In Silico Prospecting for Novel Bioactive Peptides from Seafoods: A Case Study on Pacific Oyster (Crassostrea gigas)
Abstract
1. Introduction
2. Results
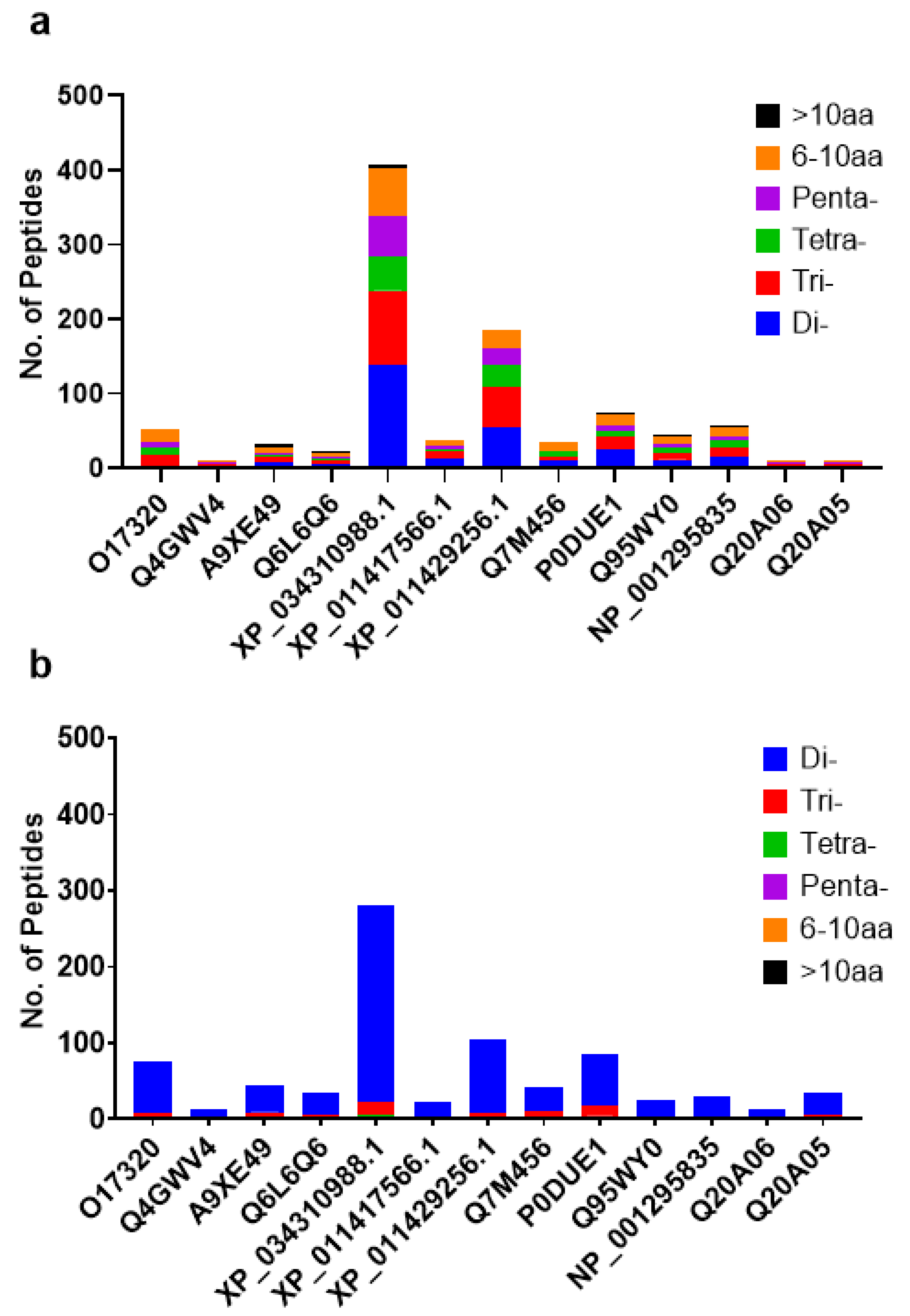
2.1. Simulated Gastrointestinal Digestion
2.2. Prospect Oyster BAPs
3. Discussion
4. Materials and Methods
4.1. Protein Selection and Retrieval
4.2. Enzyme Hydrolysis Using BIOPEP and ExPASy PeptideCutter
4.3. Toxicity, Bitterness, Stability, and Allergenicity Screening
4.4. Bioactivity and Novelty Assessments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| BAP | bioactive peptide |
| C. gigas | Crassostrea gigas |
| T2D | type 2 diabetes |
| DPP-IV | dipeptidyl peptidase-4 |
| ACE | angiotensin-converting enzyme |
| SVM | support vector machine |
| RF | Random Forest |
| ADMET | absorption, distribution, metabolism and excretion properties, and toxicities |
| QSAR | quantitative structure-activity relationship |
| GI | gastrointestinal |
References
- Chalamaiah, M.; Keskin Ulug, S.; Hong, H.; Wu, J. Regulatory Requirements of Bioactive Peptides (Protein Hydrolysates) from Food Proteins. J. Funct. Foods 2019, 58, 123–129. [Google Scholar] [CrossRef]
- Hayes, M.; Tiwari, B.K. Bioactive Carbohydrates and Peptides in Foods: An Overview of Sources, Downstream Processing Steps and Associated Bioactivities. Int. J. Mol. Sci. 2015, 16, 22485. [Google Scholar] [CrossRef] [PubMed]
- Barati, M.; Javanmardi, F.; Mousavi Jazayeri, S.M.H.; Jabbari, M.; Rahmani, J.; Barati, F.; Nickho, H.; Davoodi, S.H.; Roshanravan, N.; Mousavi Khaneghah, A. Techniques, Perspectives, and Challenges of Bioactive Peptide Generation: A Comprehensive Systematic Review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1488–1520. [Google Scholar] [CrossRef] [PubMed]
- Udenigwe, C.C.; Aluko, R.E. Food Protein-Derived Bioactive Peptides: Production, Processing, and Potential Health Benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Meisel, H. Food-Derived Peptides with Biological Activity: From Research to Food Applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef]
- Kim, S.-K.; Wijesekara, I. Development and Biological Activities of Marine-Derived Bioactive Peptides: A Review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Ngo, D.-H.; Vo, T.-S.; Ngo, D.-N.; Wijesekara, I.; Kim, S.-K. Biological Activities and Potential Health Benefits of Bioactive Peptides Derived from Marine Organisms. Int. J. Biol. Macromol. 2012, 51, 378–383. [Google Scholar] [CrossRef]
- Hao, L.; Wang, X.; Cao, Y.; Xu, J.; Xue, C. A Comprehensive Review of Oyster Peptides: Preparation, Characterisation and Bioactivities. Rev. Aquac. 2022, 14, 120–138. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.; Tian, X.; Tian, Z. Antimicrobial Capacity and Antioxidant Activity of Enzymatic Hydrolysates of Protein from Rushan Bay Oyster (Crassostrea Gigas). J. Food Process. Preserv. 2015, 39, 404–412. [Google Scholar] [CrossRef]
- Lee, H.-J.; Saravana, P.S.; Cho, Y.-N.; Haq, M.; Chun, B.-S. Extraction of Bioactive Compounds from Oyster (Crassostrea gigas) by Pressurized Hot Water Extraction. J. Supercrit. Fluids 2018, 141, 120–127. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, Y.; Cao, X.; Guo, K.; Wang, Q.; Xiao, X.; Zhai, X.; Wang, D.; Huang, Z. Enzymatic Preparation of Crassostrea Oyster Peptides and Their Promoting Effect on Male Hormone Production. J. Ethnopharmacol. 2021, 264, 113382. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.-T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A Global Review on Short Peptides: Frontiers and Perspectives. Molecules 2021, 26, 430. [Google Scholar] [CrossRef] [PubMed]
- Udenigwe, C.C. Bioinformatics Approaches, Prospects and Challenges of Food Bioactive Peptide Research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Chen, J.; Yu, X.; Chen, Q.; Wu, Q.; He, Q. Screening and Mechanisms of Novel Angiotensin-I-Converting Enzyme Inhibitory Peptides from Rabbit Meat Proteins: A Combined in Silico and in Vitro Study. Food Chem. 2022, 370, 131070. [Google Scholar] [CrossRef]
- Gomez, H.L.R.; Peralta, J.P.; Tejano, L.A.; Chang, Y.-W. In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides. Int. J. Mol. Sci. 2019, 20, 5191. [Google Scholar] [CrossRef]
- Manzoor, M.; Singh, J.; Gani, A. Exploration of Bioactive Peptides from Various Origin as Promising Nutraceutical Treasures: In Vitro, in Silico and in Vivo Studies. Food Chem. 2022, 373, 131395. [Google Scholar] [CrossRef]
- Mohd Salim, M.A.S.; Gan, C.-Y. Dual-Function Peptides Derived from Egg White Ovalbumin: Bioinformatics Identification with Validation Using in Vitro Assay. J. Funct. Foods 2020, 64, 103618. [Google Scholar] [CrossRef]
- Feng, C.; Tian, L.; Jiao, Y.; Tan, Y.; Liu, C.; Luo, Y.; Hong, H. The Effect of Steam Cooking on the Proteolysis of Pacific Oyster (Crassostrea gigas) Proteins: Digestibility, Allergenicity, and Bioactivity. Food Chem. 2022, 379, 132160. [Google Scholar] [CrossRef]
- Gülseren, İ.; Vahapoglu, B. The Stability of Food Bioactive Peptides in Blood: An Overview. Int. J. Pept. Res. Ther. 2021, 28, 2. [Google Scholar] [CrossRef]
- Keil, B. Specificity of Proteolysis; Springer: Berlin/Heidelberg, Germany, 1992; ISBN 978-3-642-48382-0. [Google Scholar]
- Arai, S.; Fukimaji, M. Enzymatic Modification of Proteins with Special Reference to Improving Their Functional Properties. In Food Enzymology; Elsevier Applied Science: London, UK, 1991; Volume 2, pp. 83–104. [Google Scholar]
- Zhang, C.; Wu, S.; Xu, D. Catalytic Mechanism of Angiotensin-Converting Enzyme and Effects of the Chloride Ion. J. Phys. Chem. B 2013, 117, 6635–6645. [Google Scholar] [CrossRef]
