Abstract
A novel series of pyrido[2,3-d]pyrimidines; pyrido[3,2-e][1,3,4]triazolo; and tetrazolo[1,5-c]pyrimidines were synthesized via different chemical transformations starting from pyrazolo[3,4-b]pyridin-6-yl)-N,N-dimethylcarbamimidic chloride 3b (prepared from the reaction of o-aminonitrile 1b and phosogen iminiumchloride). The structures of the newly synthesized compounds were elucidated based on spectroscopic data and elemental analyses. Designated compounds are subjected for molecular docking by using Auto Dock Vina software in order to evaluate the antiviral potency for the synthesized compounds against SARS-CoV-2 (2019-nCoV) main protease M pro. The antiviral activity against SARS-CoV-2 showed that tested compounds 7c, 7d, and 7e had the most promising antiviral activity with lower IC50 values compared to Lopinavir, “the commonly used protease inhibitor”. Both in silico and in vitro results are in agreement.
1. Introduction
Human health is one of the most important factors impacting economic development in the world. So, studying diseases and their potential treatment is an essential research area. The coronavirus disease 2019 (COVID-19) has imposed a great threat globally due its rapid spreading and mutation. Virus-encoded proteases are observed as potential drug targets. The main protease (Mpro) of SRAS-CoV-2 plays a crucial role in the viral maturation.
Studies have reported that the inhibition of Mpro can prevent the virus from replication [1,2,3,4,5]. At present, some medications and vaccines have been approved by the FDA for the treatment of COVID-19 but they could not completely eradicate the virus. The search for safe and effective antivirals today is urgent.
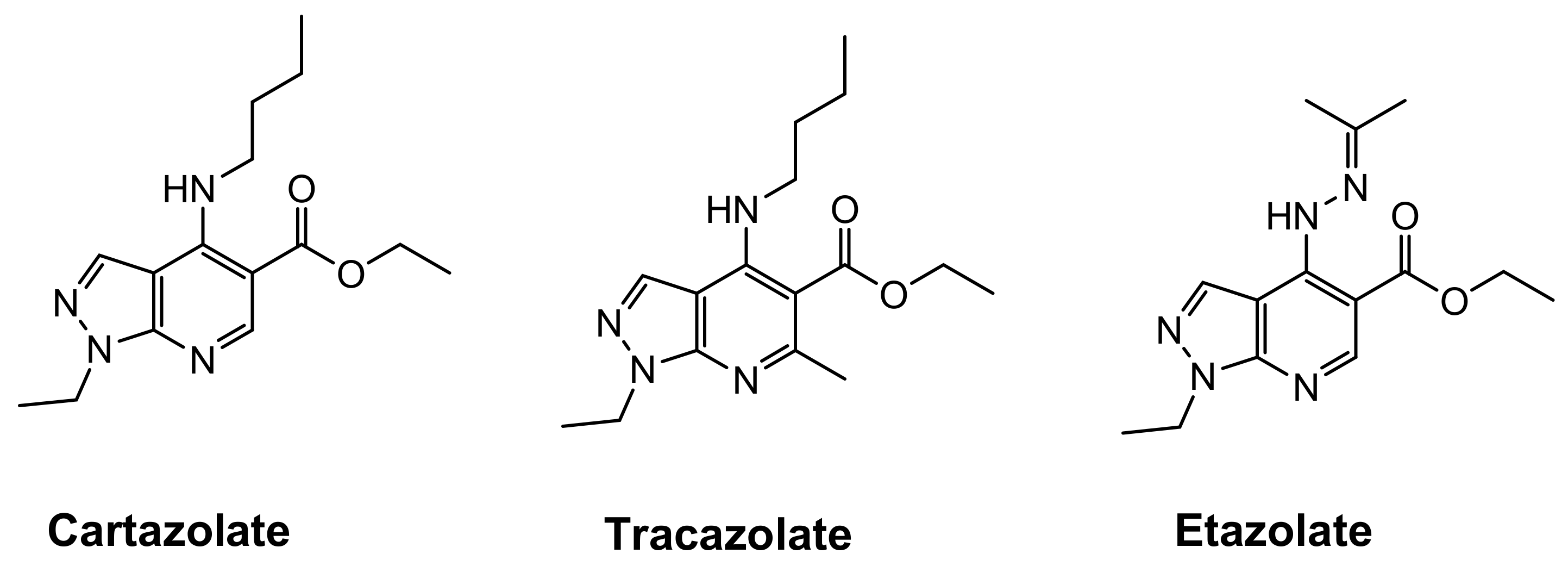
Several synthetic molecules containing heterocyclic moieties are currently known for their antiviral potentials [6,7,8,9,10,11]. Pyrazolo[3,4-b]pyridine nucleus is an important scaffold in a variety of marketed anxiolytic antidepressant drugs such as cartazolate, tracazolate and etazolate (Figure 1).
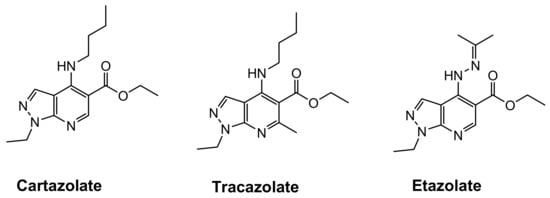
Figure 1.
Examples of marketed pyrazolopyridine drugs.
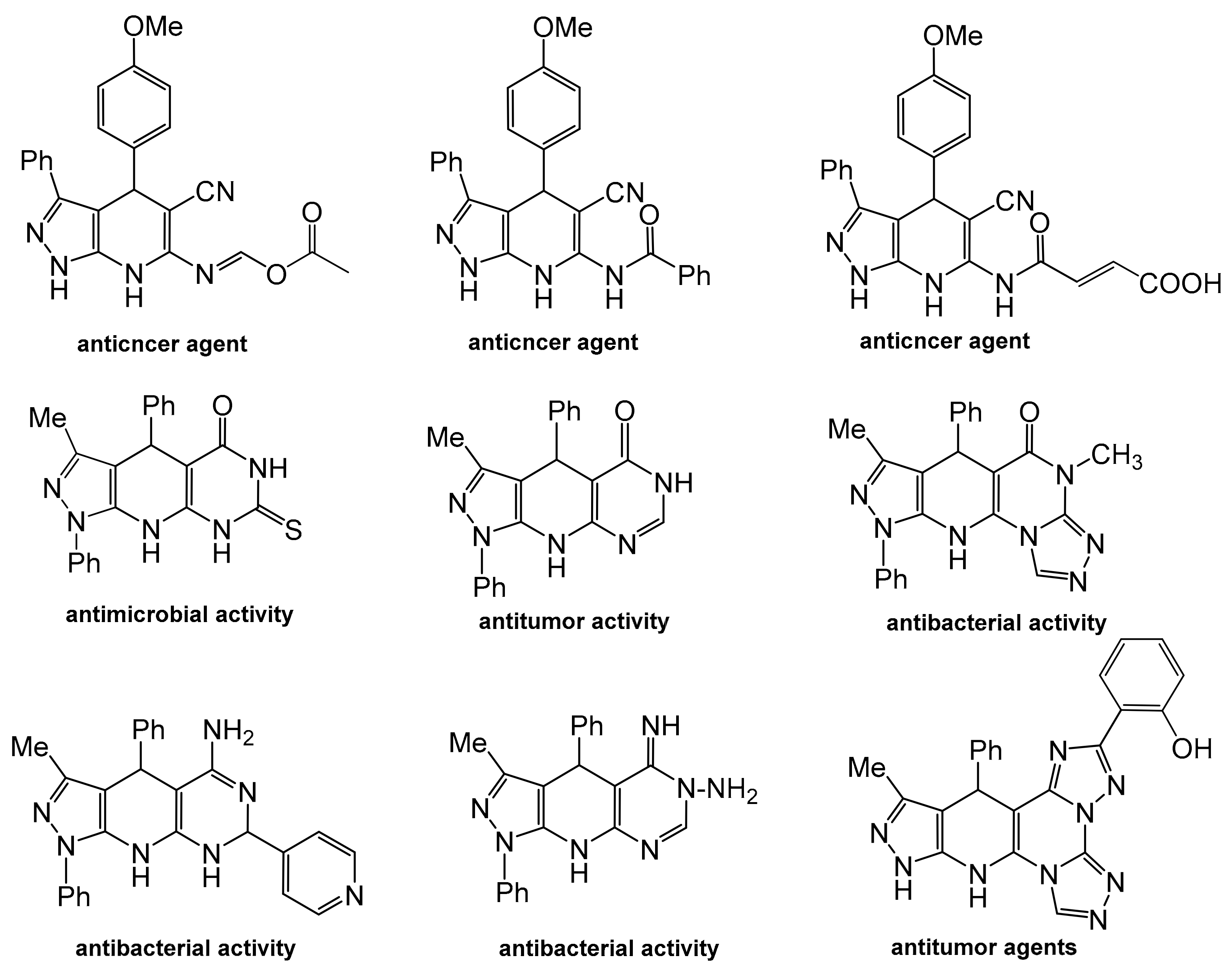
Pyrazolopyrido[2,3-d]pyrimidine, a fused hetero-tricyclic nucleus containing pyrazole, pyridine and pyrimidine rings, has attained momentary attention in the sphere of multicomponent synthetic protocol. Pyrazolo[3,4-b]pyridine nucleus is an important scaffold for synthesis of lots of bioactive compounds [12,13,14,15,16,17,18,19,20] (Figure 2). Pyrido[2,3-d]pyrimidine-derived drugs have manifested diverse pharmacological activities, particularly, anti-inflammatory, cytotoxic, antimicrobial, phosphodiesterase inhibitors and cytokine inhibitors [16,17,18,19,20,21,22].
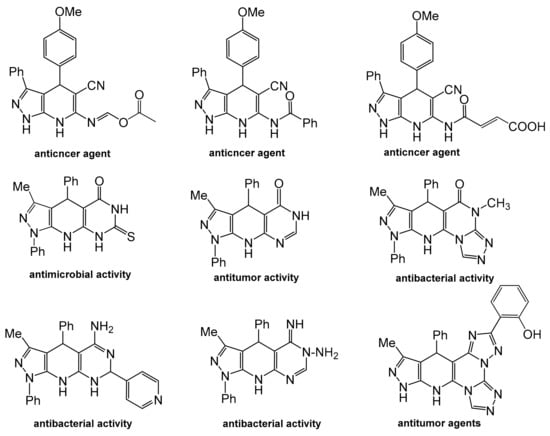
Figure 2.
Pyrazolopyridines with reported antimicrobial, antibacterial and anticancer efficacies as well as DNA-binding affinity.
These pyridine/pyrimidine core structures have been noted for their roles in many biological processes as well as in cancer pathogenesis, which make such compounds attractive scaffolds for discovery of novel drugs. In addition, applications of these derivatives have been found in various areas of medicine, such as anticancer, CNS, fungicidal, antiviral and antibacterial therapies [23,24].
The diamine analogs including pyrido[3,4-d]pyrimidine core were reported as tyrosine kinase inhibitors [25,26]. The combination of arylidene hydrazinyl moiety with pyrido[2,3-d]pyrimidin-4-one resulted in the output of unprecedented anti-microbial agents [27]. Pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidines analogs were reported to have diverse pharmacological activities, including as anticonvulsants, antiproliferative agents, anti-inflammatories and analgesic agents, antibacterial, antifungal, antimicrobial and antitumor activities [28,29,30,31]. Further, pyrazolo[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[4,3-a]pyrimidinone analogs were reported as antimicrobial agents [32].
For the abovementioned benefits and as part of our program investigating syntheses of heterocyclic compounds which have biological significance [33,34,35,36,37,38,39,40,41,42], herein, our focus will be on the synthesis, molecular docking and pharmacological evaluation of novel series of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidines and pyrazolo[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[4,3-a]pyrimidinone, which could serve as a promising scaffold in the development of novel viral protease inhibitors of SARS-CoV-2.
