yl ethanoate (5′d)
This compound was synthesized by the general procedure using precursor 4d and purified by CC with hexane/ethyl acetate 2:1. Yellow solid (112 mg, 93%), m.p. 196–197 °C. IR υmax cm−1 (film) 3379 (O-H), 1755 (C=O), and 1585 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.68 (m, 6H, 2CH3, H13, H14), 2.17 (m, 4H, 2CH2, H10, H9), 2.38 (s, 3H, CH3, H2′), 3.13 (s, 2H, CH2, H8), 3.38 (s, 2H, CH2, H5), 5.15 (t, 1H, J = 6.4 Hz, CH, H11), 5.67 (s, 1H, CH, H7), 7.31 (d, 1H, J = 8.3 Hz, CH, H22), 7.42 (s, 1H, CH, H3), 7.49 (m, 2H, 2CH, H16, H20), 7.71 (d, 1H, J = 8.3 Hz, CH, H23), 8.23 (d, 1H, J = 15.5 Hz, CH, H17), and 13.05 (s, 1H, OH, H1). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 20.8 (C2′), 24.7 (C8), 25.7 (C14), 26.1 (C10), 28.3 (C5), 37.0 (C9), 116.5 (C2), 117.8 (C3), 118.9 (C11), 123.0 (C7), 123.9 (C16), 126.0 (C8a), 127.6 (C22), 128.6 (C20), 130.2 (C23), 131.5 (C4a), 132.0 (C18), 132.5 (C12), 136.4 (C19), 136.9 (C21), 137.2 (C6), 139.7 (C17), 140.0 (C4), 159.5 (C1), 169.8 (C1′), and 192.3 (C15). Elemental analysis calculated for C27H26Cl2O4: C, 66.81; H, 5.40. Found: C, 66.87; H, 5.35.
Synthesis of (E)-2-[3-(2,3,4-Trimethoxyphenyl)prop-2-enoyl]-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-1,4-diyl diethanoate (5e)
This compound was synthesized by the general procedure using precursor 4e and purified by CC with hexane/ethyl acetate 3:1. Orange solid (121 mg, 87%), m.p. 143–144 °C. IR υmax cm−1 (film) 1766 (C=O), and 1585 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.67 (m, 6H, 2CH3, H13, H14), 2.15 (m, 4H, 2CH2, H10, H9), 2.33 (m, 6H, 2CH3, H2′, H4′), 3.22 (m, 4H, 2CH2, H8, H5), 3.91 (m, 9H, 3CH3, H24, H25, H26), 5.14 (m, 1H, CH, H11), 5.59 (s, 1H, CH, H7), 6.71 (d, 1H, J = 8.7 Hz, CH, H22), 7.19 (d, 1H, J = 16.1 Hz, CH, H16), 7.34 (d, 2H, J = 8.7 Hz, CH, H3, H23), and 7.84 (d, 1H, J = 16.1 Hz, CH, H17). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 20.8 (C2′), 20.8 (C4′), 25.3 (C8), 25.7 (C14), 26.0 (C10), 27.9 (C5), 37.0 (C9), 56.1 (C26), 60.9 (C25), 61.5 (C24), 107.6 (C22), 116.9 (C3), 120.7 (C16), 121.7 (C18), 123.8 (C7), 123.9 (C11), 124.0 (C23), 130.3 (C4a), 130.4 (C2), 132.0 (C8a), 132.6 (C12), 133.3 (C20), 140.8 (C17), 142.4 (C6), 144.5 (C4), 145.7 (C1), 153.9 (C19), 156.0 (C21), 169.0 (C1′, C3′), and 190.5 (C15). Elemental analysis calculated for C32H36O8: C, 70.06; H, 6.61. Found: C, 70.11; H, 6.57.
Synthesis of (E)-2-[3-(3,4,5-Trimethoxyphenyl)prop-2-enoyl]-6-(4-methylpent-3-en-1-yl)-5,8-dihydronaphthalen-1,4-diyl diethanoate (5f)
This compound was synthesized by the general procedure using precursor 4f and purified by CC with hexane/ethyl acetate 3:1. Orange solid (115 mg, 85%), m.p. 142–143 °C. IR υmax cm−1 (film) 1766 (C=O), and 1580 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.64 (s, 3H, CH3, H13), 1.71 (s, 3H, CH3, H14), 2.16 (m, 4H, 2CH2, H10, H9), 2.29 (s, 3H, CH3, H2′), 2.37 (s, 3H, CH3, H4′), 3.18 (m, 2H, CH2, H5), 3.28 (m, 2H, CH2, H8), 3.91 (s, 9H, 3CH3, H24, H25, H26), 5.14 (m, 1H, CH, H11), 5.60 (s, 1H, CH, H7), 6.82 (s, 2H, 2CH, H19, H23), 7.02 (d, 1H, J = 16.0 Hz, CH, H16), 7.28 (s, 1H, CH, H3), and 7.53 (d, 1H, J = 16.0 Hz, CH, H17). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 20.8 (C2′), 20.9 (C4′), 25.3 (C8), 25.7 (C14), 26.0 (C10), 27.9 (C5), 36.9 (C9), 56.2 (C24, C26), 56.3 (C25), 105.7 (C19, C23), 116.4 (C3), 120.7 (C16), 123.8 (C7), 124.5 (C11), 130.0 (C18), 130.1 (C4a), 130.5 (C2), 132.0 (C8a), 132.8 (C12), 133.4 (C6), 140.6 (C21), 144.3 (C4), 145.6 (C1), 146.2 (C17), 153.8 (C20, C22), 169.0 (C1′, C3′), and 190.7 (C15). Elemental analysis calculated for C32H36O8: C, 70.06; H, 6.61. Found: C, 70.13; H, 6.56.