- Wang, X.; Wu, S.; Xu, D.; Xie, D.; Guo, H. Inhibitor and Substrate Binding by Angiotensin-Converting Enzyme: Quantum Mechanical/Molecular Mechanical Molecular Dynamics Studies. J. Chem. Inf. Model. 2011, 51, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Thoma, R.; Löffler, B.; Stihle, M.; Huber, W.; Ruf, A.; Hennig, M. Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV. Structure 2003, 11, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Kose, A. In Silico Bioactive Peptide Prediction from The Enzymatic Hydrolysates of Edible Seaweed Rubisco Large Chain. Turk. J. Fish. Aquat. Sci. 2021, 21, 615–625. [Google Scholar] [CrossRef]
- Pripp, A.H. Initial Proteolysis of Milk Proteins and Its Effect on Formation of ACE-Inhibitory Peptides during Gastrointestinal Proteolysis: A Bioinformatic, In Silico, Approach. Eur. Food Res. Technol. 2005, 221, 712–716. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.; Guan, C.; He, D.; Liao, X.; Wang, Y.; Chen, H.; Zhang, C.; Xing, X.-H. Functional discovery and production technology for natural bioactive peptides. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2021, 37, 2166–2180. [Google Scholar] [CrossRef]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of Novel Dipeptidyl Peptidase-IV and Angiotensin-I-Converting Enzyme Inhibitory Peptides from Meat Proteins Using in Silico Analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Hong, H.; Wu, J.; Yan, X. Bioavailability of Bioactive Peptides Derived from Food Proteins across the Intestinal Epithelial Membrane: A Review. Trends Food Sci. Technol. 2019, 86, 399–411. [Google Scholar] [CrossRef]
- Aluko, R.E. Structure and Function of Plant Protein-Derived Antihypertensive Peptides. Curr. Opin. Food Sci. 2015, 4, 44–50. [Google Scholar] [CrossRef]
- Jakubczyk, A.; Karaś, M.; Rybczyńska-Tkaczyk, K.; Zielińska, E.; Zieliński, D. Current Trends of Bioactive Peptides—New Sources and Therapeutic Effect. Foods 2020, 9, 846. [Google Scholar] [CrossRef]
- Sitanggang, A.B.; Putri, J.E.; Palupi, N.S.; Hatzakis, E.; Syamsir, E.; Budijanto, S. Enzymatic Preparation of Bioactive Peptides Exhibiting ACE Inhibitory Activity from Soybean and Velvet Bean: A Systematic Review. Molecules 2021, 26, 3822. [Google Scholar] [CrossRef]
- Vermeirssen, V.; Camp, J.V.; Verstraete, W. Bioavailability of Angiotensin I Converting Enzyme Inhibitory Peptides. Br. J. Nutr. 2004, 92, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; Parthsarathy, V.; McLaughlin, C.M.; O’Keeffe, M.B.; Allsopp, P.J.; McSorley, E.M.; O’Harte, F.P.M.; FitzGerald, R.J. Atlantic Salmon (Salmo Salar) Co-Product-Derived Protein Hydrolysates: A Source of Antidiabetic Peptides. Food Res. Int. 2018, 106, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of Dipeptidyl Peptidase IV Inhibitory Peptides from Defatted Rice Bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of Dipeptidyl Peptidase IV (DPP-IV) by Proline Containing Casein-Derived Peptides. J. Funct. Foods 2013, 5, 1909–1917. [Google Scholar] [CrossRef]
- He, R.; Wang, Y.; Yang, Y.; Wang, Z.; Ju, X.; Yuan, J. Rapeseed Protein-Derived ACE Inhibitory Peptides LY, RALP and GHS Show Antioxidant and Anti-Inflammatory Effects on Spontaneously Hypertensive Rats. J. Funct. Foods 2019, 55, 211–219. [Google Scholar] [CrossRef]
- Yuan, L.; Chu, Q.; Wu, X.; Yang, B.; Zhang, W.; Jin, W.; Gao, R. Anti-Inflammatory and Antioxidant Activity of Peptides from Ethanol-Soluble Hydrolysates of Sturgeon (Acipenser schrenckii) Cartilage. Front. Nutr. 2021, 8, 689648. [Google Scholar] [CrossRef]
- Guha, S.; Majumder, K. Structural-Features of Food-Derived Bioactive Peptides with Anti-Inflammatory Activity: A Brief Review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, X.; Zhang, X.-L.; Xie, Q.-F. Purification and Identification of Anti-Inflammatory Peptides Derived from Simulated Gastrointestinal Digests of Velvet Antler Protein (Cervus elaphus Linnaeus). J. Food Drug Anal. 2016, 24, 376–384. [Google Scholar] [CrossRef]
- Chung, C.-R.; Jhong, J.-H.; Wang, Z.; Chen, S.; Wan, Y.; Horng, J.-T.; Lee, T.-Y. Characterization and Identification of Natural Antimicrobial Peptides on Different Organisms. Int. J. Mol. Sci. 2020, 21, 986. [Google Scholar] [CrossRef]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From Antimicrobial to Anticancer Peptides. A Review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef]
- Seo, M.-D.; Won, H.-S.; Kim, J.-H.; Mishig-Ochir, T.; Lee, B.-J. Antimicrobial Peptides for Therapeutic Applications: A Review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mishra, B.; Lushnikova, T.; Narayana, J.L.; Wang, G. Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin. Adv. Biosyst. 2018, 2, 1700259. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Chiangjong, W.; Lee, V.S.; Nantasenamat, C.; Hasan, M.M.; Shoombuatong, W. Improved Prediction and Characterization of Anticancer Activities of Peptides Using a Novel Flexible Scoring Card Method. Sci. Rep. 2021, 11, 3017. [Google Scholar] [CrossRef]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer Peptide: Physicochemical Property, Functional Aspect and Trend in Clinical Application (Review). Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.; Dennison, S.R.; Lea, B.; Snape, T.J.; Nicholl, I.D.; Radecka, I.; Harris, F. Anionic Antimicrobial and Anticancer Peptides from Plants. Crit. Rev. Plant Sci. 2013, 32, 303–320. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, C.; Shuo, J.; Yishan, L.; Yannan, C.; Wenjie, L.; Bin, Z. Bioinformatics Identification and Molecular Mechanism of Angiotensin-Converting Enzyme and Dipeptidyl Peptidase-IV Inhibitory Peptides from in Silico Digest of Crassostrea Gigas. Food Meas. 2022, 16, 4540–4550. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Gagnon, J.K.; Law, S.M.; Brooks III, C.L. Flexible CDOCKER: Development and Application of a Pseudo-Explicit Structure-Based Docking Method within CHARMM. J. Comput. Chem. 2016, 37, 753–762. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Fang, L.; Li, G.; Zhang, J.; Gu, R.; Cai, M.; Lu, J. Identification and Mutational Analysis of Continuous, Immunodominant Epitopes of the Major Oyster Allergen Crag 1. Clin. Immunol. 2019, 201, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Marti, P.; Truffer, R.; Stadler, M.B.; Keller-Gautschi, E.; Crameri, R.; Mari, A.; Schmid-Grendelmeier, P.; Miescher, S.M.; Stadler, B.M.; Vogel, M. Allergen Motifs and the Prediction of Allergenicity. Immunol. Lett. 2007, 109, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Ishida, M.; Shimakura, K.; Nagashima, Y.; Shiomi, K. Tropomyosin, the Major Oyster Crassostrea Gigas Allergen and Its IgE-Binding Epitopes. J. Food Sci. 1998, 63, 44–47. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Singh Chauhan, J.; Nagpal, G.; Kumar, R.; Sharma, M.; Raghava, G.P.S. An in Silico Platform for Predicting, Screening and Designing of Antihypertensive Peptides. Sci. Rep. 2015, 5, 12512. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Kanthawong, S.; Nantasenamat, C.; Hasan, M.D.M.; Shoombuatong, W. IDPPIV-SCM: A Sequence-Based Predictor for Identifying and Analyzing Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides Using a Scoring Card Method. J. Proteome Res. 2020, 19, 4125–4136. [Google Scholar] [CrossRef] [PubMed]
- Trabuco, L.G.; Lise, S.; Petsalaki, E.; Russell, R.B. PepSite: Prediction of Peptide-Binding Sites from Protein Surfaces. Nucleic Acids Res. 2012, 40, W423–W427. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.; Hasan, M.; Kurata, H. PreAIP: Computational Prediction of Anti-Inflammatory Peptides by Integrating Multiple Complementary Features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-Based Prediction of Anti-Inflammatory Peptides Using Random Forest. Front. Pharmacol. 2018, 9, 276. [Google Scholar] [CrossRef]
- Gupta, S.; Sharma, A.K.; Shastri, V.; Madhu, M.K.; Sharma, V.K. Prediction of Anti-Inflammatory Proteins/Peptides: An Insilico Approach. J. Transl. Med. 2017, 15, 7. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef]
- Huang, K.-Y.; Tseng, Y.-J.; Kao, H.-J.; Chen, C.-H.; Yang, H.-H.; Weng, S.-L. Identification of Subtypes of Anticancer Peptides Based on Sequential Features and Physicochemical Properties. Sci. Rep. 2021, 11, 13594. [Google Scholar] [CrossRef]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.-C. MACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Shtatland, T.; Guettler, D.; Kossodo, M.; Pivovarov, M.; Weissleder, R. PepBank—A Database of Peptides Based on Sequence Text Mining and Public Peptide Data Sources. BMC Bioinform. 2007, 8, 280. [Google Scholar] [CrossRef]
- Liu, F.; Baggerman, G.; Schoofs, L.; Wets, G. The Construction of a Bioactive Peptide Database in Metazoa. J. Proteome Res. 2008, 7, 4119–4131. [Google Scholar] [CrossRef]
- Zamyatnin, A.A.; Borchikov, A.S.; Vladimirov, M.G.; Voronina, O.L. The EROP-Moscow Oligopeptide Database. Nucleic Acids Res. 2006, 34, D261–D266. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, C.; Chen, H.; Xue, J.; Guo, X.; Liang, M.; Chen, M. BioPepDB: An Integrated Data Platform for Food-Derived Bioactive Peptides. Int. J. Food Sci. Nutr. 2018, 69, 963–968. [Google Scholar] [CrossRef]
- Roy, S.; Teron, R. BioDADPep: A Bioinformatics Database for Anti Diabetic Peptides. Bioinformation 2019, 15, 780–783. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Lee, H.-T.; Lee, C.-C.; Yang, J.-R.; Lai, J.Z.C.; Chang, K.Y. A Large-Scale Structural Classification of Antimicrobial Peptides. BioMed. Res. Int. 2015, 2015, e475062. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P.S. CancerPPD: A Database of Anticancer Peptides and Proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef]
- Kapoor, P.; Singh, H.; Gautam, A.; Chaudhary, K.; Kumar, R.; Raghava, G.P.S. TumorHoPe: A Database of Tumor Homing Peptides. PLoS ONE 2012, 7, e35187. [Google Scholar] [CrossRef]
| Digestion Platform | Enzyme Name | EC Number | Cleavage Residues | Additional Information and References |
|---|---|---|---|---|
| BIOPEP Enzyme Action Tool | Pepsin (pH > 2) | 3.4.23.1 | C-terminus of F, L, G, Y, A, E, Q, T, N, K, D, M; N-terminus of V, I | MEROPS A01.001 show broad cleavage specificity |
| Trypsin | 3.4.21.4 | C-terminus of K, R | P after K blocks enzyme action [20] | |
| Chymotrypsin (A) | 3.4.21.1 | C-terminus of F, Y, W, L, N, H, M | P after Y, P or M after W, P after F, P after L block enzyme action [21]. | |
| ExPASy PeptideCutter | Pepsin (pH > 2) | 3.4.23.1 | Broad specificity with preference at F, Y, W, L in position P1 or P1’ [20] | Lost specificity at pH >= 2 |
| Trypsin | 3.4.21.4 | Preferentially cleaves R and K at P1 | P at P1′ blocks enzyme action but not when K and W or R and M are at P1 and P2 at the same time. If K at P1, enzyme action is blocked when D and D, C and D, C and H, or C and Y are at P2 and P1′ respectively. When R is at P1, enzyme action is blocked with R and H, C and K, or R and R are at P2 and P1′respectively. | |
| Chymotrypsin (low specificity) | 3.4.21.1 | Preferential at F, Y, W, L, M at P1 | Exceptions: P at P1′; M or P at P1′ when is W is at P1; Y at P1′ when M is at P1; when H is at P1, D, M or W also blocks the cleavage. |
| Identifier | Toxicity | Bitterness | Stability | Novelty Finds | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide No. | Source Protein Accession Number | Peptide Sequence | # Residues | ToxinPred | iBitter-SCM | PLifePred | HLP | BIOPEP Database | PepBank | EROP-MOSCOW | BioPepDB | |||
| FASTA | Score | Descriptor | Score | Descriptor | Half-Life in Blood (s) | Intestinal Half-Life (s) | Stability | |||||||