2. Results and Discussions
2.1. Chemistry
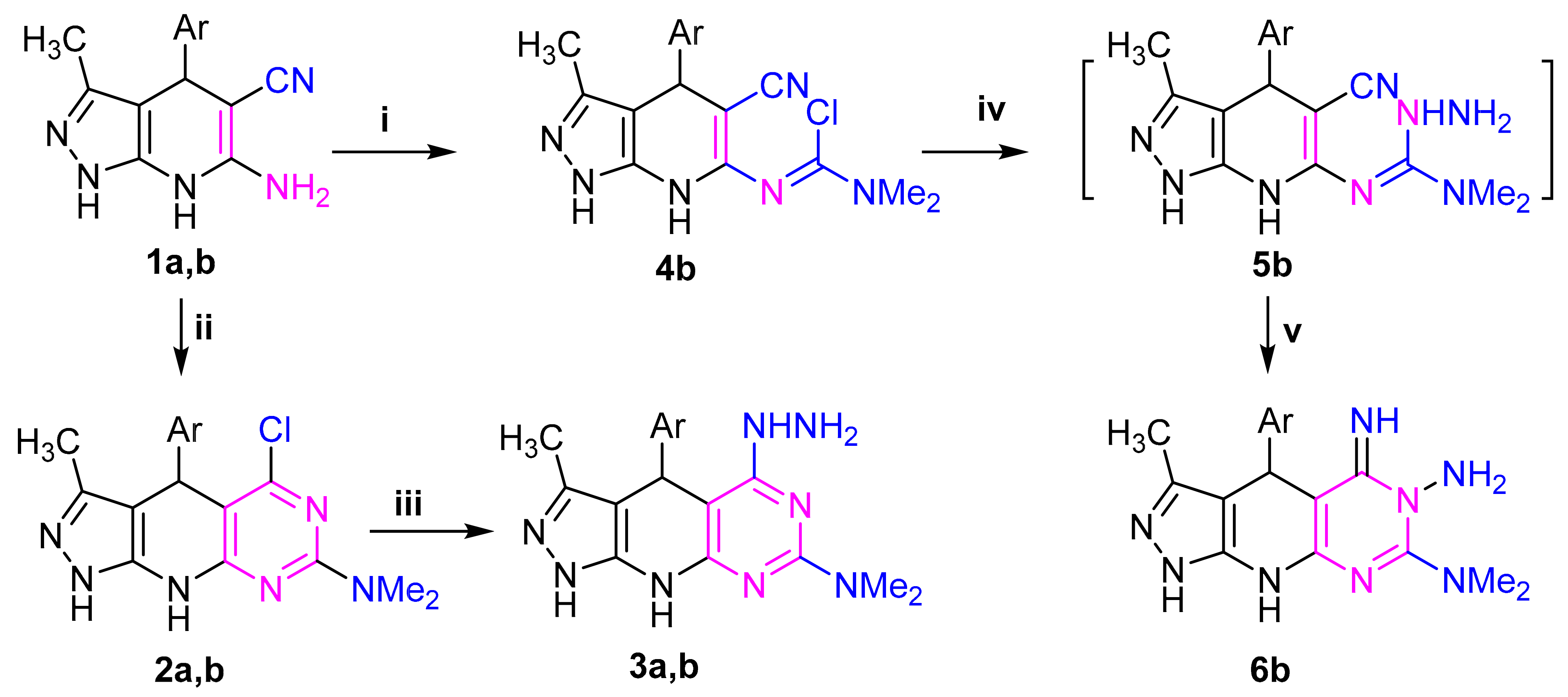
The requisite starting material 6-amino-pyrazolo[3,4-b]pyridine-5-carbonitrile 1 was used in this study. It was synthesized according to the known procedure [33] from pyrazolone, arylaldehyde and malononitrile (Scheme 1). Phosgene iminium chloride (PIC = dichloromethylene-dimethyliminium chloride) is an efficient reagent in synthetic chemistry and was used to introduce the amide chloride group into activated substrates. So, stirring of compound 1a,b with phosgenimonium chloride in 1,2-dichloroethane at room temperature provided pyridopyrimidine derivative 2a,b and Pyrazolopyridinyl derivative 4b, respectively, through the pathway outlined in Scheme 1.
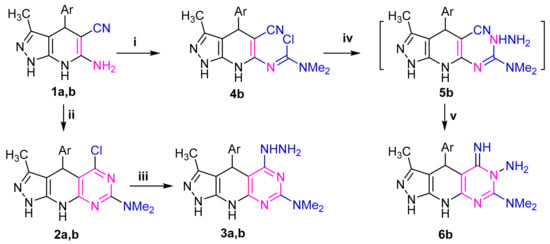
Scheme 1.
Reagents: (i) PIC (Me2N=C(Cl)2Cl), 6 °C/6h; (ii) (Me2N=C(Cl)2Cl), 25 °C/18h; (iii) H2NNH2•H2O, 25 °C/18h; (iv) H2NNH2•H2O, 5 °C/6h; (v) stirring at 25 °C/18h; Ar = (a) C6H4NO2(4), (b) thien-2-yl.
The structures of 2a,b and 4b were confirmed by elemental analysis and spectral (MS, IR and 1H NMR) data (Supplementary Materials) (c.f. experimental). Their 1H NMR spectrum in DMSO revealed in each case a characteristic signal in the region of δ 3.10–3.27 assignable to the –N(CH3)2 proton. Their IR spectra showed the absence of the characteristic band for nitrile group for compounds 2a,b which is present for compound 4b at 2217 cm−1. Additionally, a new peak at ν 735 cm−1 for the new C-Cl appeared. Stirring of the compounds 2a,b and 4b with hydrazine hydrate in ethanol afforded the corresponding 5-imino-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidine derivatives 3a,b and 5-hydrazineyl-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidine derivative 6b, respectively (Scheme 1). Both spectroscopic data and elemental analyses were consistent with the assigned structures (c.f., experimental). IR spectra of compounds 6b as an example indicated the disappearance the absorption band characteristic for CN and C-Cl groups with the appearance of peaks corresponding to -NH2 and NH groups. In the 1H NMR spectrum (Supplementary Materials) and in the presence of D2O, the signals due to the NH and NH2 groups in each compound disappeared and the signals related to methyl, aryl, and N(CH3)2 protons appeared at the expected values.
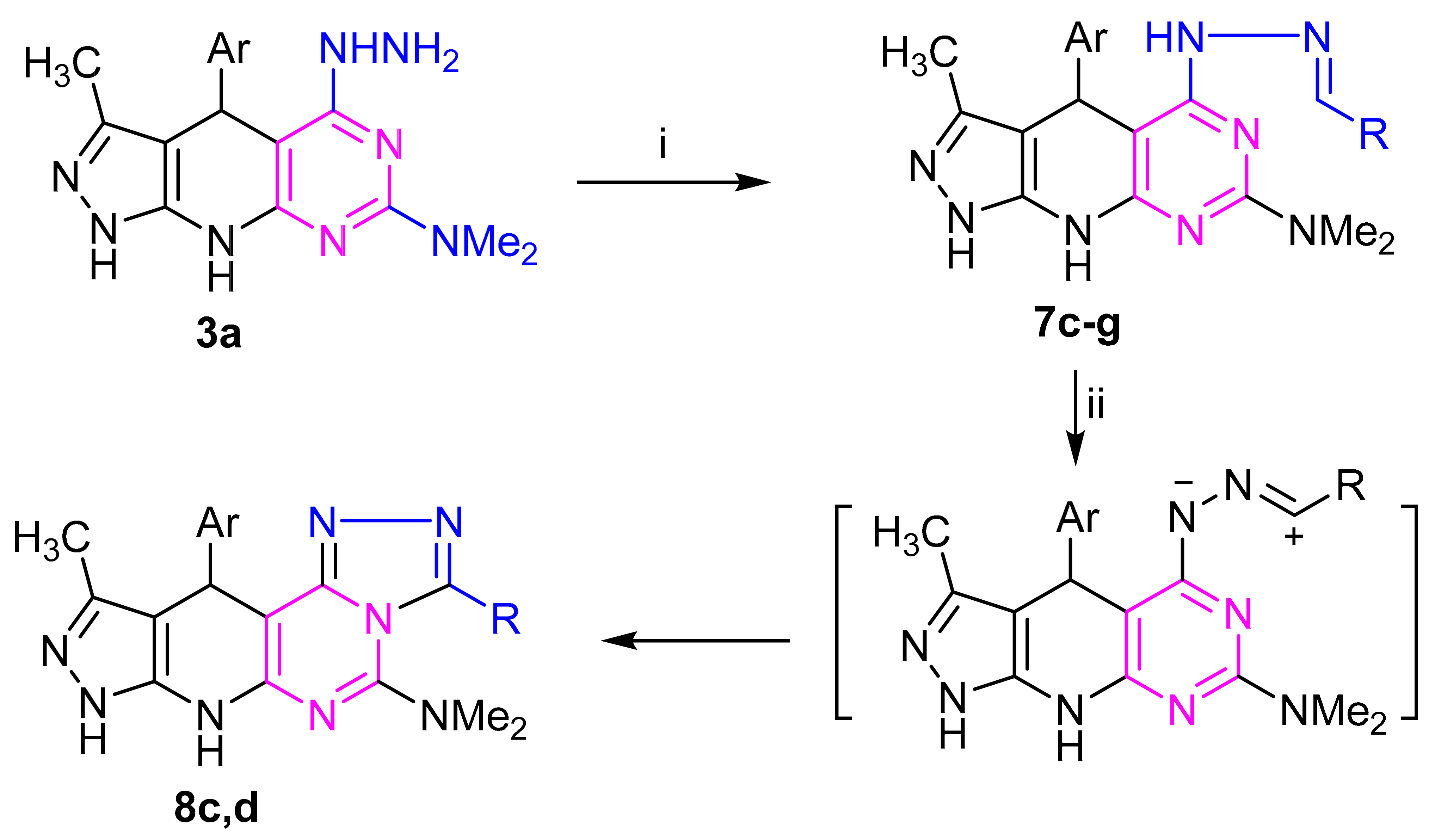
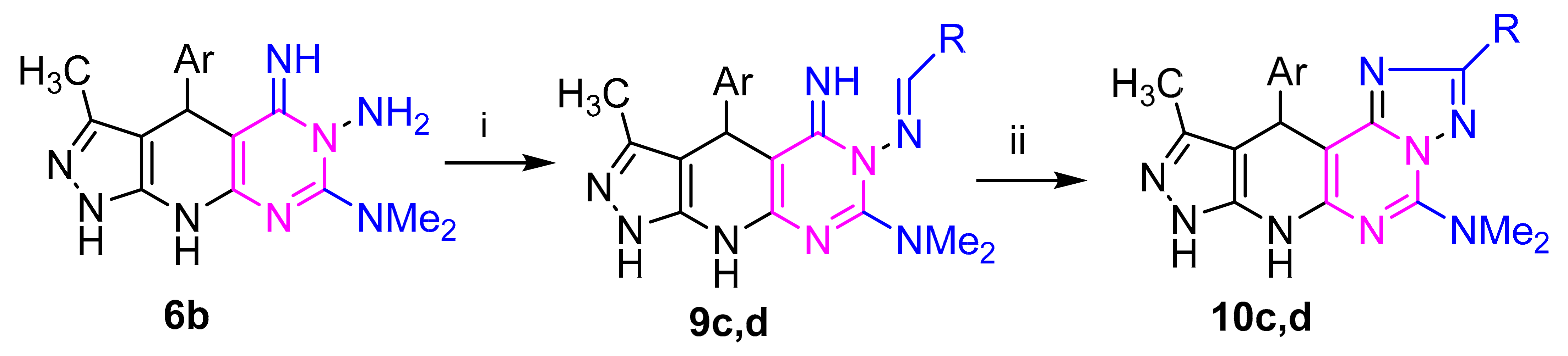
Refluxing of hydrazine derivative 3a with aldehydes and/or monosaccharaides in ethanol, with adding a few drops of acetic acid as a catalyst, gave the corresponding hydrazones derivatives 7c–g, respectively (Scheme 2). The structures of the latter products were confirmed by elemental and spectral analyses. The IR spectrum of the subsequent sugar hydrazones 7f,g as an example indicated the characteristic bands for the hydroxyl groups at (3415–3419) cm−1 in the sugar chain. The 1H NMR (c.f. experimental) spectra of compound 7g,h demonstrated the signs of H-1 methine proton showing up as doublet at δ 7.17 and 7.20 ppm, respectively. Their 13C NMR spectra (Supplementary Materials) of compound 7f revealed five resonances for sugar carbon atoms at δ 61.63, 70.59, 73.75, 75.48, 77.22 ppm. Treatment of each of the hydrazones 7c,d with iron (III) chloride in ethanol or with potassium iodide in DMSO gave, in each case, a single product as evidenced by TLC analysis. Elemental analyses and mass spectra revealed that each of such isolated products has two hydrogens less than the respective hydrazone. This finding was confirmed by the 1H NMR spectra, which indicated the absence of the –N=CH– and hydrazone–NH-N=C protons. Based on this finding, the isolated products were assigned the structure 8c,d (Scheme 2). The conversion of 7 into 8 is reminiscent of other related oxidative cyclization of aldehyde N-heteroarylhydrazones with iron(III) chloride, which have been reported to proceed via generation of the respective nitrilimines, which undergo in situ 1,5-electrocyclization to give the respective fused heterocycles [43,44]. Similarly, refluxing of amino derivative 6b with 4-chlorobenzaldehyde and 4-bromobenzaldehyde and under the same reaction conditions afforded the products 10c,d via 9c,d (Scheme 3).