General Procedure for the Synthesis and Characterization of Compounds 6a–e
2,3-dichloro-5,6-dicyanobenzoquinone (1.05 mmol) was added to a solution of the respective compound 4a–e (0.50 mmol) in CH2Cl2 (15 mL). The reaction mixture was stirred for 30 min at rt. Then, the mixture was filtered over silica gel 230–400 mesh, and the organic solution was extracted with 5% NaHCO3 solution (2 × 10 mL) and H2O (1 × 20 mL). The organic phase was dried with anhydrous Na2SO4, filtered, and evaporated under reduced pressure, obtaining the respective impure products 6a-e.
Synthesis of (E)-6-(4-Methylpent-3-en-1-yl)-2-(3-phenylprop-2-enoyl)naphthalene-1,4-dione (6a)
This compound was synthesized by the general procedure using precursor 4a and purified by recrystallization using hexane as solvent. Yellow solid (74 mg, 40%), m.p. 134–137 °C. IR υmax cm−1 (film) 1654 (C=O), and 1618 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.55 (s, 3H, CH3, H13), 1.70 (s, 3H, CH3, H14), 2.38 (m, 2H, CH2, H10), 2.82 (t, 2H, J = 7.3 Hz, CH2, H9), 5.14 (m, 1H, CH, H11), 7.19 (m, 2H, H3, H16), 7.44 (m, 3H, 3CH, H20, H21, H22), 7.64 (m, 4H, 4CH, H17, H19, H23, H7), 7.94 (d, 1H, J = 1.9 Hz, CH, H5), and 8.07 (d, 1H, J = 7.9 Hz, CH, H8). 13C NMR (CDCl3, TMS, ppm) δ 17.8 (C13), 25.7 (C14), 29.3 (C10), 36.3 (C9), 122.5 (C11), 123.6 (C16), 125.3 (C5), 127.1 (C8), 128.9 (C19, C23), 129.1 (C20, C22), 129.8 (C8a), 131.3 (C21), 131.8 (C4a), 133.4 (C12), 134.7 (C18), 134.8 (C3), 136.9 (C7), 143.4 (C2), 146.5 (C17), 150.1 (C6), 183.4 (C4), 185.3 (C1), and 190.2 (C15). Elemental analysis calculated for C25H22O3: C, 81.06; H, 5.99. Found: C, 81.00; H, 6.03.
Synthesis of (E)-2-[3-(4-Methoxyphenyl)prop-2-enoyl]-6-(4-methylpent-3-en-1-yl) naphthalene-1,4-dione (6b)
This compound was synthesized by the general procedure using precursor 4b and purified by recrystallization using hexane as solvent. Orange solid (112 mg, 56%), m.p. 110–112 °C. IR υmax cm−1 (film) 1668 (C=O), and 1600 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.55 (s, 3H, CH3, H13), 1.70 (s, 3H, CH3, H14), 2.39 (c, 2H, J = 7.6 Hz, CH2, H10), 2.82 (t, 2H, J = 7.6 Hz, CH2, H9), 3.87 (s, 3H, CH3, H24), 5.15 (t, 1H, J = 7.5 Hz, CH, H11), 6.94 (d, 2H, J = 8.6 Hz, 2CH, H20, H22), 7.07 (d, 2H, J = 15.7 Hz, 2CH, H3, H16), 7.60 (m, 4H, 4CH, H17, H19, H23, H7), 7.94 (d, 1H, J = 1.8 Hz, CH, H5), and 8.07 (d, 1H, J = 7.9 Hz, CH, H8). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 25.7 (C14), 29.4 (C10), 36.3 (C9), 55.5 (C24), 114.6 (C20, C22), 122.5 (C11), 123.1 (C16), 126.2 (C5), 126.8 (C18), 127.0 (C8), 129.8 (C8a), 130.8 (C19, C23), 131.8 (C4a), 133.4 (C12), 134.8 (C3), 136.6 (C7), 146.3 (C2), 146.6 (C17), 150.0 (C6), 162.3 (C21), 183.4 (C4), 185.4 (C1), and 190.1 (C15). Elemental analysis calculated for C26H24O4: C, 77.98; H, 6.04. Found: C, 78.02; H, 6.01.
Synthesis of (E)-6-(4-Methylpent-3-en-1-yl)-2-[3-(4-methylphenyl)prop-2-enoyl] naphthalene-1,4-dione (6c)
This compound was synthesized by the general procedure using precursor 4c and purified by recrystallization using hexane as solvent. Yellow solid (111 mg, 58%), m.p. 115–117 °C. IR υmax cm−1 (film) 1666 (C=O), and 1601 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.63 (m, 6H, 2CH3, H13, H14), 2.40 (m, 5H, CH2, CH3, H10, H24), 2.81 (t, 2H, J = 7.7 Hz, CH2, H9), 5.15 (t, 1H, J = 7.4 Hz, CH, H11), 7.20 (m, 4H, 4CH, H3, H16, H20, H22), 7.51 (d, 2H, J = 8.3 Hz, 2CH, H19, H23), 7.64 (m, 2H, 2CH, H17, H7), 7.94 (d, 1H, J = 1.7 Hz, CH, H5), and 8.07 (d, 1H, J = 7.9 Hz, CH, H8). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 21.7 (C24), 25.7 (C14), 29.3 (C10), 36.3 (C9), 122.5 (C11), 124.3 (C16), 126.2 (C5), 127.0 (C8), 129.0 (C20, C22), 129.8 (C8a), 129.8 (C19, C23), 131.4 (C12), 131.8 (C18), 133.3 (C4a), 134.7 (C3), 136.7 (C7), 142.1 (C21), 146.1 (C2), 146.8 (C17), 150.0 (C6), 183.4 (C4), 185.3 (C1), and 190.2 (C15). Elemental analysis calculated for C26H24O3: C, 81.22; H, 6.29. Found: C, 81.15; H, 6.34.