| >O1 | O17320 | NSPAM | 5 | −0.58 | Non-Toxin | 308 | non-Bitter | 832.11 | 1.553 | High | 0 | 0 | 0 | 0 |
| >O2 | P0DUE1 | PQSCR | 5 | −0.58 | Non-Toxin | 251.25 | non-Bitter | 826.31 | 1.425 | High | 0 | 0 | 0 | 0 |
| >O3 | A9XE49 | ICPSS | 5 | −0.67 | Non-Toxin | 320.25 | non-Bitter | 833.81 | 1.358 | High | 0 | 0 | 0 | 0 |
| >O4 | XP_034310988.1 | CTGAI | 5 | −0.63 | Non-Toxin | 285.25 | non-Bitter | 839.41 | 1.32 | High | 0 | 0 | 0 | 0 |
| >O5 | A9XE49 | CEPVY | 5 | −0.86 | Non-Toxin | 229 | non-Bitter | 847.51 | 1.26 | High | 0 | 0 | 0 | 0 |
| >O6 | XP_034310988.1 | QACID | 5 | −0.55 | Non-Toxin | 320.25 | non-Bitter | 840.71 | 1.477 | High | 0 | 0 | 0 | 0 |
| >O7 | P0DUE1 | SVPVL | 5 | −0.97 | Non-Toxin | 283.25 | non-Bitter | 836.51 | 1.192 | High | 0 | 0 | 0 | 0 |
| >O8 | XP_034310988.1 | QCNGV | 5 | −0.61 | Non-Toxin | 322.75 | non-Bitter | 826.41 | 1.513 | High | 0 | 0 | 0 | 0 |
| >O9 | XP_034310988.1 | STHPH | 5 | −0.73 | Non-Toxin | 308 | non-Bitter | 834.81 | 1.292 | High | 0 | 0 | 0 | 0 |
| >O10 | XP_034310988.1 | IEKPM | 5 | −0.82 | Non-Toxin | 330.25 | non-Bitter | 838.31 | 2.38 | High | 0 | 0 | 0 | 0 |
| >O11 | XP_034310988.1 | AGSVP | 5 | −0.67 | Non-Toxin | 321.5 | non-Bitter | 913.01 | 1.316 | High | 0 | 0 | 0 | 0 |
| >O12 | Q4GWV4 | CDAAT | 5 | −0.73 | Non-Toxin | 278 | non-Bitter | 835.01 | 1.239 | High | 0 | 0 | 0 | 0 |
| >O13 | Q4GWV4 | VSADM | 5 | −0.8 | Non-Toxin | 327.5 | non-Bitter | 838.01 | 1.27 | High | 0 | 0 | 0 | 0 |
| >O14 | O17320 | SSSSL | 5 | −0.8 | Non-Toxin | 312.75 | non-Bitter | 834.81 | 1.374 | High | 0 | 0 | 0 | 0 |
| >O15 | P0DUE1 | SEPNI | 5 | −0.82 | Non-Toxin | 229 | non-Bitter | 832.51 | 1.595 | High | 0 | 0 | 0 | 0 |
| >O16 | O17320 | TVPIY | 5 | −1.1 | Non-Toxin | 330.5 | non-Bitter | 823.91 | 1.402 | High | 0 | 0 | 0 | 0 |
| >O17 | P0DUE1 | SPSST | 5 | −0.88 | Non-Toxin | 325.25 | non-Bitter | 835.01 | 1.249 | High | 0 | 0 | 0 | 0 |
| >O18 | XP_034310988.1 | DAANR | 5 | −0.89 | Non-Toxin | 275.5 | non-Bitter | 835.61 | 1.251 | High | 0 | 0 | 0 | 0 |
| >O19 | Q20A06 | CDAVT | 5 | −0.81 | Non-Toxin | 322.75 | non-Bitter | 831.71 | 1.054 | High | 0 | 0 | 0 | 0 |
| >O20 | XP_034310988.1 | INQGA | 5 | −0.9 | Non-Toxin | 320 | non-Bitter | 845.01 | 1.68 | High | 0 | 0 | 0 | 0 |
| >O21 | XP_034310988.1 | QSDVR | 5 | −0.72 | Non-Toxin | 318.5 | non-Bitter | 838.71 | 1.471 | High | 0 | 0 | 0 | 0 |
| >O22 | XP_011417566.1 | IDQNR | 5 | −0.74 | Non-Toxin | 332.5 | non-Bitter | 858.61 | 1.386 | High | 0 | 0 | 0 | 0 |
| >O23 | XP_011417566.1 | DKDGK | 5 | −0.71 | Non-Toxin | 325.25 | non-Bitter | 835.31 | 1.295 | High | 0 | 0 | 0 | 0 |
| >O24 | XP_034310988.1 | SEIDR | 5 | −0.96 | Non-Toxin | 317.5 | non-Bitter | 824.71 | 1.375 | High | 0 | 0 | 0 | 0 |
| >O25 | XP_034310988.1 | DNQIK | 5 | −0.82 | Non-Toxin | 302.75 | non-Bitter | 915.21 | 2.152 | High | 0 | 0 | 0 | 0 |
| >O26 | XP_034310988.1 | AADER | 5 | −0.96 | Non-Toxin | 270.5 | non-Bitter | 865.31 | 1.306 | High | 0 | 0 | 0 | 0 |
| >O27 | XP_034310988.1 | QQQIK | 5 | −0.89 | Non-Toxin | 308.25 | non-Bitter | 834.81 | 1.405 | High | 0 | 0 | 0 | 0 |
| >O28 | XP_034310988.1 | ENQSM | 5 | −0.79 | Non-Toxin | 300.75 | non-Bitter | 840.31 | 1.834 | High | 0 | 0 | 0 | 0 |
| >O29 | XP_034310988.1 | ANDNT | 5 | −0.78 | Non-Toxin | 276 | non-Bitter | 834.31 | 1.807 | High | 0 | 0 | 0 | 0 |
| >O30 | XP_034310988.1 | GAVDK | 5 | −0.69 | Non-Toxin | 312.75 | non-Bitter | 832.31 | 1.239 | High | 0 | 0 | 0 | 0 |
| >O31 | Q95WY0 | DEAAR | 5 | −0.99 | Non-Toxin | 278 | non-Bitter | 865.31 | 1.256 | High | 0 | 0 | 0 | 0 |
| >O32 | XP_011429256.1 | DAAER | 5 | −0.95 | Non-Toxin | 268 | non-Bitter | 865.31 | 1.587 | High | 0 | 0 | 0 | 0 |
| >O33 | XP_034310988.1 | DSEQR | 5 | −1 | Non-Toxin | 327.75 | non-Bitter | 846.21 | 2.5 | High | 0 | 0 | 0 | 0 |
| >O34 | XP_034310988.1 | EGQIK | 5 | −0.77 | Non-Toxin | 269.25 | non-Bitter | 829.31 | 1.409 | High | 0 | 0 | 0 | 0 |
| >O35 | Q95WY0 | DAENR | 5 | −0.7 | Non-Toxin | 332.75 | non-Bitter | 824.81 | 1.525 | High | 0 | 0 | 0 | 0 |
| >O36 | XP_034310988.1 | VAANI | 5 | −0.96 | Non-Toxin | 256 | non-Bitter | 834.91 | 1.252 | High | 0 | 0 | 0 | 0 |
| >O37 | XP_011417566.1 | AQQQK | 5 | −0.87 | Non-Toxin | 308.5 | non-Bitter | 834.81 | 1.405 | High | 0 | 0 | 0 | 0 |
| >O38 | XP_011429256.1 | DDESR | 5 | −0.64 | Non-Toxin | 221.75 | non-Bitter | 834.31 | 1.567 | High | 0 | 0 | 0 | 0 |
| >O39 | XP_034310988.1 | EQTQP | 5 | −1.18 | Non-Toxin | 273.25 | non-Bitter | 835.11 | 1.335 | High | 0 | 0 | 0 | 0 |
| >O40 | XP_011429256.1 | SVSER | 5 | −1.02 | Non-Toxin | 300 | non-Bitter | 835.01 | 1.138 | High | 0 | 0 | 0 | 0 |
| >O41 | XP_011429256.1 | ETDIR | 5 | −0.85 | Non-Toxin | 320 | non-Bitter | 836.41 | 1.07 | High | 0 | 0 | 0 | 0 |
| >O42 | XP_011429256.1 | AAEER | 5 | −0.85 | Non-Toxin | 278 | non-Bitter | 839.41 | 2.234 | High | 0 | 0 | 0 | 0 |
| >O43 | XP_011429256.1 | EANQA | 5 | −0.96 | Non-Toxin | 307.75 | non-Bitter | 852.71 | 1.884 | High | 0 | 0 | 0 | 0 |
| >O44 | XP_011429256.1 | TEINR | 5 | −0.86 | Non-Toxin | 332.5 | non-Bitter | 830.21 | 1.509 | High | 0 | 0 | 0 | 0 |
| >O45 | XP_034310988.1 | EQAER | 5 | −1.08 | Non-Toxin | 318 | non-Bitter | 818.61 | 2.042 | High | 0 | 0 | 0 | 0 |
| >O46 | P0DUE1 | EESGK | 5 | −0.8 | Non-Toxin | 254.25 | non-Bitter | 832.51 | 1.8 | High | 0 | 0 | 0 | 0 |
| >O47 | XP_011429256.1 | DAETK | 5 | −0.83 | Non-Toxin | 330 | non-Bitter | 834.81 | 1.67 | High | 0 | 0 | 0 | 0 |
| >O48 | XP_034310988.1 | AEVTR | 5 | −0.71 | Non-Toxin | 330.25 | non-Bitter | 847.31 | 1.154 | High | 0 | 0 | 0 | 0 |
| >O49 | XP_011429256.1 | VQVDD | 5 | −0.81 | Non-Toxin | 261.75 | non-Bitter | 834.81 | 1.804 | High | 0 | 0 | 0 | 0 |
