Computational Simulation of Colorectal Cancer Biomarker Particle Mobility in a 3D Model
Abstract
1. Introduction
2. Results
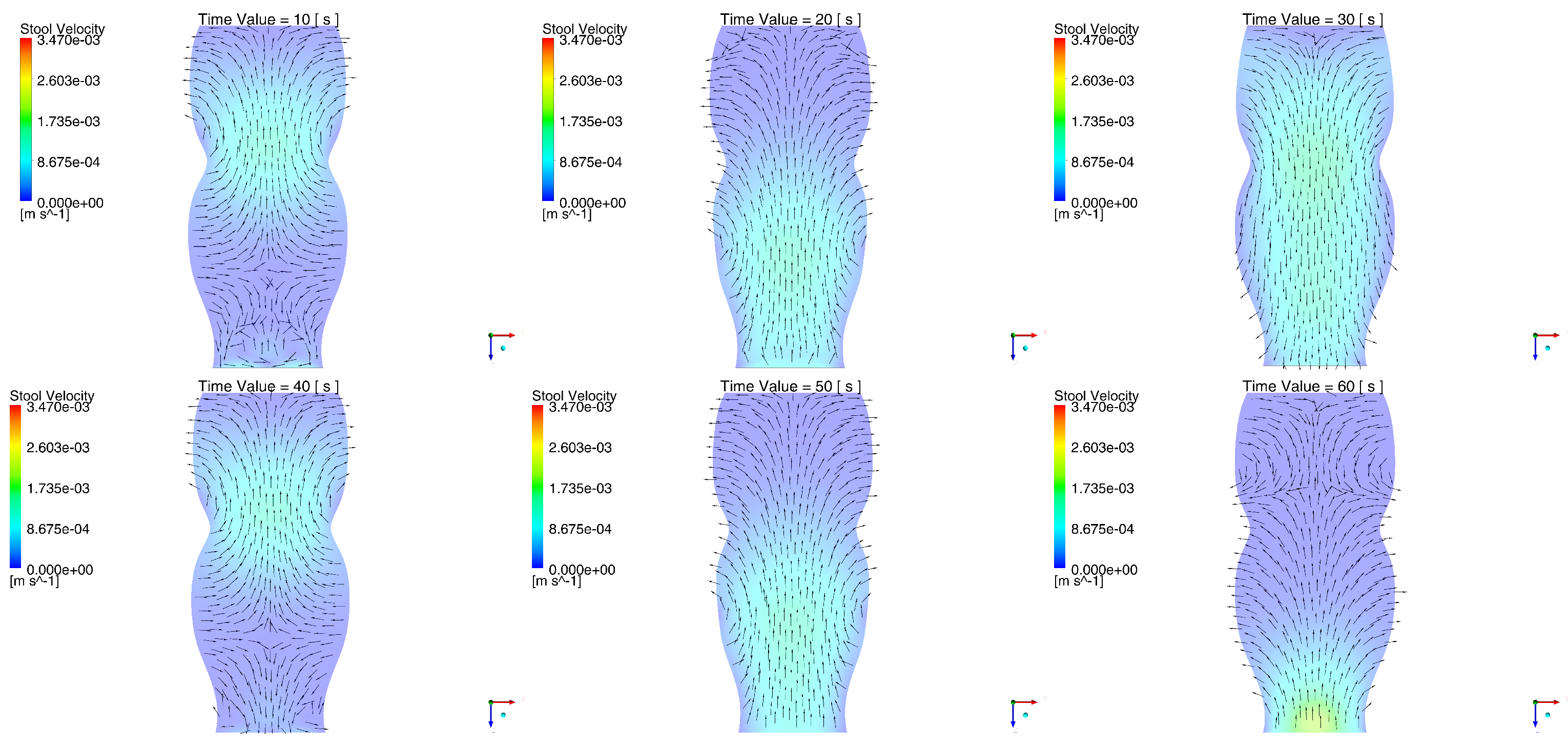
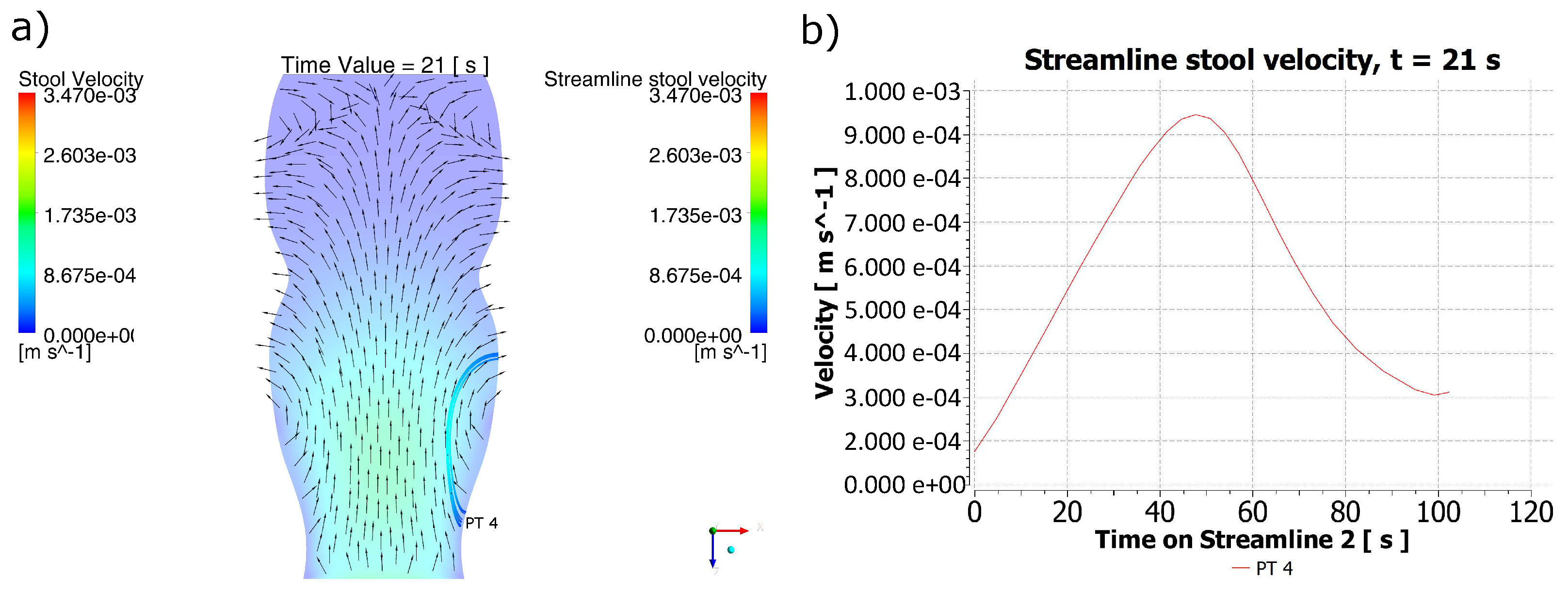
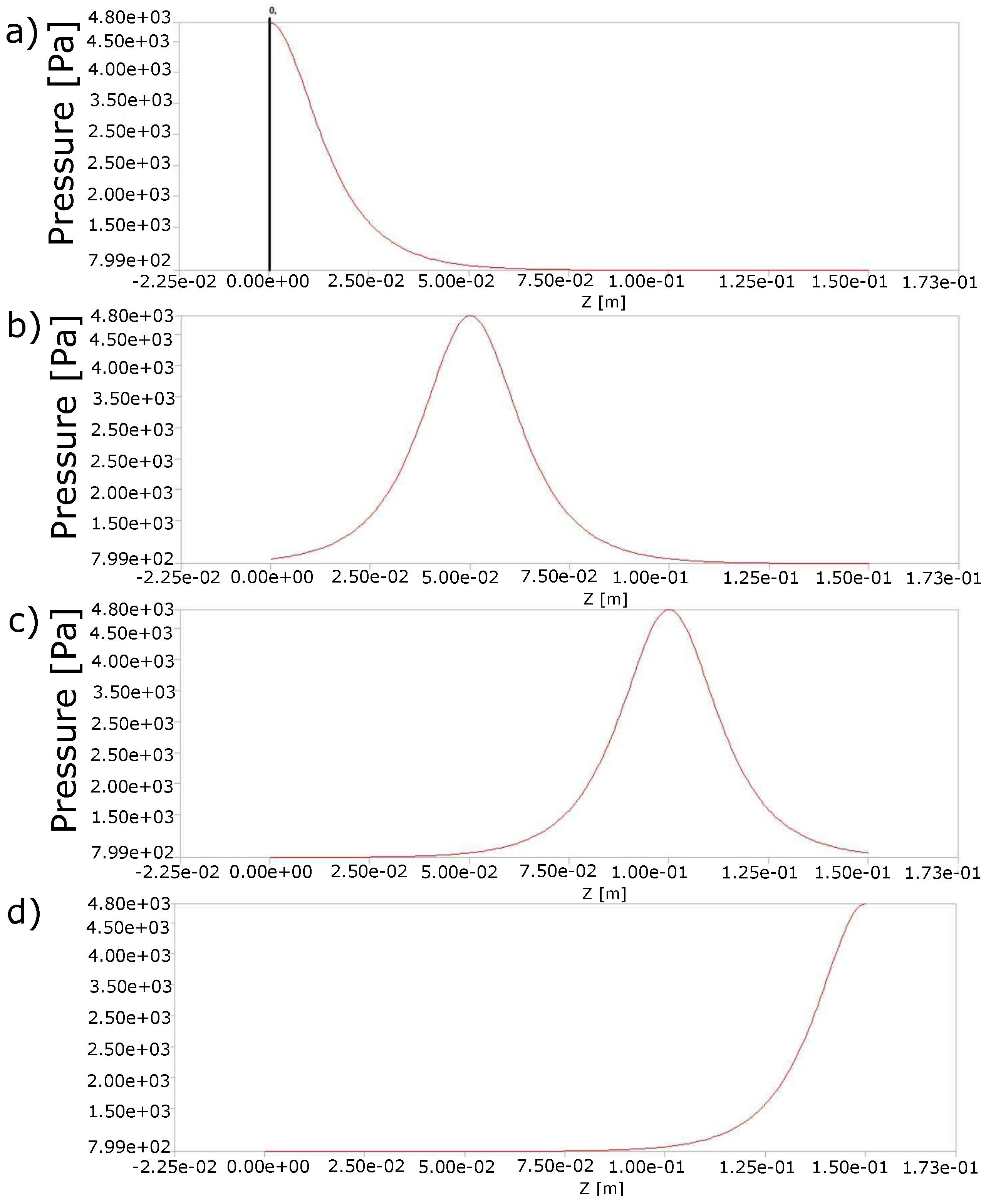