Synthesis of (E)-2-[3-(2,4-Dichlorophenyl)prop-2-enoyl]-6-(4-methylpent-3-en-1-yl) naphthalene-1,4-dione (6d)
This compound was synthesized by the general procedure using precursor 4d and purified by recrystallization using hexane as solvent. Orange solid (92 mg, 42%), m.p. 86–88 °C. IR υmax cm−1 (film) 1668 (C=O), and 1624 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.55 (s, 3H, CH3, H13), 1.69 (s, 3H, CH3, H14), 2.39 (m, 2H, CH2, H10), 2.82 (m, 2H, CH2, H9), 5.14 (t, 1H, J = 7.4 Hz, CH, H11), 7.20 (d, 1H, J = 2.8 Hz, CH, H20), 7.29 (m, 2H, 2CH, H3, H22), 7.47 (m, 2H, 2CH, H7, H16), 7.66 (m, 2H, 2CH, H23, H5), and 8.00 (m, 2H, 2CH, H17, H8). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 25.7 (C14), 29.3 (C10), 36.3 (C9), 122.4 (C11), 126.3 (C16), 127.0 (C22), 127.5 (C20), 127.7 (C5), 128.7 (C8), 129.7 (C8a), 130.2 (C3), 131.1 (C18), 131.8 (C21), 133.4 (C12), 134.8 (C7), 136.4 (C4a), 137.3 (C19), 137.6 (C23), 140.1 (C17), 145.4 (C2), 150.2 (C6), 183.5 (C4), 185.1 (C1), and 189.5 (C15). Elemental analysis calculated for C25H20Cl2O3: C, 68.35; H, 4.59. Found: C, 68.43; H, 4.55.
Synthesis of (E)-2-[3-(2,3,4-Trimethoxyphenyl)prop-2-enoyl]-6-(4-methylpent-3-en-1-yl) naphthalene-1,4-dione (6e)
This compound was synthesized by the general procedure, using precursor 4e and purified by recrystallization using hexane as solvent. Red solid (92 mg, 40%), m.p. 118–120 °C. IR υmax cm−1 (film) 1666 (C=O), and 1596 (C=C). 1H NMR (CDCl3, TMS, ppm) δ 1.56 (s, 3H, CH3, H13), 1.70 (s, 3H, CH3, H14), 2.37 (m, 2H, CH2, H10), 2.82 (m, 2H, CH2, H9), 3.91 (m, 9H, 3CH3, H24, H25, H26), 5.15 (m, 1H, CH, H11), 6.73 (d, 1H, J = 8.8 Hz, CH, H22), 7.11 (s, 1H, CH, H3), 7.18 (d, 1H, J = 16.5 Hz, CH, H16), 7.38 (d, 1H, J = 8.8 Hz, CH, H23), 7.62 (dd, 1H, J = 8.3 Hz, J = 1.9 Hz, CH, H7), 7.9 (d, 1H, J = 16.5 Hz, CH, H17), 7.94 (d, 1H, J = 1.8 Hz, CH, H5), and 8.06 (d, 1H, J = 8.1 Hz, CH, H8). 13C NMR (CDCl3, TMS, ppm) δ 17.7 (C13), 25.7 (C14), 29.3 (C10), 36.3 (C9), 56.2 (C26), 60.9 (C26), 61.5 (C24), 107.7 (C22), 121.2 (C18), 122.5 (C11), 124.2 (C16), 124.3 (C23), 126.2 (C5), 127.0 (C8), 129.8 (C8a), 131.8 (C3), 133.3 (C12), 134.7 (C7), 136.5 (C4a), 142.4 (C17), 142.5 (C2), 146.5 (C20), 149.9 (C6), 154.1 (C19), 156.6 (C21), 183.4 (C4), 185.4 (C1), and 190.3 (C15). Elemental analysis calculated for C28H28O6: C, 73.03; H, 6.13. Found: C, 73.07; H, 6.04.
3.2. Antiproliferative Assay
MCF-7 (human breast adenocarcinoma) and HT-29 (human colon adenocarcinoma) cell lines were obtained from the American Type Culture Collection (ATCC). Cells were subcultured, and antiproliferative assays were carried out, following the procedure that we have previously described [38]. Doxorubicin was included in all evaluations as a reference drug.
3.3. Computational Details
3.3.1. Ligand Preparation
The 3D structure of each compound was prepared using Chem Draw Ultra version 12.0, as previously described [38]. Hydrophobicity index (cLogP), drug-likeness values, and toxicity risks were predicted through DataWarrior algorithms [68,69].
3.3.2. In Silico ADME Prediction
Pharmacokinetics parameters were calculasted using QikProp (QP) version 4.3 of the Schrodinger suite based on Lipinski’s rule of five and its extensions, as previously described [38].
3.3.3. Macromolecule Selection and Retrieval
The crystal structure of 14 selected proteins (Table S1), including growth factor receptors, transcription regulators, and enzymes (such as reductases, oxidases, and kinases) were retrieved from the Protein Data Bank [70]. They are overexpressed in some malignancies, including breast and colon adenocarcinoma, as described in the literature [50,51,52,53,55,56,57,61,62,63,71,72,73,74,75,76].
3.3.4. Molecular Docking of Ligand–Protein Interaction
We resorted to virtual screening using Autodock Vina, a target-specific scoring method useful for virtual screening [77]. All chalcone–naphthoquinone/hydroquinone hybrids were docked into the set of proteins of known 3D structure to identify those potentially inhibited by these compounds. Both ligands and proteins were prepared using AutoDock Tools version 1.5.7 (ADT), as previously described [38,78,79]. Finally, the binding site and energies of each compound were predicted into each receptor using Autodock Vina [77]. The graphic analysis of the molecular coupling studies was performed using Visual Molecular Dynamics 1.9 (VMD) [80] and Discovery Studio Biovia [81].