| >O50 | Q95WY0 | EETIR | 5 | −0.66 | Non-Toxin | 327.5 | non-Bitter | 832.91 | 1.541 | High | 0 | 0 | 0 | 0 |
| >O51 | XP_034310988.1 | ITDEA | 5 | −0.94 | Non-Toxin | 332.5 | non-Bitter | 829.81 | 1.499 | High | 0 | 0 | 0 | 0 |
| >O52 | XP_034310988.1 | AAESE | 5 | −0.89 | Non-Toxin | 197 | non-Bitter | 833.61 | 1.548 | High | 0 | 0 | 0 | 0 |
| >O53 | XP_034310988.1 | EAEAK | 5 | −0.79 | Non-Toxin | 322.5 | non-Bitter | 835.71 | 1.455 | High | 0 | 0 | 0 | 0 |
| >O54 | XP_034310988.1 | AETQK | 5 | −0.77 | Non-Toxin | 313 | non-Bitter | 856.11 | 1.544 | High | 0 | 0 | 0 | 0 |
| >O55 | Q95WY0 | EEASK | 5 | −0.73 | Non-Toxin | 328.75 | non-Bitter | 831.11 | 1.462 | High | 0 | 0 | 0 | 0 |
| >O56 | XP_034310988.1 | TESTK | 5 | −1.01 | Non-Toxin | 269 | non-Bitter | 817.31 | 2.175 | High | 0 | 0 | 0 | 0 |
| >O57 | A9XE49 | CSGCVP | 6 | −0.72 | Non-Toxin | 305.8 | non-Bitter | 834.71 | 1.502 | High | 0 | 0 | 0 | 0 |
| >O58 | Q20A05 | GCPGDQ | 6 | −0.64 | Non-Toxin | 327.2 | non-Bitter | 844.71 | 3.646 | High | 0 | 0 | 0 | 0 |
| >O59 | XP_034310988.1 | SVTPSF | 6 | −0.95 | Non-Toxin | 312.4 | non-Bitter | 830.41 | 1.106 | High | 0 | 0 | 0 | 0 |
| >O60 | O17320 | CDVDIR | 6 | −1.1 | Non-Toxin | 323 | non-Bitter | 828.71 | 1.446 | High | 0 | 0 | 0 | 0 |
| >O61 | XP_034310988.1 | CIIPNE | 6 | −0.41 | Non-Toxin | 272.8 | non-Bitter | 834.21 | 1.451 | High | 0 | 0 | 0 | 0 |
| >O62 | XP_034310988.1 | CVAINP | 6 | −0.63 | Non-Toxin | 272 | non-Bitter | 828.71 | 1.164 | High | 0 | 0 | 0 | 0 |
| >O63 | XP_034310988.1 | DDIQQM | 6 | −1.08 | Non-Toxin | 259.6 | non-Bitter | 835.61 | 1.138 | High | 0 | 0 | 0 | 0 |
| >O64 | A9XE49 | AAAVHM | 6 | −0.94 | Non-Toxin | 232 | non-Bitter | 834.91 | 1.269 | High | 0 | 0 | 0 | 0 |
| >O65 | XP_034310988.1 | QQPAER | 6 | −1.37 | Non-Toxin | 276 | non-Bitter | 835.61 | 1.478 | High | 0 | 0 | 0 | 0 |
| >O66 | XP_011429256.1 | ASADAK | 6 | −0.72 | Non-Toxin | 312 | non-Bitter | 834.91 | 1.164 | High | 0 | 0 | 0 | 0 |
| >O67 | O17320 | ESSGIH | 6 | −0.87 | Non-Toxin | 274.6 | non-Bitter | 833.51 | 1.314 | High | 0 | 0 | 0 | 0 |
| >O68 | P0DUE1 | ICNEIK | 6 | −0.7 | Non-Toxin | 329.2 | non-Bitter | 836.11 | 1.33 | High | 0 | 0 | 0 | 0 |
| >O69 | XP_034310988.1 | QADEDR | 6 | −0.99 | Non-Toxin | 323.2 | non-Bitter | 839.91 | 1.874 | High | 0 | 0 | 0 | 0 |
| >O70 | XP_034310988.1 | VSSSSM | 6 | −0.76 | Non-Toxin | 315 | non-Bitter | 834.91 | 1.376 | High | 0 | 0 | 0 | 0 |
| >O71 | XP_011429256.1 | AAAQAA | 6 | −0.92 | Non-Toxin | 190.6 | non-Bitter | 834.81 | 1.337 | High | 0 | 0 | 0 | 0 |
| >O72 | XP_011429256.1 | EEASGM | 6 | −0.94 | Non-Toxin | 320 | non-Bitter | 830.11 | 1.463 | High | 0 | 0 | 0 | 0 |
| >O73 | XP_034310988.1 | ADQIDQ | 6 | −0.69 | Non-Toxin | 321 | non-Bitter | 835.91 | 1.355 | High | 0 | 0 | 0 | 0 |
| >O74 | Q95WY0 | QECQTK | 6 | −0.54 | Non-Toxin | 304.6 | non-Bitter | 838.01 | 1.592 | High | 0 | 0 | 0 | 0 |
| >O75 | XP_011429256.1 | ETAANM | 6 | −0.91 | Non-Toxin | 293.2 | non-Bitter | 835.71 | 1.302 | High | 0 | 0 | 0 | 0 |
| >O76 | XP_034310988.1 | SQQIEK | 6 | −1.04 | Non-Toxin | 292.6 | non-Bitter | 848.01 | 2.262 | High | 0 | 0 | 0 | 0 |
| >O77 | XP_011429256.1 | AAEVNR | 6 | −0.88 | Non-Toxin | 289.6 | non-Bitter | 852.51 | 1.335 | High | 0 | 0 | 0 | 0 |
| >O78 | P0DUE1 | AVDDSH | 6 | −0.77 | Non-Toxin | 285.4 | non-Bitter | 832.71 | 2.198 | High | 0 | 0 | 0 | 0 |
| >O79 | XP_011429256.1 | GETSQR | 6 | −1.06 | Non-Toxin | 300.4 | non-Bitter | 828.91 | 1.534 | High | 0 | 0 | 0 | 0 |
| >O80 | P0DUE1 | DSVESR | 6 | −0.78 | Non-Toxin | 317.2 | non-Bitter | 834.11 | 2.269 | High | 0 | 0 | 0 | 0 |
| >O81 | XP_011417566.1 | AATSNV | 6 | −0.98 | Non-Toxin | 255.2 | non-Bitter | 825.21 | 1.4 | High | 0 | 0 | 0 | 0 |
| >O82 | O17320 | VAIQAV | 6 | −1.12 | Non-Toxin | 280.6 | non-Bitter | 835.01 | 1.432 | High | 0 | 0 | 0 | 0 |
| >O83 | XP_034310988.1 | SAQVSS | 6 | −0.59 | Non-Toxin | 294.4 | non-Bitter | 842.51 | 1.323 | High | 0 | 0 | 0 | 0 |
| >O84 | XP_034310988.1 | SQESTD | 6 | −0.95 | Non-Toxin | 257.6 | non-Bitter | 828.21 | 1.728 | High | 0 | 0 | 0 | 0 |
| >O85 | XP_034310988.1 | IDECEE | 6 | −0.06 | Non-Toxin | 298 | non-Bitter | 834.81 | 1.455 | High | 0 | 0 | 0 | 0 |
| >O86 | XP_011429256.1 | QAANES | 6 | −1.03 | Non-Toxin | 226.8 | non-Bitter | 836.41 | 1.3 | High | 0 | 0 | 0 | 0 |
| >O87 | Q95WY0 | EAAEAK | 6 | −0.88 | Non-Toxin | 283.4 | non-Bitter | 837.01 | 1.426 | High | 0 | 0 | 0 | 0 |
| >O88 | XP_011429256.1 | QVQVDD | 6 | −0.85 | Non-Toxin | 255.6 | non-Bitter | 834.81 | 1.518 | High | 0 | 0 | 0 | 0 |
| >O89 | Q95WY0 | ATEAER | 6 | −1.16 | Non-Toxin | 319 | non-Bitter | 853.11 | 1.869 | High | 0 | 0 | 0 | 0 |
| >O90 | XP_034310988.1 | EDAEER | 6 | −0.99 | Non-Toxin | 333 | non-Bitter | 832.01 | 1.855 | High | 0 | 0 | 0 | 0 |
| >O91 | XP_034310988.1 | QAEEDK | 6 | −0.81 | Non-Toxin | 331.2 | non-Bitter | 847.91 | 2.09 | High | 0 | 0 | 0 | 0 |
| >O92 | Q95WY0 | AITEVD | 6 | −1.17 | Non-Toxin | 301.4 | non-Bitter | 831.91 | 1.789 | High | 0 | 0 | 0 | 0 |
| >O93 | XP_034310988.1 | EAQVSS | 6 | −0.89 | Non-Toxin | 300.4 | non-Bitter | 837.51 | 1.275 | High | 0 | 0 | 0 | 0 |
| >O94 | XP_034310988.1 | ESENDE | 6 | −0.78 | Non-Toxin | 272.4 | non-Bitter | 835.11 | 1.929 | High | 0 | 0 | 0 | 0 |
| >O95 | Q95WY0 | QTATEK | 6 | −1.13 | Non-Toxin | 306.4 | non-Bitter | 831.31 | 1.662 | High | 0 | 0 | 0 | 0 |
| >O96 | XP_011429256.1 | VINTEK | 6 | −0.82 | Non-Toxin | 297.8 | non-Bitter | 850.91 | 1.557 | High | 0 | 0 | 0 | 0 |
| >O97 | XP_034310988.1 | EEAEAA | 6 | −0.9 | Non-Toxin | 299.4 | non-Bitter | 834.91 | 1.561 | High | 0 | 0 | 0 | 0 |
| >O98 | XP_034310988.1 | QTEQDS | 6 | −1.33 | Non-Toxin | 320.8 | non-Bitter | 834.11 | 2.184 | High | 0 | 0 | 0 | 0 |
| >O99 | O17320 | TTTAER | 6 | −0.92 | Non-Toxin | 320.8 | non-Bitter | 834.71 | 1.464 | High | 0 | 0 | 0 | 0 |