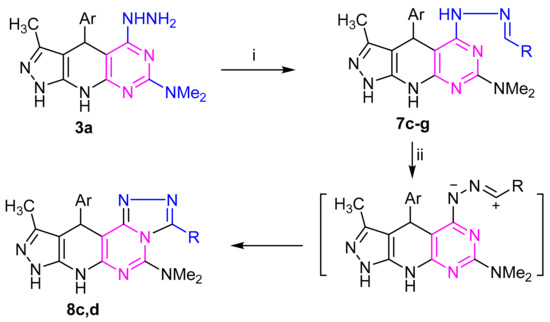
Scheme 2.
Reagents: (i) RCHO: R= (c) C6H4Cl (4), (d) C6H4Br (4), (e) C6H4Me (4), (f) glucosyl, (g) xylosyl; (ii) FeCl3/EtOH or KI/DMSO; Ar = (a) C6H4NO2 (4).
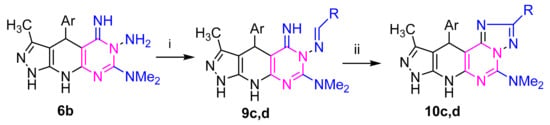
Scheme 3.
Reagents: (i) RCHO: R = (c) C6H4Cl(4), (d) C6H4Br(4); (ii) FeCl3/EtOH or KI/DMSO; Ar= (b) thien-2-yl.
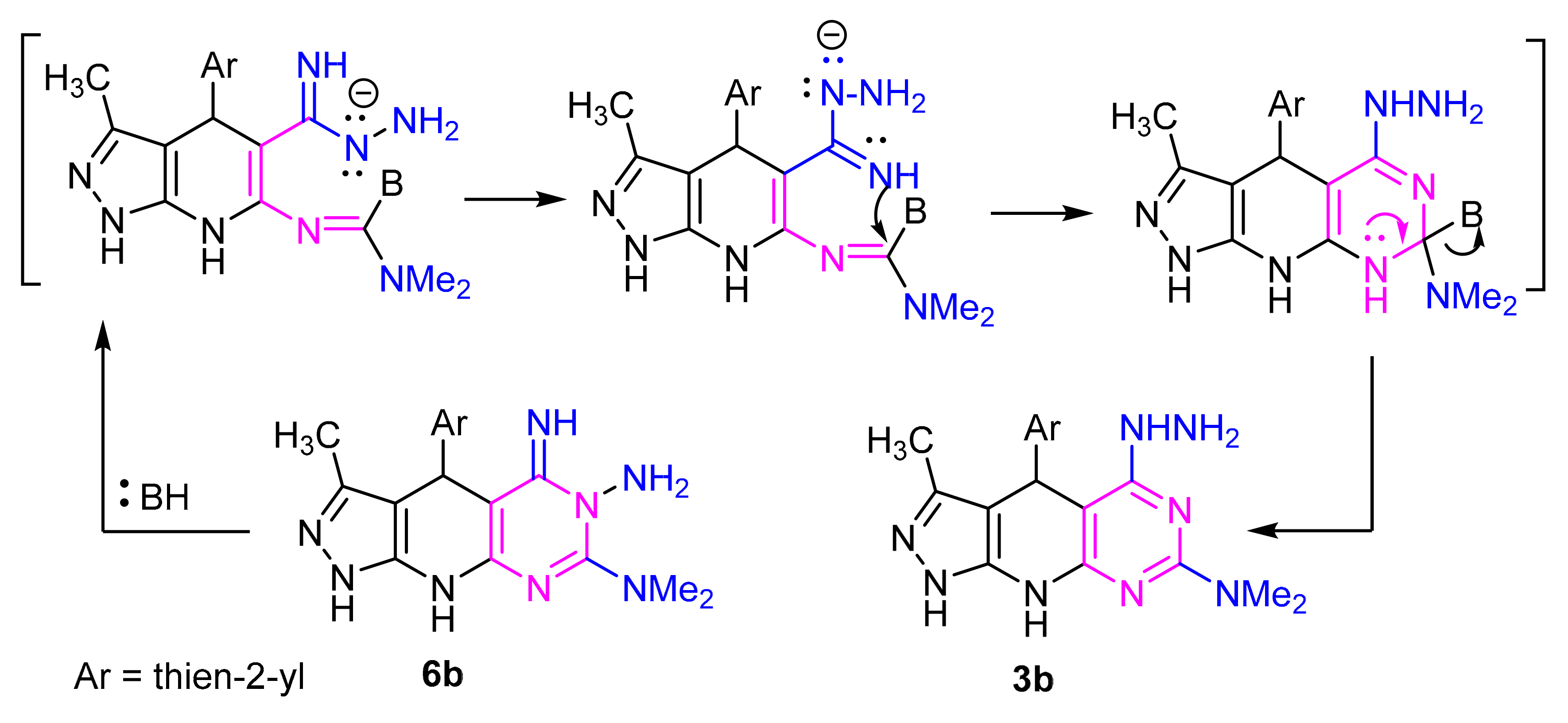
When the pyrazolo[4,3-e][1,2,4]triazolo[4,3-c]pyrimidine derivative 6b was heated in ethanol in the presence of sodium acetate, it gave a product which proved identical in all respects (mp, mixed mp, IR and 1H NMR spectra) with 3b. It has been found that compound 6b isomerized to the thermodynamically more stable pyrazolo[4,3-e][1,2,4] [1,5-c]pyrimidine derivative 3b through tandem ring opening and ring closure reactions (Scheme 4). This rearrangement is consistent with those reported in some earlier reports [45,46].
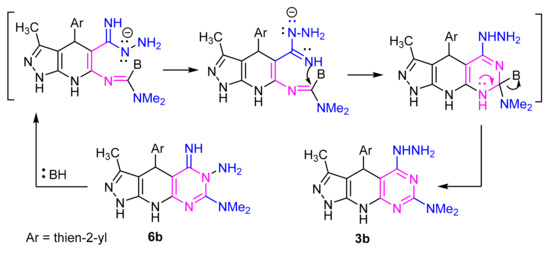
Scheme 4.
Mechanism Dimroth rearrangement.
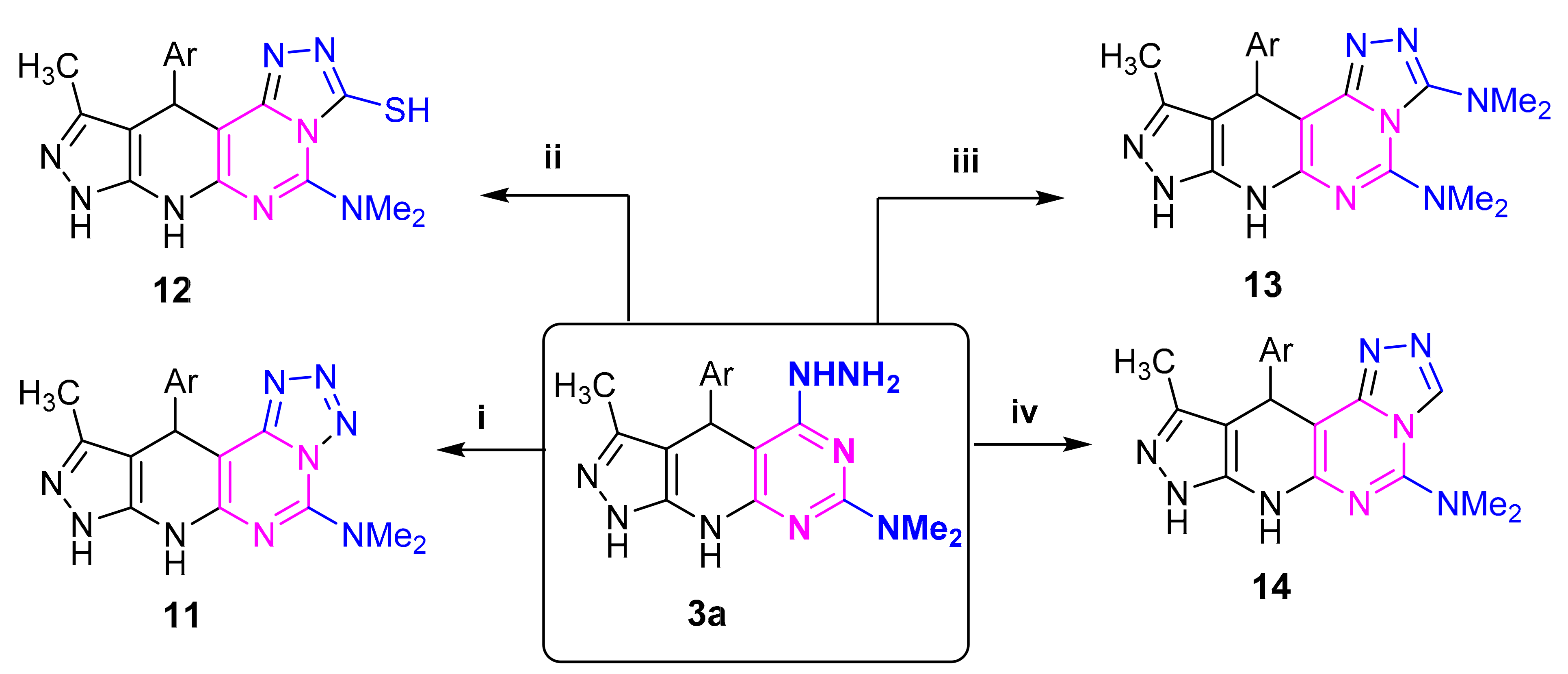
Compound 3a was utilized for synthesis of several new compounds. So, product 11 was obtained by stirring 3a with sodium nitrite in acetic acid. Compound 3a reacted with carbon disulfide to yield triazolopyrimidinethione 12. Compound 3a was converted into compound 13 by reaction with triethyl orthoformate. Dimethyldichloromethylenirninium chloride condensed with 3a to yield 14 (Scheme 5). Structural assignments of all compounds 11–14 were based on their elemental analyses and spectroscopic data (c.f., experimental).
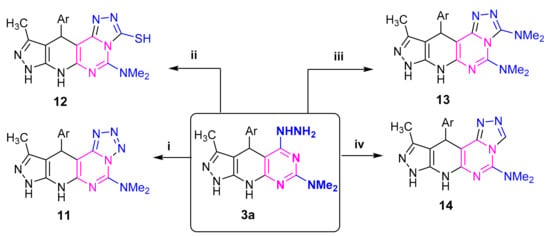
Scheme 5.
Reagents: (i) NaNO2/AcOH (ii) CS2; (iii) HC(OEt)3 (iv) Me2N=C(Cl)2Cl; Ar = C6H4NO2 (p).