4. Conclusions
In this study, a novel series of chalcone-1,4-naphthoquinones/benzohydroquinones (CNQs and CBHQs) was synthesized from the precursor 2-acetyl-5,8-dihydro-6-(4-methyl-3-pentenyl)-1,4-naphthohydroquinone. The synthesis process involved protecting the hydroxyl group at C-4 of the precursor to eliminate its acidic properties associated with the phenolic 4-OH group. This step was necessary to proceed with the Claisen–Schmidt condensation reaction. In general, CNQs 6 exhibited superior pIC50 values compared to those of CBHQs 4 and 5, except for CBHQs 5e and 5f, which are diacetylated. This suggests that the coplanar structure of the naphthoquinone system and an appropriate level of lipophilicity, facilitating cell membrane penetration, favor the antineoplastic activity of the newly synthesized hybrid derivatives against both the MCF-7 and HT-29 cancer cell lines. It can also be inferred that the precursor derivatives of the 1,4-benzohydroquinone system, obtainable through the enzymatic hydrolysis of the respective diacetylated derivatives, are suitable due to the presence of hydroxyl groups, which enhance the binding energy when interacting with target proteins through hydrogen bonds. From a theoretical perspective, the binding energy of cancer-related proteins with CNQs and CBHQs was generally higher for kinases such as cMET, TRKA, and HER2, with ΔGbin values ranging from −11.1 to −7.2 kcal/mol. In this context, the synthesized cytotoxic hybrids (SCHs) are potential multi-kinase inhibitors and could serve as promising candidates for further research in the development of novel multi-target anticancer agents. However, experimental validation of the predictions and theoretical results for SCHs is essential before proceeding with acute toxicity and efficacy preclinical assays. Furthermore, the favorable predictions for physicochemical and pharmacokinetic parameters for most SCHs, aligning well with previous in vitro anti-proliferative results, underscore their potential as promising candidates for antineoplastic drug development.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28207172/s1, Figure S1: 1H NMR spectrum of compound 2. Figure S2: 13C NMR spectrum of compound 2. Figure S3: 1H NMR spectrum of compound 3a. Figure S4: 1H NMR spectrum of compound 3b. Figure S5: 1H NMR spectrum of compound 3c. Figure S6: 1H NMR spectrum of compound 3d. Figure S7: 1H NMR spectrum of compound 3e. Figure S8: 1H NMR spectrum of compound 3f. Figure S9: 1H NMR spectrum of compound 4a. Figure S10: 1H NMR spectrum of compound 4b. Figure S11: 1H NMR spectrum of compound 4c. Figure S12: 1H NMR spectrum of compound 4d. Figure S13: 1H NMR spectrum of compound 4e. Figure S14: 1H NMR spectrum of compound 4f. Figure S15: 13C NMR spectrum of compound 3a. Figure S16: 13C NMR spectrum of compound 3b. Figure S17: 13C NMR spectrum of compound 3c. Figure S18: 13C NMR spectrum of compound 3d. Figure S19: 13C NMR spectrum of compound 3e. Figure S20: 13C NMR spectrum of compound 3f. Figure S21: 13C NMR spectrum of compound 4a. Figure S22: 13C NMR spectrum of compound 4b. Figure S23: 13C NMR spectrum of compound 4c. Figure S24: 13C NMR spectrum of compound 4d. Figure S25: 13C NMR spectrum of compound 4e. Figure S26: 13C NMR spectrum of compound 4f. Figure S27: 1H NMR spectrum of compound 5′b. Figure S28: 1H NMR spectrum of compound 5′c. Figure S29: 1H NMR spectrum of compound 5′d. Figure S30: 1H NMR spectrum of compound 5e. Figure S31: 1H NMR spectrum of compound 5f. Figure S32: 1H NMR spectrum of compound 6a. Figure S33: 1H NMR spectrum of compound 6b. Figure S34: 1H NMR spectrum of compound 6c. Figure S35: 1H NMR spectrum of compound 6d. Figure S36: 1H NMR spectrum of compound 6e. Figure S37: 13C NMR spectrum of compound 5′b. Figure S38: 13C NMR spectrum of compound 5′c. Figure S39: 13C NMR spectrum of compound 5′d. Figure S40: 13C NMR spectrum of compound 5e. Figure S41: 13C NMR spectrum of compound 5f. Figure S42: 13C NMR spectrum of compound 6a. Figure S43: 13C NMR spectrum of compound 6b. Figure S44: 13C NMR spectrum of compound 6c. Figure S45: 13C NMR spectrum of compound 6d. Figure S46: 13C NMR spectrum of compound 6e. Figure S47: Plotted 2D maps of H-bonds and hydrophobic interactions of CNQ 5e and 5f with c-MET residues. Figure S48: Plot 2 D-maps of H-bonds and hydrophobic interactions of 5e, 5f, and 6a–c,e with TRKA residues. Figure S49: Plotted 2D maps of H-bonds and hydrophobic interactions of CNQ 5e, 5f, and 6a–c,e with HER2 residues. Figure S50: A: Overlapping of the docking poses for CNQ hybrid 6c (green), ligand 2 (yellow), and larotrectinib (grey) into TRKA. Superimposition of the docking poses for B: 6c and ligand 1, and C: 6c and larotrectinib. Figure S51: A: Overlapping of the docking poses for CNQ hybrid 6c (green), ligand 2 (yellow), and erlotinib (grey) into HER2. Superimposition of the docking poses for B: 6c and ligand 1, and C: 6c and erlotinib. Table S1: Predicted binding free energy values (∆Gbin kcal/mol) of synthesized cytotoxic hybrids with selected proteins overexpressed in cancer. Table S2: Comparison (ΔGbin, kcal/mol) of chalcones-1,4-Naphthoquinone/Hydroquinone hybrids with kinase proteins overexpressed in cancer. Table S3: Predicted binding free energy values (ΔGbin, kcal/mol) and binding site contacts of chalcones hybrids with amino acids of MEK1, TPK, and EGFR. Table S4: Physical and pharmacokinetic data predicted by QikPropa extensions for compounds 5e, 5f, and 6a–c,e. Table S5: Evaluation parameters of Lipinski’s rule of five and its extensions for compounds 5e, 5f, and 6a–c,e.