| >O100 | P0DUE1 | SPSSTNM | 7 | −0.83 | Non-Toxin | 328.5 | non-Bitter | 836.01 | 1.249 | High | 0 | 0 | 0 | 0 |
| >O101 | A9XE49 | ICPSSIK | 7 | −0.8 | Non-Toxin | 313.67 | non-Bitter | 830.41 | 1.358 | High | 0 | 0 | 0 | 0 |
| >O102 | XP_034310988.1 | DAEIANM | 7 | −0.56 | Non-Toxin | 320 | non-Bitter | 830.61 | 1.398 | High | 0 | 0 | 0 | 0 |
| >O103 | XP_011429256.1 | EGDIAAM | 7 | −0.92 | Non-Toxin | 282 | non-Bitter | 846.91 | 1.856 | High | 0 | 0 | 0 | 0 |
| >O104 | XP_011417566.1 | GSTPDDK | 7 | −0.57 | Non-Toxin | 304.5 | non-Bitter | 884.11 | 1.373 | High | 0 | 0 | 0 | 0 |
| >O105 | XP_034310988.1 | ESDIQAM | 7 | −1.01 | Non-Toxin | 278.5 | non-Bitter | 875.81 | 1.265 | High | 0 | 0 | 0 | 0 |
| >O106 | XP_011429256.1 | DSDIQTK | 7 | −1.39 | Non-Toxin | 315.33 | non-Bitter | 875.11 | 2.027 | High | 0 | 0 | 0 | 0 |
| >O107 | Q7M456 | IPDSVVG | 7 | −1.04 | Non-Toxin | 314.33 | non-Bitter | 828.41 | 2.082 | High | 0 | 0 | 0 | 0 |
| >O108 | XP_034310988.1 | AVADAAR | 7 | −0.94 | Non-Toxin | 284 | non-Bitter | 835.01 | 1.225 | High | 0 | 0 | 0 | 0 |
| >O109 | O17320 | GDEDIAA | 7 | −0.88 | Non-Toxin | 283.67 | non-Bitter | 836.91 | 2.021 | High | 0 | 0 | 0 | 0 |
| >O110 | XP_034310988.1 | SSVSVTR | 7 | −0.77 | Non-Toxin | 323.33 | non-Bitter | 835.21 | 1.211 | High | 0 | 0 | 0 | 0 |
| >O111 | P0DUE1 | SDTPVTS | 7 | −1 | Non-Toxin | 290.33 | non-Bitter | 807.21 | 1.628 | High | 0 | 0 | 0 | 0 |
| >O112 | XP_034310988.1 | ANTEVQM | 7 | −0.92 | Non-Toxin | 252 | non-Bitter | 829.71 | 1.641 | High | 0 | 0 | 0 | 0 |
| >O113 | XP_034310988.1 | GDEITVK | 7 | −0.86 | Non-Toxin | 318.33 | non-Bitter | 833.81 | 2.4 | High | 0 | 0 | 0 | 0 |
| >O114 | XP_011417566.1 | GSSSEEA | 7 | −0.49 | Non-Toxin | 318.33 | non-Bitter | 831.41 | 1.297 | High | 0 | 0 | 0 | 0 |
| >O115 | XP_034310988.1 | TESIIAK | 7 | −0.73 | Non-Toxin | 296.33 | non-Bitter | 846.31 | 2.392 | High | 0 | 0 | 0 | 0 |
| >O116 | XP_011429256.1 | ESTEASM | 7 | −0.88 | Non-Toxin | 283.5 | non-Bitter | 830.91 | 2.031 | High | 0 | 0 | 0 | 0 |
| >O117 | XP_034310988.1 | EEAEAQA | 7 | −0.84 | Non-Toxin | 323.67 | non-Bitter | 837.51 | 1.561 | High | 0 | 0 | 0 | 0 |
| >O118 | XP_034310988.1 | EEEQESK | 7 | −0.68 | Non-Toxin | 312.83 | non-Bitter | 834.91 | 1.716 | High | 0 | 0 | 0 | 0 |
| >O119 | XP_034310988.1 | GPSSNPNF | 8 | −0.73 | Non-Toxin | 286.57 | non-Bitter | 832.41 | 1.282 | High | 0 | 0 | 0 | 0 |
| >O120 | P0DUE1 | VDDLPPPL | 8 | −0.72 | Non-Toxin | 261.14 | non-Bitter | 835.51 | 2.204 | High | 0 | 0 | 0 | 0 |
| >O121 | XP_034310988.1 | APNAIPQG | 8 | −0.61 | Non-Toxin | 280.71 | non-Bitter | 833.71 | 1.519 | High | 0 | 0 | 0 | 0 |
| >O122 | A9XE49 | QEGCTCVR | 8 | −0.63 | Non-Toxin | 324.71 | non-Bitter | 806.11 | 1.643 | High | 0 | 0 | 0 | 0 |
| >O123 | Q7M456 | YPPVHDNN | 8 | −0.59 | Non-Toxin | 296.57 | non-Bitter | 832.41 | 1.606 | High | 0 | 0 | 0 | 0 |
| >O124 | XP_034310988.1 | QPGVIDAA | 8 | −1.31 | Non-Toxin | 318.14 | non-Bitter | 864.71 | 2.056 | High | 0 | 0 | 0 | 0 |
| >O125 | O17320 | DSGDGVSH | 8 | −1.05 | Non-Toxin | 312.29 | non-Bitter | 823.31 | 3.856 | High | 0 | 0 | 0 | 0 |
| >O126 | O17320 | VVDNGSGM | 8 | −0.57 | Non-Toxin | 326.71 | non-Bitter | 808.81 | 1.377 | High | 0 | 0 | 0 | 0 |
| >O127 | Q7M456 | PSSDTESK | 8 | −0.89 | Non-Toxin | 290.29 | non-Bitter | 846.41 | 1.411 | High | 0 | 0 | 0 | 0 |
| >O128 | Q6L6Q6 | QQGCNVNS | 8 | −0.45 | Non-Toxin | 306 | non-Bitter | 830.51 | 1.425 | High | 0 | 0 | 0 | 0 |
| >O129 | XP_034310988.1 | IAGADIET | 8 | −0.65 | Non-Toxin | 320.86 | non-Bitter | 842.61 | 1.209 | High | 0 | 0 | 0 | 0 |
| >O130 | NP_001295835 | SVVANNIK | 8 | −0.97 | Non-Toxin | 318.57 | non-Bitter | 818.21 | 1.147 | High | 0 | 0 | 0 | 0 |
| >O131 | XP_011429256.1 | DEEIDSIR | 8 | −0.82 | Non-Toxin | 325 | non-Bitter | 833.01 | 1.544 | High | 0 | 0 | 0 | 0 |
| >O132 | XP_034310988.1 | TSVSSSSM | 8 | −0.68 | Non-Toxin | 317.86 | non-Bitter | 835.51 | 1.094 | High | 0 | 0 | 0 | 0 |
| >O133 | XP_034310988.1 | QTDTANEM | 8 | −0.54 | Non-Toxin | 324.57 | non-Bitter | 828.51 | 1.376 | High | 0 | 0 | 0 | 0 |
| >O134 | P0DUE1 | DSIADESS | 8 | −0.71 | Non-Toxin | 283 | non-Bitter | 832.91 | 3.6 | High | 0 | 0 | 0 | 0 |
| >O135 | O17320 | VGDEAQSK | 8 | −0.63 | Non-Toxin | 299.71 | non-Bitter | 902.11 | 2.04 | High | 0 | 0 | 0 | 0 |
| >O136 | XP_034310988.1 | DEEDAAAD | 8 | −1.04 | Non-Toxin | 282.71 | non-Bitter | 835.01 | 1.586 | High | 0 | 0 | 0 | 0 |
| >O137 | O17320 | TTAASSSS | 8 | −0.65 | Non-Toxin | 285.86 | non-Bitter | 834.91 | 1.159 | High | 0 | 0 | 0 | 0 |
| >O138 | Q4GWV4 | CTCTDCNGK | 9 | −0.5 | Non-Toxin | 321 | non-Bitter | 819.11 | 1.392 | High | 0 | 0 | 0 | 0 |
| >O139 | O17320 | VAPEEHPVL | 9 | −0.59 | Non-Toxin | 311.75 | non-Bitter | 828.01 | 2.088 | High | 0 | 0 | 0 | 0 |
| >O140 | XP_034310988.1 | ASQDEVIAR | 9 | −1.14 | Non-Toxin | 309.12 | non-Bitter | 937.71 | 1.243 | High | 0 | 0 | 0 | 0 |
| >O141 | Q7M456 | EVSETTCPR | 9 | −1.11 | Non-Toxin | 312.25 | non-Bitter | 800.31 | 1.3 | High | 0 | 0 | 0 | 0 |
| >O142 | XP_011429256.1 | GTSPSTQNR | 9 | −1.05 | Non-Toxin | 330.25 | non-Bitter | 816.21 | 2.136 | High | 0 | 0 | 0 | 0 |
| >O143 | XP_034310988.1 | ITGESGAGK | 9 | −1.28 | Non-Toxin | 278.5 | non-Bitter | 1058.51 | 1.721 | High | 0 | 0 | 0 | 0 |
| >O144 | XP_034310988.1 | VGAEIQSSK | 9 | −0.92 | Non-Toxin | 308.25 | non-Bitter | 873.21 | 1.519 | High | 0 | 0 | 0 | 0 |
| >O145 | XP_034310988.1 | EESQDSIEQ | 9 | −1.5 | Non-Toxin | 295.5 | non-Bitter | 838.81 | 1.576 | High | 0 | 0 | 0 | 0 |
| >O146 | XP_011429256.1 | EEESESASN | 9 | −0.6 | Non-Toxin | 240.75 | non-Bitter | 834.81 | 1.517 | High | 0 | 0 | 0 | 0 |
| >O147 | XP_034310988.1 | QTQIEEEQR | 9 | −1.08 | Non-Toxin | 326.25 | non-Bitter | 835.51 | 2.215 | High | 0 | 0 | 0 | 0 |