2.2. Antiviral Assay
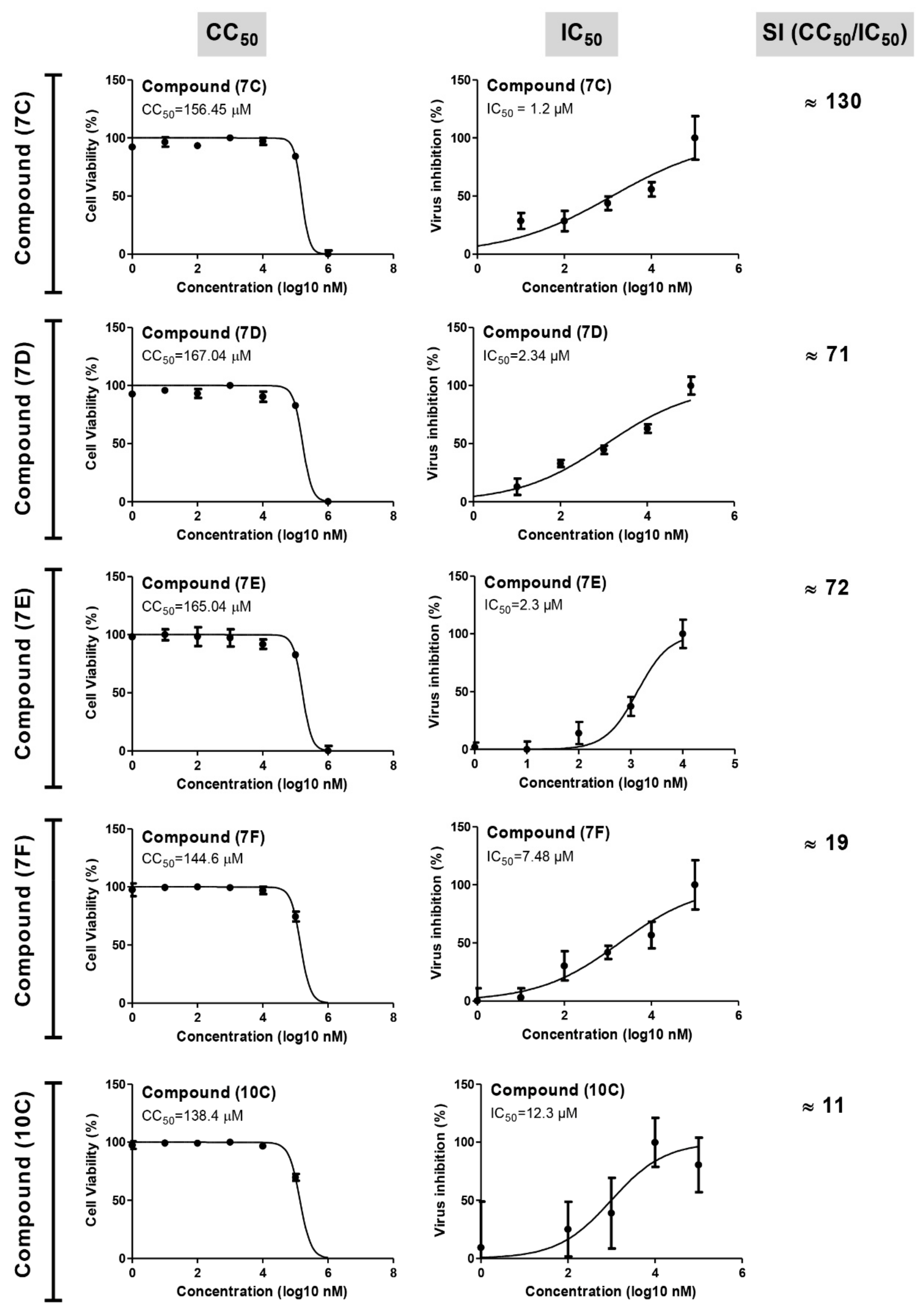
To experimentally evaluate the antiviral activity of the compounds showing promising activity based on the virtual analysis, the cytotoxicity (CC50) and antiviral activity (IC50) were evaluated (Figure 3 and Table 1). Compound 7c (IC50 = 1.2), 7d (IC50 = 2.34) and 7E (IC50 = 2.3) displayed promising antiviral activity against SARS-CoV-2 (Alpha strain, isolate hCoV-19/Egypt/NRC-3/2020 SARS-CoV-2 “NRC-03-nhCoV” virus) [47] with wider selectivity indices (SI = 71–130). Compounds 7f and 10c showed a moderate potency and a lower selectivity indices (SI = 11–19) (Figure 3). Interestingly, the compounds 7c–7e showed better IC50 and SI values when compared to the commonly used protease inhibitor [48], namely, Lopinavir (IC50 = 5.246, SI = 8.57).
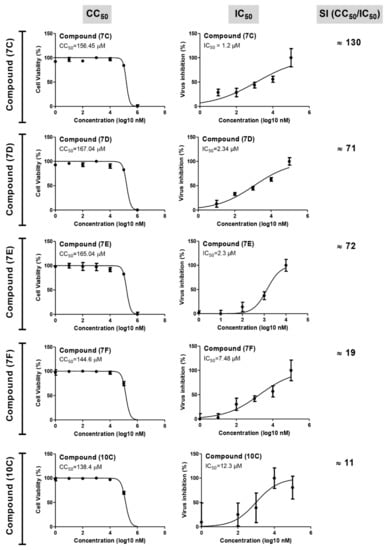
Figure 3.
Cell viability and virus inhibition assays for the novel tested anti-viral drugs in Vero-E6 cells. Various dilutions of the drugs were applied to the 90% confluent cell monolayers and assayed after 72 h with MTT assay.

Table 1.
The half maximal cytotoxic concentrations “CC50” and the half maximal inhibitory concentrations “IC50” were calculated using nonlinear regression analysis on GraphPad Prism software (version 5.01, GraphPad Software, Boston, MA, USA) by plotting log inhibitor versus normalized response (variable slope). The selectivity index (SI) values were calculated for each compound by dividing its CC50 against its IC50.
Based on anti-viral assay, we would endorse compound 7c for further detailed investigation, as it showed the best selectivity index (130), which is even better than the Lopinavir drug.
2.3. Molecular Docking
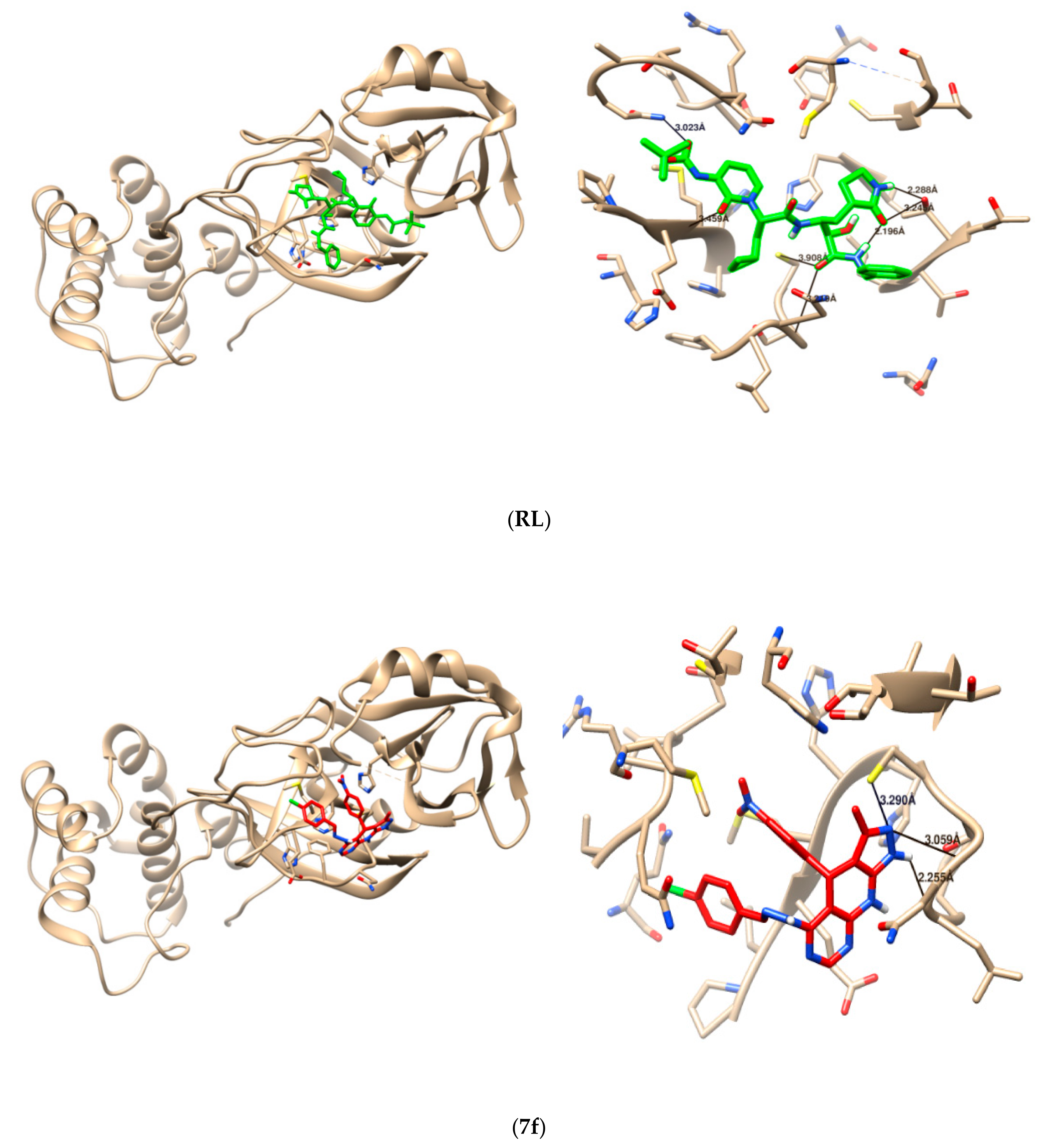
To pick up the mode of action of the tested compounds, a molecular-docking study was utilized to determine the binding modes against SARS-CoV-2 main protease “Mpro”, which are significant targets to create anti-SARS-CoV2 agents. These targets were chosen dependent on their key roles in viral protein formation; therefore, targeting these proteins give potential advantages in killing the virus. The co-crystalized ligand “{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanyl-idene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-amino]-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate” was re-docked to guarantee the validity of the docking parameters and methods using Auto Dock vina to represent the position and orientation of the ligand recognized in the crystal structure. The distinction of RMSD value between co-crystalized ligands to the original co-crystal ligand was <2 Å, which affirmed the accuracy of the docking protocols and parameters. As reference ligand, the co-crystalized ligand interacted with the active site of the Mpro protein via 5 hydrogen bonds, with the active amino acid residues (THR26, GLU166, CSY145, CYS145, SER144) with bond distance (3.139, 3.299, 2.890, 2.959, 3.023, respectively), and the value of the free binding energy was –7.5 Kcal/mol (Table 2 and Table 3) (Figure 3). Comparing our tested compounds based on their binding to Mpro protein, 6 compounds (7c, 7d, 7e, 7f, 10c, and 10d) showed preferable binding affinity to the Mpro protein more than the co-crystalized ligand did, which was evidenced by the lower values of their free binding energy (−8.4, −8.3, −8.5, −8.0, −8.1, and −8.1 Kcal/mol, respectively), and by the hydrogen bond interaction with the key amino acid residues in the active site of the Mpro protein (Table 2 and Table 3) (Figure 4). Based on the docking results, we have selected the most promising tested compounds, and ordered them according to their activities (7e > 7c > 7d > 7f >10c = 10d). Therefore, these compounds may have interesting applications against the Alpha variant of SARS-CoV-2. It is encouraging to expand this series via the synthesis of more analogues in an optically pure form, and to test them all experimentally.

Table 2.
The results of molecular docking of tested compounds against SARS-CoV-2 main protease “Mpro”.

Table 3.
The results of Hydrogen bond interaction of tested compounds against SARS-CoV-2 main protease “Mpro”.
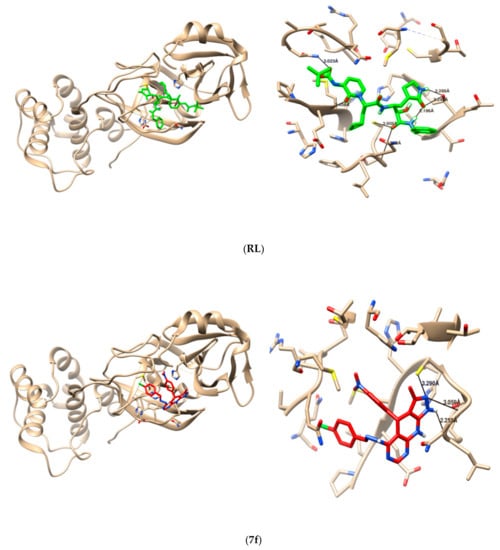
Figure 4.
3D interaction (left), and hydrogen bond formation (right), between reference ligand (RL), and tested compound 7f with 6Y2F protein.