Author Contributions
Conceived, designed, and supervised all the experiments: A.O., A.M. and J.M.; Analyzed the in silico data and performed the theoretical calculations: W.A. and J.M.; Performed statistical analysis: J.M.; Supervised the bioassays and analyzed the in vitro data: A.M.; Contributed ideas and analyzed the data: A.O. and A.M.; Wrote the manuscript: J.M., W.A. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
We are grateful for the financial support from the Comisión Nacional de Investigación Científica y Tecnológica CONICYT of Chile (Project FONDECYT 1100316) and from the Dirección de Investigación de la Vicerrectoría de Investigación y Estudios Avanzados, Pontificia Universidad Católica de Valparaíso, Chile (Projects DI 039.471/2020, DI 039.338/2022 and DI 125.750/2023). J. Maldonado is grateful for the support of the Doctoral Fellowship CONICYT- PCHA/Doctorado Nacional/2014-21140688.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest, financial or otherwise.
Sample Availability
Not applicable.
References
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 7 July 2023).
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bisio, A.; Pedrelli, F.; D’Ambola, M.; Labanca, F.; Schito, A.M.; Govaerts, R.; De Tommasi, N.; Milella, L. Quinone Diterpenes from Salvia Species: Chemistry, Botany, and Biological Activity. Phytochem. Rev. 2019, 18, 665–842. [Google Scholar] [CrossRef]
- Manickam, M.; Boggu, P.R.; Cho, J.; Nam, Y.J.; Lee, S.J.; Jung, S.-H. Investigation of Chemical Reactivity of 2-Alkoxy-1,4-Naphthoquinones and Their Anticancer Activity. Bioorg. Med. Chem. Lett. 2018, 28, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Zorzanelli, B.C.; Ouverney, G.; Pauli, F.P.; da Fonseca, A.C.C.; de Almeida, E.C.P.; de Carvalho, D.G.; Possik, P.A.; Rabelo, V.W.-H.; Abreu, P.A.; Pontes, B.; et al. Pro-Apoptotic Antitumoral Effect of Novel Acridine-Core Naphthoquinone Compounds against Oral Squamous Cell Carcinoma. Molecules 2022, 27, 5148. [Google Scholar] [CrossRef]
- Sallustio, B.C.; Boddy, A.V. Is There Scope for Better Individualisation of Anthracycline Cancer Chemotherapy? Br. J. Clin. Pharmacol. 2021, 87, 295–305. [Google Scholar] [CrossRef]
- Liang, X.; Wu, Q.; Luan, S.; Yin, Z.; He, C.; Yin, L.; Zou, Y.; Yuan, Z.; Li, L.; Song, X.; et al. A Comprehensive Review of Topoisomerase Inhibitors as Anticancer Agents in the Past Decade. Eur. J. Med. Chem. 2019, 171, 129–168. [Google Scholar] [CrossRef]
- Kuete, V.; Donfack, A.R.N.; Mbaveng, A.T.; Zeino, M.; Tane, P.; Efferth, T. Cytotoxicity of Anthraquinones from the Roots of Pentas Schimperi towards Multi-Factorial Drug-Resistant Cancer Cells. Investig. New Drugs 2015, 33, 861–869. [Google Scholar] [CrossRef]
- Mancini, I.; Vigna, J.; Sighel, D.; Defant, A. Hybrid Molecules Containing Naphthoquinone and Quinolinedione Scaffolds as Antineoplastic Agents. Molecules 2022, 27, 4948. [Google Scholar] [CrossRef]
- Kabakci, Z.; Käppeli, S.; Cantù, C.; Jensen, L.D.; König, C.; Toggweiler, J.; Gentili, C.; Ribaudo, G.; Zagotto, G.; Basler, K.; et al. Pharmacophore-Guided Discovery of CDC25 Inhibitors Causing Cell Cycle Arrest and Tumor Regression. Sci. Rep. 2019, 9, 1335. [Google Scholar] [CrossRef]
- Hsu, M.-J.; Chen, H.-K.; Lien, J.-C.; Huang, Y.-H.; Huang, S.-W. Suppressing VEGF-A/VEGFR-2 Signaling Contributes to the Anti-Angiogenic Effects of PPE8, a Novel Naphthoquinone-Based Compound. Cells 2022, 11, 2114. [Google Scholar] [CrossRef] [PubMed]
- Nursamsiar; Asnawi, A.; Kartasasmita, R.E.; Ibrahim, S.; Tjahjono, D.H. Synthesis, Biological Evaluation, and Docking Analysis of Methyl Hydroquinone and Bromo Methyl Hydroquinone as Potent Cyclooxygenase (COX-1 and COX-2) Inhibitors. J. Appl. Pharm. Sci. 2018, 8, 16–20. [Google Scholar] [CrossRef]
- Byeon, S.; Yi, Y.-S.; Lee, J.; Yang, W.; Kim, J.; Kim, J.; Hong, S.; Kim, J.-H.; Cho, J. Hydroquinone Exhibits In Vitro and In Vivo Anti-Cancer Activity in Cancer Cells and Mice. Int. J. Mol. Sci. 2018, 19, 903. [Google Scholar] [CrossRef]
- Rudrapal, M.; Khan, J.; Dukhyil, A.A.B.; Alarousy, R.M.I.I.; Attah, E.I.; Sharma, T.; Khairnar, S.J.; Bendale, A.R. Chalcone Scaffolds, Bioprecursors of Flavonoids: Chemistry, Bioactivities, and Pharmacokinetics. Molecules 2021, 26, 7177. [Google Scholar] [CrossRef]
- Lou, H.; Hu, L.; Lu, H.; Wei, T.; Chen, Q. Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules 2021, 26, 4522. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulou, I.; Tzani, A.; Polyzos, N.-I.; Karadendrou, M.-A.; Kritsi, E.; Pontiki, E.; Liargkova, T.; Hadjipavlou-Litina, D.; Zoumpoulakis, P.; Detsi, A. Exploring the 2′-Hydroxy-Chalcone Framework for the Development of Dual Antioxidant and Soybean Lipoxygenase Inhibitory Agents. Molecules 2021, 26, 2777. [Google Scholar] [CrossRef] [PubMed]