| >O148 | P0DUE1 | ESSDDTTVCM | 10 | −0.2 | Non-Toxin | 303.67 | non-Bitter | 801.21 | 1.398 | High | 0 | 0 | 0 | 0 |
| >O149 | NP_001295835 | QVQNDQASQR | 10 | −0.98 | Non-Toxin | 290.44 | non-Bitter | 838.51 | 2.22 | High | 0 | 0 | 0 | 0 |
| >O150 | XP_011429256.1 | QIDEAEDVAN | 10 | −0.69 | Non-Toxin | 321.78 | non-Bitter | 877.01 | 1.482 | High | 0 | 0 | 0 | 0 |
| >O151 | XP_011429256.1 | EIVTQAEDDR | 10 | −1.19 | Non-Toxin | 330.33 | non-Bitter | 852.41 | 1.067 | High | 0 | 0 | 0 | 0 |
| Bioactivity | Identifier | Activity Prediction | ||||
|---|---|---|---|---|---|---|
| Oyster Protein Source | Peptide Sequence | # Residues | Predictor | Score | ||
| Accession Number | Name | |||||
| Antihypertensive | A9XE49 | Interleukin 17-like protein (CgIL-17) | CEPVY | 5 | AHTpin | 1.44 |
| XP_034310988.1 | Myosin Heavy Chain, Striated Muscle | QQQIK | 5 | 1.44 | ||
| O17320 | Actin | VAPEEHPVL | 9 | 1.37 | ||
| XP_011417566.1 | Myosin Regulatory Light Chain B, Smooth Adductor Muscle Isoform X1 | AQQQK | 5 | 1.27 | ||
| XP_034310988.1 | Myosin Heavy Chain, Striated Muscle | STHPH | 5 | 1.02 | ||
| DPPIV-Inhibitory | XP_034310988.1 | Myosin Heavy Chain, Striated Muscle | EQTQP | 5 | iDPPIV-SCM | 418.75 |
| Q7M456 | Ribonuclease Oy (RNase Oy) (EC 3.1.27.-) | YPPVHDNN | 8 | 373 | ||
| O17320 | Actin | VAPEEHPVL | 9 | 360 | ||
| O17320 | Actin | NSPAM | 5 | 359.25 | ||
| P0DUE1 | Stimulator of interferon genes protein (TIR-STING) (Probable NAD(+) hydrolase) (EC 3.2.2.6) | SVPVL | 5 | 350 | ||
| Anti-inflammatory | Q4GWV4 | Defensin Cg-Defm (Cg-Def) (Mantle defensin) | CTCTDCNGK | 9 | PreAIP | 0.613 |
| XP_034310988.1 | Myosin Heavy Chain, Striated Muscle | QACID | 5 | PreAIP | 0.534 | |
| A9XE49 | Interleukin 17-like protein (CgIL-17) | QEGCTCVR | 8 | PreAIP | 0.521 | |
| P0DUE1 | Stimulator of interferon genes protein (TIR-STING) (Probable NAD(+) hydrolase) (EC 3.2.2.6) | ICNEIK | 6 | PreAIP | 0.491 | |
| Antiinflam | 3.356 | |||||
| Antimicrobial | XP_011429256.1 | Paramyosin Isoform X2 | EEESESASN | 9 | CAMP-SVM | 1 |
| CAMP-RF | 0.6545 | |||||
| ADAM-SVM | - | |||||
| XP_011429256.1 | Paramyosin Isoform X2 | EEASGM | 6 | CAMP-SVM | 1 | |
| CAMP-RF | 0.6395 | |||||
| ADAM-SVM | - | |||||
| XP_011429256.1 | Paramyosin Isoform X2 | ETAANM | 6 | CAMP-SVM | 1 | |
| CAMP-RF | 0.554 | |||||
| ADAM-SVM | - | |||||
| ADAM-SVM | - | |||||
| XP_011417566.1 | Myosin Regulatory Light Chain B, Smooth Adductor Muscle Isoform X1 | DKDGK | 5 | CAMP-SVM | 1 | |
| CAMP-RF | 0.5475 | |||||
| ADAM-SVM | - | |||||
| Q95WY0 | Tropomyosin (Allergen Cra g 1.03) (allergen Cra g 1) (Fragment) | SVVANNIK | 8 | DBAASPv3.0 | - | |
| ADAM-SVM | - | |||||
| Anti-cancer | A9XE49 | Interleukin 17-like protein (CgIL-17) | CSGCVP | 6 | ACPred | 0.947 |
| iDACP | 0.5196 | |||||
| mACPpred | 0.9812 | |||||
| Q20A06 | Hemocyte defensin Cg-Defh1 (Fragment) | CQSIGCR | 7 | ACPred | 0.935 | |
| iDACP | 0.5196 | |||||
| mACPpred | 0.8374 | |||||
| Q4GWV4 | Defensin Cg-Defm (Cg-Def) (Mantle defensin) | CTCTDCNGK | 9 | ACPred | 0.985 | |
| iDACP | 0.5574 | |||||
| mACPpred | 0.9253 | |||||
| Peptide Sequence | AHTpin SVM Score | Ranking/Selection Criteria | PepSite2 p-Value | Potential Binding Sites of ACE Protein | Potential Binding Residues on Peptide |
|---|---|---|---|---|---|
| CEPVY | 1.44 | AHTpin SVM Score > 1, Rank #1 | 6.94 × 10−4 | H353, H383 *, H387 *, F409, H410, E411 *, A412, I413, G414, D415, F457, K511, H513, Y523, S526, F527, Q281, A354, E384, Y520, S355, Y146, F512, A356, Y146 | C1, E2, P3, V4, Y5 |
| QQQIK | 1.44 | AHTpin SVM Score > 1, Rank #2 | 1.37 × 10−4 | Q281, H353, A354, H383 * | Q1, Q2, Q3, I4, K5 |
| EVSETTCPR | 1.43 | AHTpin SVM Score > 1, Rank #3 | 6.96 × 10−4 | Q281, H353, A354, S355, A356, H383 *, E384, H387 *, F391, E411 *, F457, K511, H513, Y520, H410, R522, W59, Y360 | E1, V2, S3, E4, T5, T6, C7, P8, R9 |
| VAPEEHPVL | 1.37 | AHTpin SVM Score > 1, Rank #4 | 2.30 × 10−4 | Y146, W279, Q281, H353, H383 *, H387 *, H410, E411 *, A412, D415, F457, F460, K511, F512, H513, Y520, Y523, S526, F527, E384, A354, S355, A356, R522 | V1, A2, P3, E4, E5, H6, P7, V8, L9 |
| PQSCR | 1.33 | AHTpin SVM Score > 1, Rank #5 | 5.62 × 10−5 | W279, Q281, H353, H383 *, E411 *, D415, F457, F460, K511, H513, Y520, Y523, S526, F527, A354, E384, H387 *, F409, H410, A412, I413, G414, Q530 | P1, Q2, S3, C4, R5 |
| AQQQK | 1.27 | AHTpin SVM Score > 1, Rank #6 | 7.32 × 10−5 | Q281, H353, A354, H383 *, E384, H387 *, E411 *, F457, K511, H513, Y520, Y523, F527, W279, F460, F512 | A1, Q2, Q3, Q4, K5 |
| GPSSNPNF | 1.17 | AHTpin SVM Score > 1, Rank #7 | 1.07 × 10−4 | W279, Q281, H353, A354, S355, H383 *, E384, H387 *, E411 *, F457, F460, K511, H513, Y520, Y523, N66, A356, W357, F391, F512, D358, Y360, F527 | G1, P2, S3, S4, N5, P6, N7, F8 |
| EQTQP | 1.14 | AHTpin SVM Score > 1, Rank #8 | 2.71 × 10−5 | Q281, H353, A354, H383 *, E384, H387 *, E411 *, F457, F460, K511, H513, Y520, F527, F512, Y523, D415, S526, S355 | E1, Q2, T3, Q4, P5 |
| STHPH | 1.02 | AHTpin SVM Score > 1, Rank #9 | 5.55 × 10−5 | Q281, H353, A354, S355, H383 *, E384, H387 *, E411 *, F457, H513, Y520, Y523, K511, W279, F460 | S1, T2, H3, P4, H5 |
| Peptide Sequence | iDPPIV-SCM Score | Ranking/Selection Criteria | PepSite2 p-Value | Potential Binding Sites of DPP-IV (Enzyme Protein) | Potential Binding Residues (Peptide) |
|---|---|---|---|---|---|
| EQTQP | 418.75 | iDPPIV-SCM Score >= 350, Rank #1 | 5.18 × 10−3 | Y48, W627, W629, V653, I703, I742, H748, I751, Y752, M755, Y547, S630 *, H740 *, G741 | E1, Q2, T3, Q4, P5 |
| TVPIY | 389.25 | iDPPIV-SCM Score >= 350, Rank #2 | 1.78 × 10−2 | Y48, W627, W629, V653, I703, I742, H748, I751, Y752, M755, G741, F357, Y547, Y666 | T1, V2, P3, I4, Y5 |