3. Structure Activity Relationships
The structure-based activity analysis (SAR) of synthesized compounds revealed the compounds having an electron-withdrawing group bonded to the phenyl ring as shown in 7c (IC50 = 1.2), 7d (IC50 = 2.34), and an electron-donating group bonded to the phenyl ring as observed in 7e (IC50 = 2.3), displaying promising antiviral activity against SARS-CoV-2 with wider selectivity indices (SI = 71–130). The presence of a sugar moiety in compounds 7f, 7g, 10f, and 10g did not result in increased antiviral activity. Compound 10c showed moderate activity.
4. Materials and Methods
4.1. General
All melting points were measured on a Gallenkamp Melting point apparatus and are uncorrected. The IR spectra were recorded on a Shimadzu FT-IR 8101 PC infrared spectrophotometer (Shimadzu, Tokyo, Japan) using KBr disks. The NMR spectra were preserved on a Varian Mercury VX-400 NMR spectrometer (Varian, Palo Alto, CA, USA). 1H NMR spectra were run at 400 MHz and 13C NMR spectra were run at 75.46 MHz in deuterated chloroform (CDCl3) or dimethyl sulfoxide (DMSO-d6) as specified in individual compound characterizations. Chemical shifts are given in parts per million and were referenced to those of the solvents. Mass spectra were recorded on a Shimadzu GCMS-QP 1000 EX mass spectrometer at 70 eV. Elemental analyses were registered on an Elementar-Vario EL (Germany) automatic analyzer.
4.2. Synthetic Procedures
4.2.1. General Procedure for Synthesis of Starting 6-Amino-3-Methyl-4-Aryl-Pyrazolo[3,4-b]Pyridine-5-Carbonitrile 1a,b
A solution of pyrazolone 1 (0.98 g, 0.01 mol), arylaldehyde (0.01 mol), and malononitrile (0.66 g, 0.01 mol) in ethanol (10 mL) containing Ammonium acetate (0.98 gm, 2% excess) or piperidine (1 mL) was heated under reflux for 5 h. The solvent was evaporated under vacuum and the remaining solids were treated with the proper solvent for crystallization. The spectroscopic data and melting points of 1a,b agreed with those reported [33].
4.2.2. General Procedure for Synthesis of Compounds 2a, 2b, and 4b
A solution of 1a, b (10 mmol) and phosogen iminiumchloride (11 mmol) in dry 1,2-dichloroethane (100 mL) was stirred at the temperature and time mentioned in the Scheme 1. The precipitated solid was collected by filtration and recrystallized from dioxane to afford the title products.
5-Chloro-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido-[2,3-d]pyrimidin-7-amine (2a)
Yield (65%) as yellow powder; mp 213-215 °C. IR (KBr, υmax, cm−1): 3428 (NH), 2927 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.09 (s, 3H, CH3), 3.23 (s, 6H, N-Me2), 5.50 (s, 1H, CH), 7.53–7.55 (m, 3H, Ar-H and NH), 8.16–8.18 (d, 2H, J = 8.6 Hz, Ar-H), 12.05 (s, 1H, NH, exchangeable with D2O). 13C NMR (DMSO-d6) δC ppm: 14.6, 32.0, 35.3, 94.8, 103.9, 105.8, 111.9, 116.5, 121.7, 127.6, 132.9, 139.3, 144.6, 147.6, 158.3, 161.3, 163.3. (m/z, %) (386, 10). Anal. Calcd. for C17H16ClN7O2 (385.81): C, 52.92; H, 4.18; Cl, 9.19; N, 25.41;. Found: C, 52.99; H, 4.27; Cl, 9.12; N, 25.32%.
5-Chloro-N,N,3-trimethyl-4-(thiophen-2-yl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido-[2,3-d]pyrimidin-7-amine (2b)
Yield (69%) as yellow powder; mp 228–232 °C. IR (KBr, υmax, cm−1): 3432 (NH), 2935 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.11 (s, 3H, CH3), 3.16 (s, 6H, N-Me2), 5.48 (s, 1H, CH), 6.89 (m, 3H, thiophene-H and NH), 7.25 (d, 1H, J = 4.9 Hz, thiophene-H), 11.90 (s, 1H, NH, exchangeable with D2O); MS (m/z, %) (346, 48). Anal. Calcd. for C15H15ClN6S (346.84): C, 51.95; H, 4.36; Cl, 10.22; N, 24.23; S, 9.24. Found: C, 51.84; H, 4.45; Cl, 10.27; N, 24.19; S, 9.28%.
N′-(5-Cyano-3-methyl-4-(thiophen-2-yl)-4,7-dihydro-1H-pyrazolo[3,4-b]pyridin-6-yl)-N,N-dimethylcarbamimidic chloride (4b)
Yield: 60%, mp 151–152 °C. IR (KBr, υmax, cm−1): 3440 (NH), 2212 (CN), 2998 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.14 (s, 3H, CH3), 3.15 (s, 6H, N-Me2), 5.46 (s, 1H,CH), 6.89 (m, 3H, thiophene-H, NH), 7.25 (d,1H, J = 4.9 Hz, thiophene-H),; 11.90 (br s, 1H, NH, exchangeable with D2O). MS (m/z, %) (346, 48). Anal. Calcd. for C15H15ClN6S (346.84): C, 51.95; H, 4.36; Cl, 10.22; N, 24.23; S, 9.24. Found: C, 51.84; H, 4.45; Cl, 10.27; N, 24.19; S, 9.28%.
4.2.3. Synthesis of 3a, 3b, 6b
A solution of 2a, b, and 4b (10 mmol) and hydrazine hydrate (11 mmol) in EtOH (40 mL) were stirring for 10 h at room temperature. The products 3a, 3b, and 6b were collected by filtration and recrystallized from dioxane.
5-Hydrazineyl-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]-pyrido[2,3-d]pyrimidin-7-amine (3a)
Yield (61%) as brown powder; mp 230–232 °C. IR (KBr, υmax, cm−1): 3430 (NH + NH2), 2923 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 1.96 (s, 3H, CH3), 3.09 (s, 6H, N-Me2), 4.36 (br, 2H, NH2, exchangeable with D2O), 5.26 (s, 1H, CH), 7.56–7.67 (m, 2H, Ar-H and NH), 7.70–8.1 (d, 2H, J = 8.6 Hz, Ar-H), 8.18 (m, 2H, Ar-H), 12.06 (s, 1H, NH, exchangeable with D2O). 13C NMR (DMSO-d6) δC ppm: 21.0, 29.1, 30.4, 98.6, 99.3, 109.8, 115.8, 124.8, 128.7, 129.4, 135.4, 139.9, 143.4, 149.0, 158.1, 160.5, 166.8. MS (m/z, %) (381, 60). Anal. Calcd. for C17H19N9O2 (381.40): C, 53.54; H, 5.02; N, 33.05. Found. C, 53.60; H, 5.08; N, 33.10%.
5-Hydrazineyl-N,N,3-trimethyl-4-(thiophen-2-yl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]-pyrido[2,3-d]pyrimidin-7-amine (3b)
Yield (65%) as brown powder; mp 236–238 °C. IR (KBr, υmax, cm−1): 3436 (NH + NH2), 2929 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.12 (s, 3H, CH3), 3.39 (s, 6H, N-Me2), 5.37 (s, 1H, CH), 5.45 (br s, 2H, NH2, exchangeable with D2O), 6.76–6.83 (m, 3H, thiophene-H and NH), 7.23 (d, 1H, J = 4.9 Hz, thiophene-H), 8.83 (br s, 1H, NH, exchangeable with D2O), 12.11 (br s, 1H, NH, exchangeable with D2O). MS (m/z, %) (342, 68). Anal. Calcd. for C15H18N8S (342.43): C, 52.61; H, 5.30; N, 32.72; S, 9.36. Found. C, 52.52; H, 5.26; N, 32.84; S, 9.39%.
5-Imino-N,N,3-trimethyl-4-(thiophen-2-yl)-1,4,5,9-tetrahydro-6H-pyrazolo[4′,3′:5,6]-pyrido[2,3-d]pyrimidine-6,7-diamine (6b)
Yield (60%) as brown powder; mp 239–241 °C. IR (KBr, υmax, cm−1): 3425 (NH + NH2), 2925 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.13 (s, 3H, CH3), 3.35 (s, 6H, N-Me2), 4.73 (s, 2H, NH2, exchangeable with D2O), 5.50 (s, 1H, CH), 6.99–7.11 (m, 3H, thiophene-H and NH), 7.35(d, 1H, J = 4.9 Hz, thiophene-H), 11.28 (s, 1H, NH, exchangeable with D2O); 12.34 (s, 1H, NH, exchangeable with D2O). 13C NMR (DMSO-d6) δC ppm: 14.8, 27.7, 30.1, 89.6, 99.0, 115.8, 124.8, 129.7, 131.8, 139.2, 139.6, 143.4, 157.8, 159.8, 160.5. MS (m/z, %) (342, 63). Anal. Calcd. for C15H18N8S (342.43): C, 52.61; H, 5.30; N, 32.72; S, 9.36. Found. C, 52.43; H, 5.10; N, 32.92; S, 9.46%.
4.2.4. General Procedure for the Synthesis of 7c–g and 9c,d
To a mixture of 1 (0.6 g, 2.5 mmol) and the appropriate aldose 2a–d (2.5 mmol) in ethanol (15 ml), a catalytic amount of glacial acetic acid (0.1 ml) was added
To a mixture of derivatives 3a or 6b (10 mmol) and the appropriate aldehyde (10 mmol) or respective monosaccharides (10 mmol) in ethanol (30 mL), a catalytic amount of glacial acetic acid (1.0 mL) was added. The reaction mixture was refluxed for eight hours. After cooling at room temperature, the precipitated solid was collected by filtration and recrystallised from the proper solvent.
5-(2-(4-Chlorobenzylidene)hydrazineyl)-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (7c)
Yield (70%) as brown powder; mp 189–191 °C. IR (KBr, υmax, cm−1): 3417 (NH), 2930 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.17 (s, 3H, CH3), 3.11 (s, 6H, N-Me2), 6.08 (s, 1H, CH), 6.85–6.89 (m, 3H, Ar-H and NH), 7.22–7.42 (d, 2H, J = 8.4 Hz, Ar-H), 7.50–7.52 (m, 2H, J = 8.4Hz, Ar-H), 7.68-7.80 (d, 2H, J = 8.4 Hz, Ar-H), 8.12 (s, 1H, N=CH), 10.52 (s, 1H, NH, exchangeable with D2O), 12.12 (s, 1H, NH, exchangeable with D2O). 13C NMR (DMSO-d6) δC ppm: 21.1, 30.1, 30.4, 89.9, 99.3, 115.8, 119.24, 119.9, 120.3, 120.7, 121.4, 124.5, 129.1, 132.9, 134.9, 136.7, 138.1, 139.9, 143.7, 147.9, 157.8, 159.8, 166.5, 167.8. MS (m/z, %) (503, 53). Anal. Calcd. for C24H22ClN9O2 (503.95): C, 57.20; H, 4.40; Cl, 7.03; N, 25.01. Found. C, 57.38; H, 4.61; Cl, 6.88; N, 24.80%.
5-(2-(4-Bromobenzylidene)hydrazineyl)-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (7d)
Yield (67%) as brown powder; mp 180–182 °C. IR (KBr, υmax, cm−1): 3422 (NH), 2940 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.13 (s, 3H, (CH3), 3.34 (s, 6H, N-Me2), 5.85 (s, 1H, CH), 7.43–7.53 (m, 3H, Ar-H and NH), 7.55–7.60 (d, 2H, J = 8.4 Hz, Ar-H), 7.67 (m, 2H, J = 8.4Hz, Ar-H), 7.92 (d, 2H, J = 8.4 Hz, Ar-H), 8.07 (s, 1H, N=CH), 10.63 (s, 1H, NH, exchangeable with D2O), 12.19 (s, 1H, NH, exchangeable with D2O). MS (m/z, %) (548, 71). Anal. Calcd. for C24H22BrN9O2 (548.41): C, 52.56; H, 4.04; Br, 14.57; N, 22.99. Found. C, 52.39; H, 4.29; Br, 14.31; N, 23.12%.