- Elkanzi, N.A.A.; Hrichi, H.; Alolayan, R.A.; Derafa, W.; Zahou, F.M.; Bakr, R.B. Synthesis of Chalcones Derivatives and Their Biological Activities: A Review. ACS Omega 2022, 7, 27769–27786. [Google Scholar] [CrossRef]
- Sahu, N.K.; S. Balbhadra, S.; Choudhary, J.; V. Kohli, D. Exploring Pharmacological Significance of Chalcone Scaffold: A Review. Curr. Med. Chem. 2012, 19, 209–225. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, J.; Chen, X.; Fu, X.; Sun, S.; Wu, Q. Chalcone Derivatives: Role in Anticancer Therapy. Biomolecules 2021, 11, 894. [Google Scholar] [CrossRef]
- Ducki, S. The Development of Chalcones as Promising Anticancer Agents. IDrugs 2007, 10, 42–46. [Google Scholar]
- Zhuang, C.; Zhang, W.; Sheng, C.; Zhang, W.; Xing, C.; Miao, Z. Chalcone: A Privileged Structure in Medicinal Chemistry. Chem. Rev. 2017, 117, 7762–7810. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; He, M.; Li, Y.; Peng, Z.; Wang, G. A Review on Synthetic Chalcone Derivatives as Tubulin Polymerisation Inhibitors. J. Enzyme Inhib. Med. Chem. 2022, 37, 9–38. [Google Scholar] [CrossRef] [PubMed]
- Ducki, S. Antimitotic Chalcones and Related Compounds as Inhibitors of Tubulin Assembly. Anticancer. Agents Med. Chem. 2009, 9, 336–347. [Google Scholar] [CrossRef]
- Michalkova, R.; Mirossay, L.; Gazdova, M.; Kello, M.; Mojzis, J. Molecular Mechanisms of Antiproliferative Effects of Natural Chalcones. Cancers 2021, 13, 2730. [Google Scholar] [CrossRef]
- Mahapatra, D.K.; Bharti, S.K.; Asati, V. Anti-Cancer Chalcones: Structural and Molecular Target Perspectives. Eur. J. Med. Chem. 2015, 98, 69–114. [Google Scholar] [CrossRef]
- Noser, A.A.; Shehadi, I.A.; Abdelmonsef, A.H.; Salem, M.M. Newly Synthesized Pyrazolinone Chalcones as Anticancer Agents via Inhibiting the PI3K/Akt/ERK1/2 Signaling Pathway. ACS Omega 2022, 7, 25265–25277. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Anand, A.; Kumar, V. Recent Developments in Biological Activities of Chalcones: A Mini Review. Eur. J. Med. Chem. 2014, 85, 758–777. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, A.; Singh, H.; Sonawane, P.; Paliwal, H.; Thareja, S.; Pathak, P.; Grishina, M.; Jaremko, M.; Emwas, A.-H.; et al. Concept of Hybrid Drugs and Recent Advancements in Anticancer Hybrids. Pharmaceuticals 2022, 15, 1071. [Google Scholar] [CrossRef]
- Nepali, K.; Sharma, S.; Sharma, M.; Bedi, P.M.S.; Dhar, K.L. Rational Approaches, Design Strategies, Structure Activity Relationship and Mechanistic Insights for Anticancer Hybrids. Eur. J. Med. Chem. 2014, 77, 422–487. [Google Scholar] [CrossRef]
- Viegas-Junior, C.; Barreiro, E.J. Carlos Alberto Manssour Fraga Molecular Hybridization: A Useful Tool in the Design of New Drug Prototypes. Curr. Med. Chem. 2007, 14, 1829–1852. [Google Scholar] [CrossRef]
- Karthikeyan, C.; Solomon, V.R.; Lee, H.; Trivedi, P. Design, Synthesis and Biological Evaluation of Some Isatin-Linked Chalcones as Novel Anti-Breast Cancer Agents: A Molecular Hybridization Approach. Biomed. Prev. Nutr. 2013, 3, 325–330. [Google Scholar] [CrossRef]
- Mohamed, M.F.A.; Abuo-Rahma, G.E.-D.A. Molecular Targets and Anticancer Activity of Quinoline–Chalcone Hybrids: Literature Review. RSC Adv. 2020, 10, 31139–31155. [Google Scholar] [CrossRef] [PubMed]
- Kurt, B.Z.; Ozten Kandas, N.; Dag, A.; Sonmez, F.; Kucukislamoglu, M. Synthesis and Biological Evaluation of Novel Coumarin-Chalcone Derivatives Containing Urea Moiety as Potential Anticancer Agents. Arab. J. Chem. 2020, 13, 1120–1129. [Google Scholar] [CrossRef]
- Gao, F.; Huang, G.; Xiao, J. Chalcone Hybrids as Potential Anticancer Agents: Current Development, Mechanism of Action, and Structure-activity Relationship. Med. Res. Rev. 2020, 40, 2049–2084. [Google Scholar] [CrossRef]
- Bahia, S.B.B.B.; Reis, W.J.; Jardim, G.A.M.; Souto, F.T.; de Simone, C.A.; Gatto, C.C.; Menna-Barreto, R.F.S.; de Castro, S.L.; Cavalcanti, B.C.; Pessoa, C.; et al. Molecular Hybridization as a Powerful Tool towards Multitarget Quinoidal Systems: Synthesis, Trypanocidal and Antitumor Activities of Naphthoquinone-Based 5-Iodo-1,4-Disubstituted-, 1,4- and 1,5-Disubstituted-1,2,3-Triazoles. Medchemcomm 2016, 7, 1555–1563. [Google Scholar] [CrossRef]