| VDDLPPPL | 385 | iDPPIV-SCM Score >= 350, Rank #3 | 6.52 × 10−2 | F357, Y547, P550, W629, Y631, Y666, Y670, G741, Y752, Y48, W627, H748 | V1, D2, D3, L4, P5, P6, P7, L8 |
| YPPVHDNN | 373 | iDPPIV-SCM Score >= 350, Rank #4 | 3.23 × 10−2 | F357, Y547, P550, C551, Y585, W629, S630 *, Y631, Y662, Y666, Y670, H740 *, Y48, W627, H748, Y752, G741 | Y1, P2, P3, V4, H5, D6, N7, N8 |
| VAPEEHPVL | 360 | iDPPIV-SCM Score >= 350, Rank #5 | 8.93 × 10−2 | F357, Y547, P550, W627, W629, S630 *, Y631, Y662, Y666, Y670, H740 *, Y752, Y48, M733, W734, Y735, H750, G741, H748 | V1, A2, P3, E4, E5, H6, P7, V8, L9 |
| NSPAM | 359.25 | iDPPIV-SCM Score >= 350, Rank #6 | 1.22 × 10−2 | Y48, W627, W629, H740 *, Y752, H748, S630 *, Y631, V656, W659, Y662, Y666, Y547 | N1, S2, P3, A4, M5 |
| SVPVL | 350 | iDPPIV-SCM Score >= 350, Rank #7 | 8.85 × 10−3 | Y48, W627, W629, H740 *, Y752, S630 *, G741, H748 | S1, V2, P3, V4, L5 |
| Database Source | Accession # a | Protein Name | Length (Residues) | Mass (Da) |
|---|---|---|---|---|
| Swiss-Prot | Q4GWV4 | Defensin Cg-Defm (Cg-Def) (Mantle defensin) | 65 | 7008 |
| P0DUE1 | Stimulator of interferon genes protein (TIR-STING) (Probable NAD(+) hydrolase) (EC 3.2.2.6) | 415 | 47,031 | |
| Q6L6Q6 | Lysozyme (EC 3.2.1.17) (1,4-beta-N-acetylmuramidase) (Invertebrate-type lysozyme) | 137 | 15,274 | |
| Q20A05 | Hemocyte defensin Cg-Defh2 (Fragment) | 60 | 6439 | |
| Q20A06 | Hemocyte defensin Cg-Defh1 (Fragment) | 60 | 6587 | |
| Q7M456 | Ribonuclease Oy (RNase Oy) (EC 3.1.27.-) | 213 | 24,360 | |
| Q95WY0 | Tropomyosin (Allergen Cra g 1.03) (allergen Cra g 1) (Fragment) | 233 | 26,867 | |
| A9XE49 | Interleukin 17-like protein (CgIL-17) | 200 | 21,551 | |
| O17320 | Actin | 376 | 41,792 | |
| NCBI RefSeq | XP_034310988.1 | Myosin Heavy Chain, Striated Muscle | 1986 | 222,660 |
| XP_011429256.1 | Paramyosin Isoform X2 | 886 | 102,210 | |
| NP_001295835 | Tropomyosin Isoform X1 | 284 | 33,020 | |
| XP_011417566.1 | Myosin Regulatory Light Chain B, Smooth Adductor Muscle Isoform X1 | 166 | 20,580 |
| Platform Name | Purpose | URL | Citation |
|---|---|---|---|
| AHTpin | Anti-hypertensive peptide prediction | http://crdd.osdd.net/raghava/ahtpin/, accessed on 15 January 2022 | [56] |
| iDPPIV-SCM | DPP-IV inhibitory peptide prediction | http://camt.pythonanywhere.com/iDPPIV-SCM/, accessed on 15 January 2022 | [57] |
| PepSite2 | Peptide-protein binding interaction modeling | http://pepsite2.russelllab.org/, accessed on 5 May 2022 | [58] |
| PreAIP | Anti-inflammatory peptide prediction | http://kurata14.bio.kyutech.ac.jp/PreAIP/, accessed on 18 January 2022 | [59] |
| AIPpred | Anti-inflammatory peptide prediction | http://www.thegleelab.org/AIPpred/, accessed on 1 March 2022 | [60] |
| Antiinflam | Anti-inflammatory peptide prediction | http://metagenomics.iiserb.ac.in/antiinflam/, accessed on 1 March 2022 | [61] |
| CAMP-R3 | Antimicrobial peptide prediction | http://www.camp.bicnirrh.res.in/predict/, accessed on 20 January 2022 | [62] |
| ACPred | Anti-cancer peptide prediction | http://codes.bio/acpred/, accessed on 11 March 2022 | [63] |
| iDACP | Anti-cancer peptide prediction | http://mer.hc.mmh.org.tw/iDACP/, accessed on 11 March 2022 | [64] |
| mACPpred | Anti-cancer peptide prediction | http://www.thegleelab.org/mACPpred/ACP.html, accessed on 13 March 2022 | [65] |
| BIOPEP-UVM | Comprehensive BAP database | https://biochemia.uwm.edu.pl/en/biopep-uwm-2, accessed on 1 February 2022 | [66] |
| PepBank | Comprehensive BAP database | http://pepbank.mgh.harvard.edu, accessed on 10 March 2022 | [67] |
| PeptideDB | Comprehensive BAP database | http://www.peptides.be, accessed on 10 March 2022 | [68] |
| EROP-Moscow | Comprehensive BAP database | http://erop.inbi.ras.ru/index.html, accessed on 10 March 2022 | [69] |
| BioPepDB | Comprehensive BAP database | http://bis.zju.edu.cn/biopepdbr/index.php, accessed on 11 April 2022 | [70] |
| AHTpDB | Anti-hypertensive peptide database | http://crdd.osdd.net/raghava/ahtpdb. accessed on 15 January 2022 | [56] |
| BioDADPep | Anti-diabetic peptide database | https://omicsbase.com/BioDADPep. accessed on 11 April 2022 | [71] |
| APD3 | Antimicrobial peptide database | https://aps.unmc.edu, accessed on 20 January 2022 | [72] |
| ADAM | Antimicrobial peptide database | http://bioinformatics.cs.ntou.edu.tw/adam, accessed on 12 May 2022 | [73] |
| DBAASP v3.0 | Antimicrobial peptide database | https://dbaasp.org, accessed on 12 May 2022 | [74] |
| CancerPPD | Anti-cancer peptide database | http://crdd.osdd.net/raghava/cancerppd, accessed on 27 March 2022 | [75] |
| TumorHoPe | Anti-cancer peptide database | https://webs.iiitd.edu.in/raghava/tumorhope/pepsearch.php, accessed on 27 March 2022 | [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Mendez, R.L.; Kwon, J.Y. In Silico Prospecting for Novel Bioactive Peptides from Seafoods: A Case Study on Pacific Oyster (Crassostrea gigas). Molecules 2023, 28, 651. https://doi.org/10.3390/molecules28020651
Zhou L, Mendez RL, Kwon JY. In Silico Prospecting for Novel Bioactive Peptides from Seafoods: A Case Study on Pacific Oyster (Crassostrea gigas). Molecules. 2023; 28(2):651. https://doi.org/10.3390/molecules28020651
Chicago/Turabian StyleZhou, Leyi, Rufa L. Mendez, and Jung Yeon Kwon. 2023. "In Silico Prospecting for Novel Bioactive Peptides from Seafoods: A Case Study on Pacific Oyster (Crassostrea gigas)" Molecules 28, no. 2: 651. https://doi.org/10.3390/molecules28020651
APA StyleZhou, L., Mendez, R. L., & Kwon, J. Y. (2023). In Silico Prospecting for Novel Bioactive Peptides from Seafoods: A Case Study on Pacific Oyster (Crassostrea gigas). Molecules, 28(2), 651. https://doi.org/10.3390/molecules28020651