(E)-N,N,3-Trimethyl-5-(2-(4-methylbenzylidene)hydrazineyl)-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (7e)
Yield (66%) as brown powder; mp 188–190 °C. IR (KBr, υmax, cm−1): 3420 (NH), 2945 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.17 (s, 3H, CH3), 2.43 (s, 3H, CH3), 3.37 (s, 6H, N-Me2), 5.91 (s, 1H, CH), 7.31–7.35 (m, 3H, Ar-H and NH), 7.49 (d, 2H, J = 8.4 Hz, Ar-H), 7.71 (d, 2H, J = 8.4 Hz, Ar-H), 7.95 (d, 2H, J = 8.4 Hz, Ar-H), 8.09 (s, 1H, N=CH), 10.56 (s, 1H, NH, exchangeable with D2O), 12.06 (s, 1H, NH, exchangeable with D2O). MS (m/z, %) (483, 66). Anal. Calcd. for C25H25N9O (483.54): C, 62.10; H, 5.21; N, 26.07; O, 6.62. Found. C, 62.29; H, 5.08; N, 26.27%.
5-(2-(4-Glucosylhydrazinyl)hydrazineyl)-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (7f).
Yield (75%) as brown powder; mp 173–175 °C. IR (KBr, υmax, cm−1): 3419-3347 (OH + NH). 1H NMR (DMSO-d6) δH ppm: 2.13 (s, 3H, CH3), 3.10 (s, 6H, N-Me2), 3.37 (m, 2H, H-6′ and H-6′′), 3.41 (m, 1H, H-5′), 4.27–4.29 (d, 1H, H-4′), 4.44–4.51 (m, 2H, H-3′ and H-2′), 4.85–4.92 (m, 1H, OH), 5.53 (m, 1H, OH), 5.55 (s, 1H, CH), 5.90 (m, 1H, OH), 6.21 (m, 1H, OH), 6.43 (br s, 1H, NH, exchangeable with D2O), 6.59 (m, 1H, OH), 6.99–7.00 (d, 1H, J = 7.6 Hz, H-1), 7.10–7.12 (m, 3H, Ar-H and NH), 7.32–7.36 (d, 2H, J = 8.4 Hz Ar-H), 12.29 (s, 1H, NH, exchangeable with D2O). 13C NMR (DMSO-d6) δC ppm: 10.2, 34.7, 36.9, 37.1, 61.6, 70.7, 72.4, 75.5, 77.2, 92.7, 97.3, 99.6, 101.9, 124.7, 124.9, 125.0, 127.1, 136.8, 149.5, 150.8, 159.8, 162.1, 164.9. Anal. Calcd. for C23H29N9O7 (543.54): C, 50.82; H, 5.38; N, 23.19. Found. C, 50.20; H, 5.51; N, 23.10%.
5-(2-(4-Xylosylhydrazinyl)hydrazineyl)-N,N,3-trimethyl-4-(4-nitrophenyl)-4,9-dihydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (7g)
Yield (72%) as brown powder; mp 165–167 °C. IR (KBr, υmax, cm−1): 3415-3340 (OH + NH). 1H NMR (DMSO-d6) δH ppm: 2.10 (s, 3H, CH3), 3.33 (s, 6H, N-Me2), 3.54–4.03 (m, 2H, H-5′ and H-5′′), 4.29–4.45 (m, 2H, H-3′ and H-4′), 4.95 (m, 1H, H-2′), 5.13 (m, 1H, OH), 5.54 (s, 1H, CH), 5.85 (m, 1H, OH), 5.94 (m, 1H, OH), 6.10 (s, 1H, NH, exchangeable with D2O), 6.25 (m, 1H, OH), 7.11 (d, 1H, J = 7.6 Hz, H-1′), 7.34–7.39 (m, 3H, Ar-CH and NH), 7.59–7.71 (d, 2H, J = 8.4 Hz, Ar-CH), 11.99 (br s, 1H, NH, exchangeable with D2O); 13C NMR (DMSO-d6) δC ppm: 14.6, 29.3, 36.6, 37.3, 66.1, 70.1, 75.1, 77.1, 93.2, 98.5, 100.5, 102.5, 124.6, 125.3, 129.9, 134.9, 139.2, 145.9, 152.3, 161.7, 162.7, 165.1. Anal. Calcd. for C22H27N9O6 (513.52): C, 51.46; H, 5.30; N, 24.55. Found. C, 51.66; H, 5.10; N, 24.50%.
4.2.5. General Procedure for the Synthesis of 8c,d and 10c,d
Method A: Derivatives (7c,d) or (9c,d) (5mmol) were dissolved in DMF (15 mL) and (5 mmol) of potassium iodide; the mixture was stirred and heated at 110 °C for 20 h. The reaction was allowed to cool and the precipitate isolated by filtration and washed with a little methanol and recrystallized from dioxane.
Method B: 2 M solution of iron (III) chloride in ethanol (2 mL) was added dropwise to a boiling solution of (7c,d) or (9c,d) (10 mmol) in ethanol (50 mL). Heating was continued for 20 min and the mixture was then kept overnight at room temperature. The reaction was allowed to cool and the precipitate isolated by filtration and washed with a little methanol and recrystallized from dioxane.
3-(4-Chlorophenyl)-N,N,10-trimethyl-11-(4-nitrophenyl)-8,11-dihydro-7H-pyrazolo-4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-5-amine (8c)
Yield (63%) as brown powder; mp 250–252 °C. IR (KBr, υmax, cm−1): 3430 (NH). 1H NMR (DMSO-d6) δH ppm: 2.31 (s, 3H, CH3), 3.36 (s, 6H, N-Me2), 5.84 (s, 1H, CH), 7.33–7.53 (m, 3H, Ar-CH and NH), 7.74 (d, 2H, J = 8.4 Hz, Ar-CH), 8.09 (d, 2H, J = 8.4 Hz, Ar-CH), 8.18 (d, 2H, J = 8.4 Hz, Ar-CH), 12.03 (s, 1H, NH, exchangeable with D2O). Anal. Calcd. for C24H20ClN9O2 (501.94): C, 57.43; H, 4.02; Cl, 7.06; N, 25.12. Found: C, 57.60; H, 4.20; Cl, 6.90; N, 25.05%.
3-(4-Bromophenyl)-N,N,10-trimethyl-11-(4-nitrophenyl)-8,11-dihydro-7H-pyrazolo-[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-5-amine (8d)
Yield (70%) as brown powder; mp 247–249 °C. IR (KBr, υmax, cm−1): 3420 (NH). 1H NMR (DMSO-d6) δH ppm: 2.29 (s, 3H, CH3), 3.38 (s, 6H, N-Me2), 5.80 (s, 1H,CH), 7.50–7.55 (m, 3H, Ar-CH, NH), 7.70 (d, 2H, J = 8.4 Hz, Ar-CH), 8.00 (d, 2H, J = 8.4 Hz, Ar-CH), 8.20 (d, 2H, J = 8.4 Hz, Ar-CH), 12.20 (s, 1H, NH, exchangeable with D2O). Anal. Calcd. for C24H20BrN9O2 (546.39): C, 52.76; H, 3.69; Br, 14.62; N, 23.07. Found: C, 52.70; H, 3.59; Br, 14.55; N, 23.30%.
(E)-6-((4-Chlorobenzylidene)amino)-5-imino-N,N,3-trimethyl-4-(thiophen-2-yl)-4,5,6,9-tetrahydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (9c)
Yield (68%) as brown powder; mp 183–185 °C. IR (KBr, υmax, cm−1): 3436 (NH), 2936 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.13 (s, 3H, CH3), 3.39 (s, 6H, N-Me2), 6.03 (s, 1H, CH), 7.19–7.27 (m, 3H, thiophene-H, NH), 7.47 (d, 1H, J = 4.9 Hz, thiophene-H), 7.54 (d, 2H, J = 8.4 Hz, Ar-H), 7.64 (d, 2H, J = 8.4 Hz, Ar-H), 8.07 (s, 1H, N=CH), 11.05 (s, 1H, NH, exchangeable with D2O), 12.12 (s, 1H, NH, exchangeable with D2O). MS (m/z, %) (464, 59). Anal. Calcd. for C22H21ClN8S (464.98): C, 56.83; H, 4.55; Cl, 7.62; N, 24.10; S, 6.89. Found. C, 56.58; H, 4.78; Cl, 7.47; N, 24.230; S, 6.80%.
(E)-6-((4-Bromobenzylidene)amino)-5-imino-N,N,3-trimethyl-4-(thiophen-2-yl)-4,5,6,9-tetrahydro-1H-pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-7-amine (9d)
Yield (60%) as brown powder; mp 199–201 °C. IR (KBr, υmax, cm−1): 3410 (NH), 2923 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.18 (s, 3H, CH3), 3.40 (s, 6H, N-Me2), 6.03 (s, 1H, CH), 6.89–6.92 (m, 3H, thiophene-H and NH), 7.30 (d, 1H, J = 4.9 Hz, thiophene-H), 7.58 (d, 2H, J = 8.4 Hz, Ar-H), 7.70 (d, 2H, J =8.4 Hz, Ar-H), 8.23 (s, 1H, N=CH), 10.50 (s, 1H, NH, exchangeable with D2O), 12.00 (s, 1H, NH, exchangeable with D2O). MS (m/z, %) (509, 41). Anal. Calcd. for C22H21BrN8S (509.43): C, 51.87; H, 4.16; Br, 15.68; N, 22.00; S, 6.29. Found. C, 51.57; H, 4.40; Br, 15.40; N, 22.31; S, 6.21%.
2-(4-Chlorophenyl)-N,N,10-trimethyl-11-(thiophen-2-yl)-8,11-dihydro-7H-pyrazolo-[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine (10c)
Yield (60%) as brown powder; mp 219–221 °C. IR (KBr, υmax, cm−1): 3448 (NH). 1H NMR (DMSO-d6) δH ppm: 2.13 (s, 3H, CH3), 3.41 (s, 6H, N-Me2), 5.80 (s, 1H, CH), 6.79–7.01 (m, 3H, thiophene-H and NH), 7.23 (d, 1H, J = 4.9 Hz, thiophene-H), 7.51 (d, 2H, J = 8.4 Hz, Ar-H), 7.69 (d, 2H, J = 8.4 Hz, Ar-H), 11.94 (s, 1H, NH, exchangeable with D2O). MS (m/z, %) (462, 18). Anal. Calcd. for C22H19ClN8S (462.96): C, 57.08; H, 4.14; Cl, 7.66; N, 24.20; S, 6.93. Found: C, 57.20; H, 4.30; Cl, 7.60; N, 24.06; S, 6.89%.
2-(4-Bromophenyl)-N,N,10-trimethyl-11-(thiophen-2-yl)-8,11-dihydro-7H-pyrazolo-[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine (10d)
Yield (67%) as brown powder; mp 222–224 °C. IR (KBr, υmax, cm−1): 3440 (NH). 1H NMR (DMSO-d6) δH ppm: 2.12 (s, 3H, CH3), 3.47 (s, 6H, N-Me2), 5.82 (s, 1H, CH), 6.75-6.80 (m, 3H, thiophene-H and NH), 7.27 (d, 1H, J = 4.9 Hz, thiophene-H), 7.59 (d, 2H, J = 8.4 Hz, Ar-H), 7.73 (d, 2H, J = 8.4 Hz, Ar-H), 11.97 (s, 1H, NH, exchangeable with D2O). Anal. Calcd. for C22H19BrN8S (507.41): C, 52.08; H, 3.77; Br, 15.75; N, 22.08; S, 6.32. Found: C, 52.18; H, 3.80; Br, 15.89; N, 21.88; S, 6.11%.
N,N,10-Trimethyl-11-(4-nitrophenyl)-8,11-dihydro-7H-pyrazolo[4′,3′:5,6]-pyrido[3,2-e]tetrazolo[1,5-c]pyrimidin-5-amine (11)
A solution of 3a (4.01 g, 10 mmol) and sodium nitrite (0.69 g, mmol) in acetic acid (50 mL) was stirred at room temperature for 24 h. Cold distilled water was added and the precipitate was collected by filtration and crystallized from dioxane. Yield (70%) as yellow powder; mp 225–227 °C; IR ν 3425 cm−1 (NH), 2928 cm−1 (CH alkyl); 1H NMR (DMSO-d6) δH ppm: 2.15 (s, 3H, CH3), 3.43 (s, 6H, N-Me2), 5.52 (s, 1H, CH), 7.66–7.75 (m, 3H, Ar-H and NH), 8.20 (d, 2H, J = 8.3 Hz, Ar-H), 12.13 (s, 1H, NH, exchangeable with D2O). Anal. Calcd. for C17H16N10O2 (392.38): C, 52.04; H, 4.11; N, 35.70. Found. C, 52.21; H, 3.91; N, 35.61%.