- Kadela-Tomanek, M.; Jastrzębska, M.; Chrobak, E.; Bębenek, E.; Latocha, M. Hybrids of 1,4-Quinone with Quinoline Derivatives: Synthesis, Biological Activity, and Molecular Docking with DT-Diaphorase (NQO1). Molecules 2022, 27, 6206. [Google Scholar] [CrossRef]
- Maldonado, J.; Acevedo, W.; Molinari, A.; Oliva, A.; Knox, M.; San Feliciano, A. Synthesis, In Vitro Evaluation and Molecular Docking Studies of Novel Naphthoisoxazolequinone Carboxamide Hybrids as Potential Antitumor Agents. Polycycl. Aromat. Compd. 2022, 43, 4960–4983. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; Arismendi, M.; Imbarack, E.; Gálvez, C.; Maldonado, J.; Feliciano, A.S. The Synthesis of Some Fused Pyrazolo-1,4-Naphthoquinones. J. Heterocycl. Chem. 2015, 52, 620–622. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; Arismendi-Macuer, M.; Guzmán, L.; Fuentealba, M.; Knox, M.; Vinet, R.; San Feliciano, A. New 1H-Benzo[f]Indazole-4,9-Diones Conjugated with C-Protected Amino Acids and Other Derivatives: Synthesis and in Vitro Antiproliferative Evaluation. Molecules 2015, 20, 21924–21938. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; del Corral, J.M.M.; Castro, M.A.; Araya, C.; García-Grávalos, M.D.; San Feliciano, A. Cytotoxic–Antineoplastic Activity of Acetyl Derivatives of Prenylnaphthohydroquinone. Farm. 2004, 59, 651–656. [Google Scholar] [CrossRef]
- Cooper, S.C.; Sammes, P.G. Acyl Rearrangements in Acylbenzoquinone Cycloadducts. J. Chem. Soc. Perkin Trans. 1984, 1, 2407–2414. [Google Scholar] [CrossRef]
- Andrade, J.T.; Santos, F.R.S.; Lima, W.G.; Sousa, C.D.F.; Oliveira, L.S.F.M.; Ribeiro, R.I.M.A.; Gomes, A.J.P.S.; Araújo, M.G.F.; Villar, J.A.F.P.; Ferreira, J.M.S. Design, Synthesis, Biological Activity and Structure-Activity Relationship Studies of Chalcone Derivatives as Potential Anti-Candida Agents. J. Antibiot. 2018, 71, 702–712. [Google Scholar] [CrossRef]
- Hsieh, H.K.; Lee, T.H.; Wang, J.P.; Wang, J.J.; Lin, C.N. Synthesis and Anti-Inflammatory Effect of Chalcones and Related Compounds. Pharm. Res. 1998, 15, 39–46. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; Ojeda, C.; Miguel del Corral, J.M.; Castro, M.A.; Cuevas, C.; San Feliciano, A. New Cytotoxic-Antineoplastic Prenyl-1,2-Naphthohydroquinone Derivatives. Bioorg. Med. Chem. 2005, 13, 6645–6650. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, T.; Mihis, A.G. Two Important Anticancer Mechanisms of Natural and Synthetic Chalcones. Int. J. Mol. Sci. 2022, 23, 11595. [Google Scholar] [CrossRef] [PubMed]
- Gul, H.I.; Yamali, C.; Gunesacar, G.; Sakagami, H.; Okudaira, N.; Uesawa, Y.; Kagaya, H. Cytotoxicity, Apoptosis, and QSAR Studies of Phenothiazine Derived Methoxylated Chalcones as Anticancer Drug Candidates. Med. Chem. Res. 2018, 27, 2366–2378. [Google Scholar] [CrossRef]
- Sharma, R.; Kumar, R.; Kodwani, R.; Kapoor, S.; Khare, A.; Bansal, R.; Khurana, S.; Singh, S.; Thomas, J.; Roy, B.; et al. A Review on Mechanisms of Anti Tumor Activity of Chalcones. Anticancer. Agents Med. Chem. 2015, 16, 200–211. [Google Scholar] [CrossRef]
- Albright, C.F.; Graciani, N.; Han, W.; Yue, E.; Stein, R.; Lai, Z.; Diamond, M.; Dowling, R.; Grimminger, L.; Zhang, S.-Y.; et al. Matrix Metalloproteinase–Activated Doxorubicin Prodrugs Inhibit HT1080 Xenograft Growth Better than Doxorubicin with Less Toxicity. Mol. Cancer Ther. 2005, 4, 751–760. [Google Scholar] [CrossRef]
- Negi, R.R.; Rana, S.V.; Gupta, V.; Gupta, R.; Chadha, V.D.; Prasad, K.K.; Dhawan, D.K. Over-Expression of Cyclooxygenase-2 in Colorectal Cancer Patients. Asian Pacific J. Cancer Prev. 2019, 20, 1675–1681. [Google Scholar] [CrossRef]
- Singh, B.; Berry, J.A.; Shoher, A.; Ramakrishnan, V.; Lucci, A. COX-2 Overexpression Increases Motility and Invasion of Breast Cancer Cells. Int. J. Oncol. 2005, 26, 1393–1399. [Google Scholar]
- Guo, S.; Colbert, L.S.; Fuller, M.; Zhang, Y.; Gonzalez-Perez, R.R. Vascular Endothelial Growth Factor Receptor-2 in Breast Cancer. Biochim. Biophys. Acta—Rev. Cancer 2010, 1806, 108–121. [Google Scholar] [CrossRef]
- Ali, S.; Coombes, R.C. Estrogen Receptor Alpha in Human Breast Cancer: Occurrence and Significance. J. Mammary Gland Biol. Neoplasia 2000, 5, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Pines, G. The ERBB Network: At Last, Cancer Therapy Meets Systems Biology. Nat. Rev. Cancer 2012, 12, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, S.; Ko, S.; In, Y.; Moon, H.-G.; Ahn, S.K.; Kim, M.K.; Lee, M.; Hwang, J.-H.; Ju, Y.S.; et al. Recurrent Fusion Transcripts Detected by Whole-Transcriptome Sequencing of 120 Primary Breast Cancer Samples. Genes, Chromosom. Cancer 2015, 54, 681–691. [Google Scholar] [CrossRef]