5-(Dimethylamino)-10-methyl-11-(4-nitrophenyl)-2,7,8,11-tetrahydro-3H-pyrazolo-[4′,3′:5,6]pyrido[3,2-e][1,2,4]triazolo[4,3-c]pyrimidine-3-thione (12)
To a warm ethanolic sodium hydroxide solution (prepared by refluxing (0.40 g, 10 mol) of NaOH in abs. EtOH (50 mL)) was added (10 mmol) of compound 3a and carbon disulfide (15 mmol). The mixture was heated on a water bath for 6 h, then allowed to cool, poured into water, and neutralized with diluted HCl. The solid product was collected by filtration and crystallized from benzene. Yield (62%) as brown powder; mp 210–212 °C. IR (KBr, υmax, cm−1): 3430 (NH), 2934 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.19 (s, 3H, CH3), 3.38 (s, 6H, N-Me2), 5.60 (s, 1H, CH), 7.61–7.68 (m, 3H, Ar-H and NH), 8.16 (d, 2H, J = 8.3 Hz, Ar-H), 8.23 (s, 1H, -SH), 12.10 (br s, 1H, NH, exchangeable with D2O); (M-1, %) (422, 2). Anal. Calcd. for C18H17N9O2S (423.46): C, 51.06; H, 4.05; N, 29.77; S, 7.57. Found. C, 51.20; H, 4.19; N, 29.52; S, 7.30%.
N,N,10-Trimethyl-11-(4-nitrophenyl)-8,11-dihydro-7H-pyrazolo[4′,3′:5,6]pyrido[3,2-e]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine (13)
A solution of 3a (10 mmol) in triethyl orthoformate (25 mL) was stirred at 70 °C for 9 h and then cooled overnight. The solid product, so-formed, was collected by filtration and crystallized from methanol.Yield (67%) as brown powder; mp 215–217 °C. IR (KBr, υmax, cm−1): 3408 (NH), 2947 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.12 (s, 3H, CH3), 3.44 (s, 6H, N-Me2), 5.61 (s, 1H, CH), 7.57-7.60 (m, 3H, Ar-H and NH), 8.10 (d, 2H, J = 8.3 Hz, Ar-H), 8.55 (s, 1H, CH),12.10 (s, 1H, NH, exchangeable with D2O). (m/z, %) (391, 4). Anal. Calcd. for C18H17N9O2 (391.40): C, 55.24; H, 4.38; N, 32.21. Found. C, 55.15; H, 4.20; N, 32.10%.
N3,N3,N5,N5,10-Pentamethyl-11-(4-nitrophenyl)-8,11-dihydro-7H-pyrazolo[4′,3′:5,6]-pyrido[3,2-e][1,2,4]triazolo[4,3-c]pyrimidine-3,5-diamine (14)
A solution of 3a (10 mmol) in 1,2-dichloroethane (10 mL) was added to a stirred suspension of phosgene iminium chloride (10 mmol) in 1,2-dichloroethane (30 mL) at room temperature. The mixture was then refluxed for 4 h. The solid product was collected by filtration, washed with saturated solution of NaHCO3, H2O, dried and crystallized from ethanol. Yield (60%) as brown powder; mp 203–205 °C. IR (KBr, υmax, cm−1): 3419 (NH), 2946 (CH alkyl). 1H NMR (DMSO-d6) δH ppm: 2.17 (s, 3H, CH3), 3.36 (s, 6H, N-Me2), 3.38 (s, 6H, N-(CH3)2), 5.59 (s, 1H,CH), 7.68–7.75 (m,3H, Ar-H,NH), 8.19 (d, 2H, J = 8.3 Hz, Ar-H), 12.14 (s, 1H, NH, exchangeable with D2O); (m/z, %) (434, 9). Anal. Calcd. for C20H22N10O2 (434.46): C, 55.29; H, 5.10; N, 32.24. Found. C, 55.179; H, 5.22; N, 32.13%.
4.3. Cytotoxic Concentration 50 (CC50) and Viral Inhibitory Concentration 50 (IC50) Calculation
The assay was performed according to the procedure that was previously described (Feoktistova, M., Geserick, P., Leverkus, M. 2016. Crystal Violet Assay for Determining Viability of Cultured Cells. In Cold Spring Harb Protoc, pdb.prot087379) with minor modifications. Vero E6 cells were seeded into 96-well plates in 100 µL of high glucose Dulbecco’s modified Eagle’s medium (DMEM) containing medium 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C in 5% CO2. After 24 h (90–100% confluence monolayer of Vero E6), each compound was diluted into varying concentrations in a separate U-shape 96-well plate (with a range of concentration from 10 µg/mL to 1 ng/mL) using DMEM containing 2% FBS (maintenance medium). A volume of 100 µL of each dilution was transferred into a new U-shape 96-well plate and supplemented with 100 TCID50 in 100 µL maintenance medium. In parallel, the wells dedicated for CC50 calculation were supplemented with 100 µL maintenance medium without virus. Aliquots of 100 µL of infection media containing 100 TCID50 were used as virus control. After 1 h of incubation, 100 µL of each well was transferred to the corresponding wells into the 96-well plates containing Vero E6 cultures. The plates were incubated for 72 h, the cell monolayers were washed with PBS and subjected to cell fixation using 100 µL of 10% formalin for 1 h. Subsequently, the plates are washed well 3 times with 1 × PBS and dried well before staining with 50 µL (0.5%) crystal violet to each well ((0.5 g crystal violet powder (Sigma-Aldrich), 80 mL distilled H2O and 20 mL methanol)) for 30 min. The plates were then washed well with rinsed water and air-dried at room temperature for 2 to 24 h. To distain crystal violet, 200 µL methanol was added to each well, and the plate was incubated with its lid on a bench rocker (20 oscillations/minute) for 20 min at room temperature. Finally, the optical density of each well at λ 590 nm (OD590) was measured with a plate reader. The average OD of each dilution without or with virus was compared to control cells and control virus wells to calculate CC50 and IC50 values using nonlinear regression in GraphPad Prism 5.01.
4.4. Molecular Docking Study
The structures of all tested compounds were modeled using the Chemsketch software (http://www.acdlabs.com/resources/freeware/, accessed on 15 December 2021). The structures were optimized and energy minimized using the VEGAZZ software [49]. The optimized compounds were used to perform molecular docking. The three-dimensional structures of the molecular target were obtained from Protein Data Bank (PDB) (www.rcsb.org, accessed on 15 December 2021): SARS-CoV-2 (2019-nCoV) main protease M pro (PDB: 6Y2F, https://www.rcsb.org/structure/6Y2F, accessed on 15 December 2021). The steps for receptor preparation included the removal of heteroatoms (water and ions), the addition of polar hydrogen, and the assignment of charge. The active sites were defined using grid boxes of appropriate sizes around the bound cocrystal ligands. The docking study was performed using Autodock vina [50] and Chimera for visualization [51].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules28020739/s1, 1H and 13C NMR and MS spectra for the synthesized compounds: Chart S1: 1H NMR of starting material, Chart S2: 1H NMR of starting material, Chart S3: 1H NMR of Compound 2a, Chart S4: 13C NMR of Compound 2a, Chart S5: MS of Compound 2a, Chart S6: 1H NMR of Compound 3a, Chart S7: 13C NMR of Compound 3a, Chart S8: 1H NMR of Compound 6b, Chart S9: 13C NMR of Compound 6b, Chart S10: 1H NMR of Compound 7c, Chart S11: 13C NMR of Compound 7c, Chart S12: 1H NMR of Compound 7f, Chart S13: 13C NMR of Compound 7f, Chart S14: 13C NMR of Compound 7g, Chart S15: 1H NMR of Compound 7g, Chart S16: 1H NMR of Compound 8e, Chart S17: 13C NMR of Compound 8e, Chart S18: MS of Compound 11, Chart S19: MS of Compound 12, Chart S20: MS of Compound 13, Chart S21: MS of Compound 14.
Author Contributions
Conceptualization, Z.M.A., R.R.K., N.A.H., A.A.E.-S. and A.A.H.; methodology, Z.M.A., R.R.K., N.A.H., A.A.E.-S. and A.A.H.; software, M.A.T. and A.M.; validation, Z.M.A., R.R.K., M.A.T. and A.A.H.; formal analysis, Z.M.A., R.R.K., N.A.H., A.A.E.-S., M.A.T. and A.A.H.; investigation, N.A.H., A.A.E.-S. and A.A.H.; resources, N.A.H. and A.A.E.-S.; data curation, Z.M.A., R.R.K., N.A.H., A.A.E.-S. and A.A.H.; writing—original draft preparation, Z.M.A., R.R.K., A.A.E.-S., M.A.T., A.M. and A.A.H.; writing—review and editing, N.A.H., M.A.T. and A.A.H.; visualization: M.A.T., A.M., A.A.E.-S. and N.A.H.; supervision: N.A.H.; project administration, Z.M.A. and A.A.E.-S.; funding acquisition: Z.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudia Arabia, under grant No. (G-447-247-14422). The authors, therefore, acknowledge with thanks DSR and the technical and financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and the Supplementary Materials.
Acknowledgments
The authors are thankful to their respective institutions for providing the necessary facility to complete this work. The authors offered their thanks to the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudia Arabia for funding this research work through the project number (G-447-247-14422).
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the synthesized compounds are available from the authors.
References
- Wang, C.; Wang, Z.; Wang, G.; Lau, J.Y.-N.; Zhang, K.; Li, W. COVID-19 in early 2021: Current status and looking forward. Signal Transduct Target Ther. 2021, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, M.Y.; Wang, W.; Song, G.Z.; Hu, Y.; Tao, W.Z.; Tain, Y.Y.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Cao, R.; Xu, M.; Wu, Y.; Shang, W.; Wang, X.; Zhang, H.; Jiang, X.; Sun, Y.; et al. Comparative Antiviral Efficacy of Viral Protease Inhibitors against the Novel SARS-CoV-2 In Vitro. Virol. Sin. 2020, 35, 776–784. [Google Scholar] [CrossRef]
- Anand, K. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, W.; Peng, C.; Ma, X.; He, X.; Zhang, H.; Zhou, M. Discovery of novel pyrazolopyrimidine derivatives as potent mTOR/HDAC bi-functional inhibitors via pharmacophore-merging strategy. Bioorg. Med. Chem. Lett. 2021, 49, 128286. [Google Scholar] [CrossRef]
- Tuyen, N.T.; Maged, H. Synthesis and Applications of Nitrogen-Containing Heterocycles as Antiviral Agents. Molecules 2022, 27, 2700. [Google Scholar] [CrossRef]
- Ghasemi, L.; Hasanzadeh, E.M.; Abbasi, A.; Behzad, M. Synthesis and crystal structures of new mixed-ligand Schiff base complexes containing N-donor heterocyclic co-ligands: Molecular docking and pharmacophore modeling studies on the main proteases of SARS-CoV-2 virus (COVID-19 disease). Polyhedron 2022, 220, 115825. [Google Scholar] [CrossRef]
- Artem’ev, G.A.; Rusinov, V.L.; Kopchuk, D.S.; Savchuk, M.I.; Santra, S.; Ulomsky, E.N.; Zyryanov, G.V.; Majee, A.; Du, W.; Charushin, V.N.; et al. Synthetic approaches to 1,2,4-triazolo [5,1-c][1,2,4]triazin-7-ones as basic heterocyclic structures of the antiviral drug Riamilovir (“Triazavirin”) active against SARS-CoV-2 (COVID-19). Org. Biomol. Chem. 2022, 20, 1828–1837. [Google Scholar] [CrossRef]
- Horchani, M.; Heise, N.V.; Csuk, R.; Ben Jannet, H.; Harrath, A.; Romdhane, A. Synthesis and In Silico Docking Study towards M-Pro of Novel Heterocyclic Compounds Derived from Pyrazolopyrimidinone as Putative SARS-CoV-2 Inhibitors. Molecules 2022, 27, 5303. [Google Scholar] [CrossRef]
- Dorababu, A. Pyrazolopyrimidines as attractive pharmacophores in efficient drug design: A recent update. Arch. Pharm. 2022, 355, 2200154. [Google Scholar] [CrossRef] [PubMed]
- El-Gohary, S.N.; Gabr, T.M.; Shaaban, I.M. Synthesis, molecular modeling and biological evaluation of new pyrazolo [3,4-b]pyridine analogs as potential antimicrobial, antiquorum-sensing and anticancer agents. Bioorg. Chem. 2019, 89, 102976. [Google Scholar] [CrossRef] [PubMed]
- Shejale, S.R.; Rajput, C.G.; Bansode, S.S.; Kondawar, M.S. Synthesis, characterisation, evaluation of antimicrobial & antifungal activity of novel pyrazolopyrimidine & pyrazolopyridine derivatives. Int. J. Pharm. Chem. 2017, 7, 144–148. [Google Scholar] [CrossRef]
- El-Gohary, S.N.; Shaaban, I.M. Design, synthesis, antimicrobial, antiquorum-sensing and antitumor evaluation of new series of pyrazolopyridine derivatives. Eur. J. Med. Chem. 2018, 157, 729–742. [Google Scholar] [CrossRef]
- El-Gohary, S.N.; Shaaban, I.M. New pyrazolopyridine analogs: Synthesis, antimicrobial, antiquorum-sensing and antitumor screening. Eur. J. Med. Chem. 2018, 152, 126–136. [Google Scholar] [CrossRef]
- Salem, S.M.; Ali, M.A.M. Novel pyrazolo[3,4-b]pyridine derivatives: Synthesis, characterization, antimicrobial and antiproliferative profile. Biol. Pharm. Bull. 2016, 39, 473–483. [Google Scholar] [CrossRef]
- Eissa, H.I.; El-Naggar, M.A.; El-Hashash, A.M. Design, synthesis, molecular modeling and biological evaluation of novel-1H-pyrazolo [3,4-b]pyridine derivatives as potential anticancer agents. Bioorg. Chem. 2016, 67, 43–56. [Google Scholar] [CrossRef]
- Arias, A.D.; Montagut, A.M.; Puig de la Bellacasa, R.; Estrada-Tejedor, R.; Teixidó, J.; Borrell, J.I. 1H-Pyrazolo [3,4-b]pyridines: Synthesis and Biomedical Applications. Molecules 2022, 27, 2237. [Google Scholar] [CrossRef]
- Karrouchi, K.; Radi, S.; Ramli, Y.; Taoufik, J.; Mabkhot, Y.N.; Al-aizari, F.A.; Ansar, M. Synthesis and Pharmacological Activities of Pyrazole Derivatives. Molecules 2018, 23, 134. [Google Scholar] [CrossRef]
- Hamama, S.W.; El-Gohary, G.H.; Soliman, M.; Zoorob, H.H. A versatile synthesis, PM3-semiempirical, antibacterial, and antitumor evaluation of some bioactive pyrazoles. J. Heterocycl. Chem. 2012, 49, 543–554. [Google Scholar] [CrossRef]
- Yadav, P.; Shah, K. An overview on synthetic and pharmaceutical prospective of pyrido [2,3-d]pyrimidines scaffold. Chem. Biol. Drug Des. 2020, 97, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Yousif, M.N.M.; El-Gazzar, A.B.A.; El-Enany, M.M. Synthesis and Biological Evaluation of Pyrido(2,3-d)pyrimidines. Mini-Rev. Org. Chem. 2021, 18, 43–54. [Google Scholar] [CrossRef]
- Prachayasittikul, S.; Pingaew, R.; Worachartcheewan, A.; Sinthupoom, N.; Prachayasittikul, V.; Ruchirawat, S.; Prachayasittikul, V. Roles of Pyridine and Pyrimidine Derivatives as Privileged Scaffolds in Anticancer Agents. Mini-Rev. Med. Chem. 2017, 17, 869–901. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, Y.S.; Larson, S.B.; Willis, R.C.; Robins, R.K.; Revankar, G.R. Synthesis and biological evaluation of certain C-4 substituted pyrazolo[3,4-b]pyridine nucleosides. J. Med. Chem. 1989, 32, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Monier, M.; Abdel-Latif, D.; El-Mekabaty, A.; Mert, B.D.; Elattar, K.M. Advances in the Chemistry of 6-6 Bicyclic Systems: Chemistry of Pyrido[3,4-d]pyrimidines. Curr. Org. Chem. 2019, 16, 812–854. [Google Scholar] [CrossRef]
- El-Remaily, A.A.M.; El Hady, M.O.; Abo Zaid, S.H.; Abd El-Raheem, M.M.E. Synthesis and in vitro antibacterial activity of some novel fused pyridopyrimidine derivatives. J. Heterocycl. Chem. 2016, 53, 1304–1309. [Google Scholar] [CrossRef]
- Abdelhameed, M.R.; Darwesh, M.O.; El-Shahat, M. Synthesis of arylidene hydrazinylpyrido[2,3-d]pyrimidin-4-ones as potent anti-microbial agents. Heliyon. 2020, 6, e04956. [Google Scholar] [CrossRef]
- Acosta, P.; Insuasty, B.; Ortiz, A.; Abonia, R.; Sortino, M.; Zacchino, A.S.; Quirog, J. Solvent-free microwave-assisted synthesis of novel pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidines with potential antifungal activity. Arab. J. Chem. 2016, 9, 481–492. [Google Scholar] [CrossRef]
- Bazgir, A.; Khanaposhtani, M.M.; Soorki, A.A. One-pot synthesis and antibacterial activities of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidine-dione derivatives. Bioorg. Med. Chem. Lett. 2008, 18, 5800–5803. [Google Scholar] [CrossRef]
- El-Emary, I.T.; Abd-El-Mohsen, A.S. Multi-component one-pot synthesis and antimicrobial activities 3-methyl-1,4-diphenyl-7-thioxo-4,6,8,9-tetrahydropyrazolo [5,4-b]pyrimidino [5,4-e]pyridine-5-one and related derivatives. Molecules 2012, 17, 14464–14483. [Google Scholar] [CrossRef]
- Javahershenas, R.; Khalafy, J. One-pot, four component synthesis of pyrazolo[4’,3’:5,6]pyrido[2,3-d]pyrimidines derivatives. Asian J. Green Chem. 2018, 2, 318–329. [Google Scholar] [CrossRef]
- El-Gohary, S.N.; Hawasa, S.S.; Gabra, M.T.; Shaaband, M.I.; El-Ashmawy, M.B. New series of fused pyrazolopyridines: Synthesis, molecular modeling, antimicrobial, antiquorum-sensing and antitumor activities. Bioorg. Chem. 2019, 92, 103109. [Google Scholar] [CrossRef]
- Mohamed, R.N.; Khaireldin, Y.N.; Fahmy, F.A.; El-Sayed, A.A. Facile synthesis of fused nitrogen containing heterocycles as anticancer agents. Der Pharma Chem. 2010, 2, 400–417. [Google Scholar]
- El-Sayed, A.A.; Pedersen, B.E.; Khaireldin, Y.N. Thermal Stability of Modified i-Motif Oligonucleotides with Naphthalimide Intercalating Nucleic Acids. Helv. Chim. Acta. 2016, 99, 14–19. [Google Scholar] [CrossRef]
- El-Sayed, A.A.; Pedersen, B.E.; Khaireldin, A.N. Studying the influence of the pyrene intercalator TINA on the stability of DNA i-motifs. Nucleosides Nucleotides Nucleic Acids 2012, 31, 872–879. [Google Scholar] [CrossRef]
- El-Sayed, A.A.; El-Saidi, M.M.T.; Reham, R.K. Unexpected Reactions of Azido-p-Benzoquinone Derivatives towards Lawesson’s Reagent & Molecular Docking Study as a promising anticancer agent. Egypt. J. Chem. 2019, 62, 315–326. [Google Scholar] [CrossRef]
- Khattab, R.R.; Hassan, A.A.; Kutkatd, O.M.; Abuzeid, K.M.; Hassan, N.A. Synthesis and Antiviral Activity of Novel Thieno [2,3-d]pyrimidine Hydrazones and Their C-Nucleosides. Russ. J. Gen. Chem. 2019, 89, 1707–1717. [Google Scholar] [CrossRef]
- Wasfy, A.F.A.; Hassan, A.A.; Khattab, R.R.; Abu-Zied, M.K.; Awad, M.H.; Al Otaibi, F.; Hassan, A.N. Synthesis of Some New Thioglycosides Derived from Thieno[2,3-d]pyrimidine Derivatives and Their Anticancer and Antioxidant Activity. Res. J. Pharm. Biol. Chem. Sci. 2018, 9, 77–88. [Google Scholar]
- Hassan, N.A.; Abu-Zeid, K.M.; El-Gazzar, A.B. New routes to pyrano[2,3-d]pyrimidine derivatives from β-enamino nitrile and phosgene iminium chloride. Heterocycl. Comm. 2006, 12, 73–78. [Google Scholar] [CrossRef]
- El-Sayed, H.A.; Moustafa, A.H.; Hassan, A.A.; El-Seadawy, N.A.M.; Pasha, S.H.; Shmiess, N.A.M.; Awad, H.M.; Hassan, N.A. Microwave synthesis, anti-oxidant and anti-tumor activity of some nucleosides derived 2-oxonicotinonitrile. Synth. Commun. 2019, 49, 3465–3474. [Google Scholar] [CrossRef]
- Khattab, R.R.; Hassan, A.A.; Elganzor, H.H.; Tashkandi, N.Y.; Awad, H.M.; Nossier, E.S.; El-Sayed, W.A.; Hassan, N.A. Click Chemistry Based Synthesis, Cytotoxic Activity and Molecular Docking of Novel Triazole-Thienopyrimidine Hybride Glycosides Targeting EGFR. J. Enzyme Inhib. Med. Chem. 2020, 36, 405. [Google Scholar] [CrossRef]
- Basiony, E.A.; Hassan, A.A.; Al-Amshany, Z.M.; Abd-Rabou, A.A.; Abdel-Rahman, A.A.H.; Hassan, N.A.; El-Sayed, W.A. Synthesis and Cytotoxic Activity of New Thiazolopyrimidine Sugar Hydrazones and Their Derived Acyclic Nucleoside Analogues. Molecules 2020, 25, 399. [Google Scholar] [CrossRef] [PubMed]
- Rashad, A.E.; Shamroukh, A.H.; Abdel-Megeid, R.E.; Ali, H.S. Synthesis and Isomerization of Some Novel Pyrazolopyrimidine and Pyrazolotriazolopyrimidine Derivatives. Molecules 2014, 19, 5459–5469. [Google Scholar] [CrossRef] [PubMed]
- Shawali, A.S.; Hassaneen, H.M.; Shurrab, N.K. A new strategy for the synthesis of pyrazolo[4,3-e][1,2,4]triazolo[4,3-c]pyrimidines and pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidines. Tetrahedron 2008, 64, 10339–10343. [Google Scholar] [CrossRef]
- Shaban, M.A.; Nasr, A.Z. Synthesis of condensed 1,2,4-triazol[3,4-z] heterocycles. Adv. Heterocycl. Chem. 1999, 49, 277. [Google Scholar]
- Shawali, A.S.; Hassaneen, H.M.; Shurrab, N.K. Aconvenient synthesis of novel series of 4-cyclohexyl-2-substituted [1,2,4] triazolo [1,5-a] quinazolin-5(4H)-ones. Novel isoomers of H1 antihistaminic acitve agents. Heterocycles 2008, 75, 1479. [Google Scholar] [CrossRef]
- Kandeil, A.; Mostafa, A.; El-Shesheny, R.; Shehata, M.; Roshdy, W.H.; Ahmed, S.S.; Gomaa, M.; Taweel, A.E.; Kayed, A.E.; Mahmoud, S.H. Coding-Complete Genome Sequences of Two SARS-CoV-2 Isolates from Egypt. Microbiol. Resour. Announc. 2020, 9, e00489-20. [Google Scholar] [CrossRef]
- Andrés, P.; Blandine, P.; Julia, D.; Thomas, J.; Aurélien, T.; Victoria, D.; Pauline, B.; Bruno, L.; Manuel, R.; Olivier, T. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020, 181, 104878. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA—An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 2004, 18, 167–173. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).