- Ahmad, D.A.J.; Negm, O.H.; Alabdullah, M.L.; Mirza, S.; Hamed, M.R.; Band, V.; Green, A.R.; Ellis, I.O.; Rakha, E.A. Clinicopathological and Prognostic Significance of Mitogen-Activated Protein Kinases (MAPK) in Breast Cancers. Breast Cancer Res. Treat. 2016, 159, 457–467. [Google Scholar] [CrossRef]
- Pashirzad, M.; Khorasanian, R.; Fard, M.M.; Arjmand, M.-H.; Langari, H.; Khazaei, M.; Soleimanpour, S.; Rezayi, M.; Ferns, G.A.; Hassanian, S.M.; et al. The Therapeutic Potential of MAPK/ERK Inhibitors in the Treatment of Colorectal Cancer. Curr. Cancer Drug Targets 2021, 21, 932–943. [Google Scholar] [CrossRef]
- Abourehab, M.A.S.; Alqahtani, A.M.; Youssif, B.G.M.; Gouda, A.M. Globally Approved EGFR Inhibitors: Insights into Their Syntheses, Target Kinases, Biological Activities, Receptor Interactions, and Metabolism. Molecules 2021, 26, 6677. [Google Scholar] [CrossRef]
- Liu, F.; Wei, Y.; Zhang, H.; Jiang, J.; Zhang, P.; Chu, Q. NTRK Fusion in Non-Small Cell Lung Cancer: Diagnosis, Therapy, and TRK Inhibitor Resistance. Front. Oncol. 2022, 12, 864666. [Google Scholar] [CrossRef]
- Li, S. Anlotinib: A Novel Targeted Drug for Bone and Soft Tissue Sarcoma. Front. Oncol. 2021, 11, 664853. [Google Scholar] [CrossRef]
- Sylvester, P.W. Targeting Met Mediated Epithelial-mesenchymal Transition in the Treatment of Breast Cancer. Clin. Transl. Med. 2014, 3, 30. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, B.P. Epithelial-Mesenchymal Transition in Breast Cancer Progression and Metastasis. Chin. J. Cancer 2011, 30, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, G.; Smith, C.; Goddard, L.; Jordan, N.; McClelland, R.; Barrett-Lee, P.; Nicholson, R.I.; Hiscox, S. Targeting Focal Adhesion Kinase in ER+/HER2+ Breast Cancer Improves Trastuzumab Response. Endocr. Relat. Cancer 2013, 20, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Brylinski, M. Aromatic Interactions at the Ligand-Protein Interface: Implications for the Development of Docking Scoring Functions. Chem. Biol. Drug Des. 2018, 91, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Liu, D. Suppression of C-MET Overcomes Erlotinib Resistance in Tongue Cancer Cells. Onco. Targets. Ther. 2018, 11, 5499–5508. [Google Scholar] [CrossRef]
- Han, S.-Y. TRK Inhibitors: Tissue-Agnostic Anti-Cancer Drugs. Pharmaceuticals 2021, 14, 632. [Google Scholar] [CrossRef]
- Schaefer, G.; Shao, L.; Totpal, K.; Akita, R.W. Erlotinib Directly Inhibits HER2 Kinase Activation and Downstream Signaling Events in Intact Cells Lacking Epidermal Growth Factor Receptor Expression. Cancer Res. 2007, 67, 1228–1238. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Duffy, E.M. Prediction of Drug Solubility from Structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Nakano, M.; Fukami, T.; Gotoh, S.; Nakajima, M. A-to-I RNA Editing Up-Regulates Human Dihydrofolate Reductase in Breast Cancer. J. Biol. Chem. 2017, 292, 4873–4884. [Google Scholar] [CrossRef]
- Lei, H.; Deng, C.-X. Fibroblast Growth Factor Receptor 2 Signaling in Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Haldosén, L.-A.; Zhao, C.; Dahlman-Wright, K. Estrogen Receptor Beta in Breast Cancer. Mol. Cell. Endocrinol. 2014, 382, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhu, G.; Li, Y.; Padia, R.N.; Dong, Z.; Pan, Z.K.; Liu, K.; Huang, S. Extracellular Signal–Regulated Kinase Signaling Pathway Regulates Breast Cancer Cell Migration by Maintaining Slug Expression. Cancer Res. 2009, 69, 9228–9235. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Song, M.; Song, J. Expression and Significance of ERK Protein in Human Breast Carcinoma. Chinese J. Cancer Res. 2004, 16, 269–273. [Google Scholar] [CrossRef]
- Sultan, A.S.; Brim, H.; Sherif, Z.A. Co-Overexpression of Janus Kinase 2 and Signal Transducer and Activator of Transcription 5a Promotes Differentiation of Mammary Cancer Cells through Reversal of Epithelial–Mesenchymal Transition. Cancer Sci. 2008, 99, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; Arismendi-Macuer, M.; Guzmán, L.; Acevedo, W.; Aguayo, D.; Vinet, R.; San Feliciano, A. Antiproliferative Benzoindazolequinones as Potential Cyclooxygenase-2 Inhibitors. Molecules 2019, 24, 2261. [Google Scholar] [CrossRef]
- Acevedo, W.; González-Nilo, F.; Agosin, E. Docking and Molecular Dynamics of Steviol Glycoside–Human Bitter Receptor Interactions. J. Agric. Food Chem. 2016, 64, 7585–7596. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer; V20.1.0, Vol19295; Dassault Systèmes: San Diego, CA, USA, 2019. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).