2.1. Behavior of the Stool during the Passing of the Contraction Waves
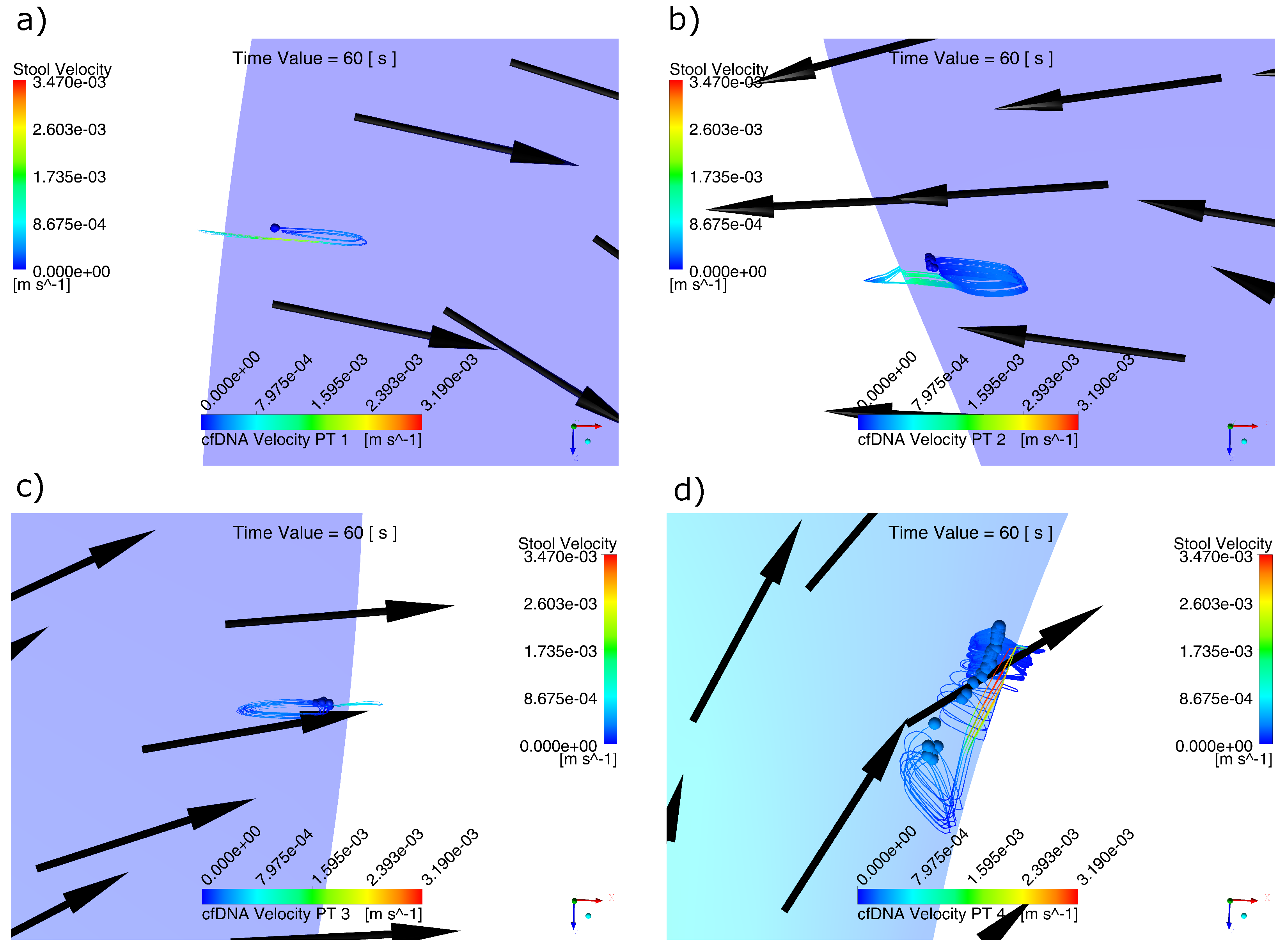
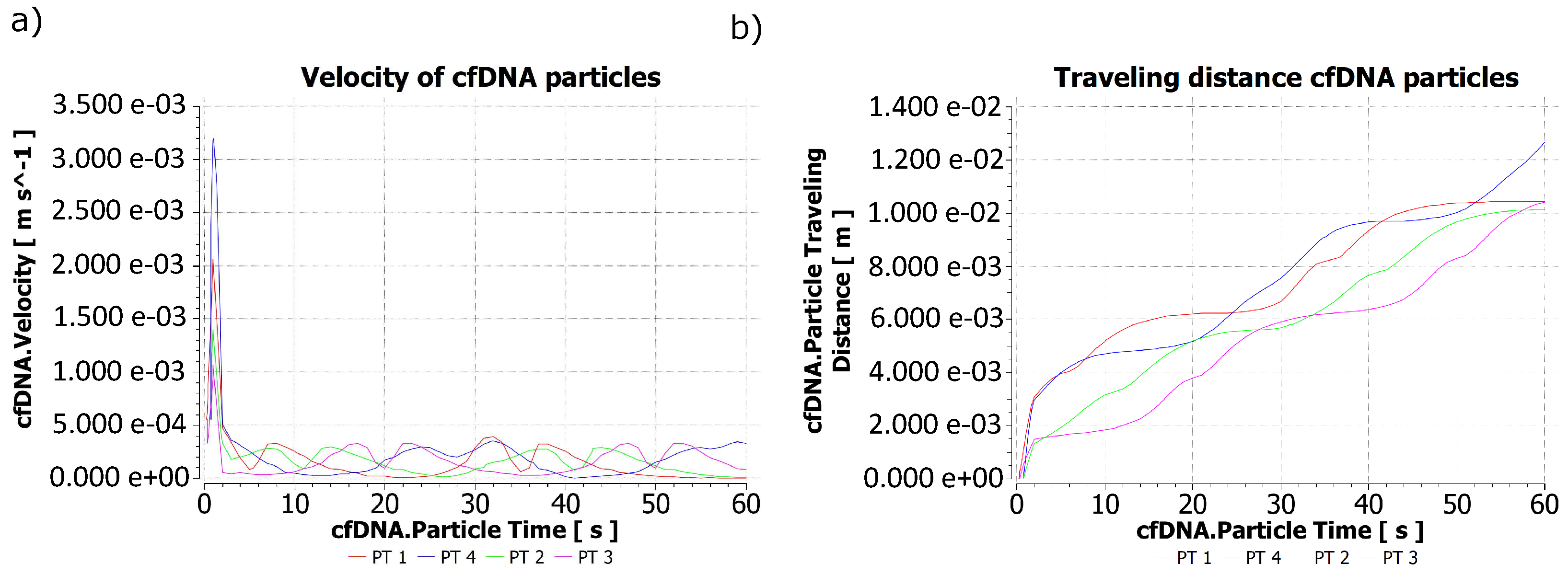
2.2. Behavior of Biomarker Particles during the Passing of the Contraction Waves
3. Discussion
4. Materials and Methods
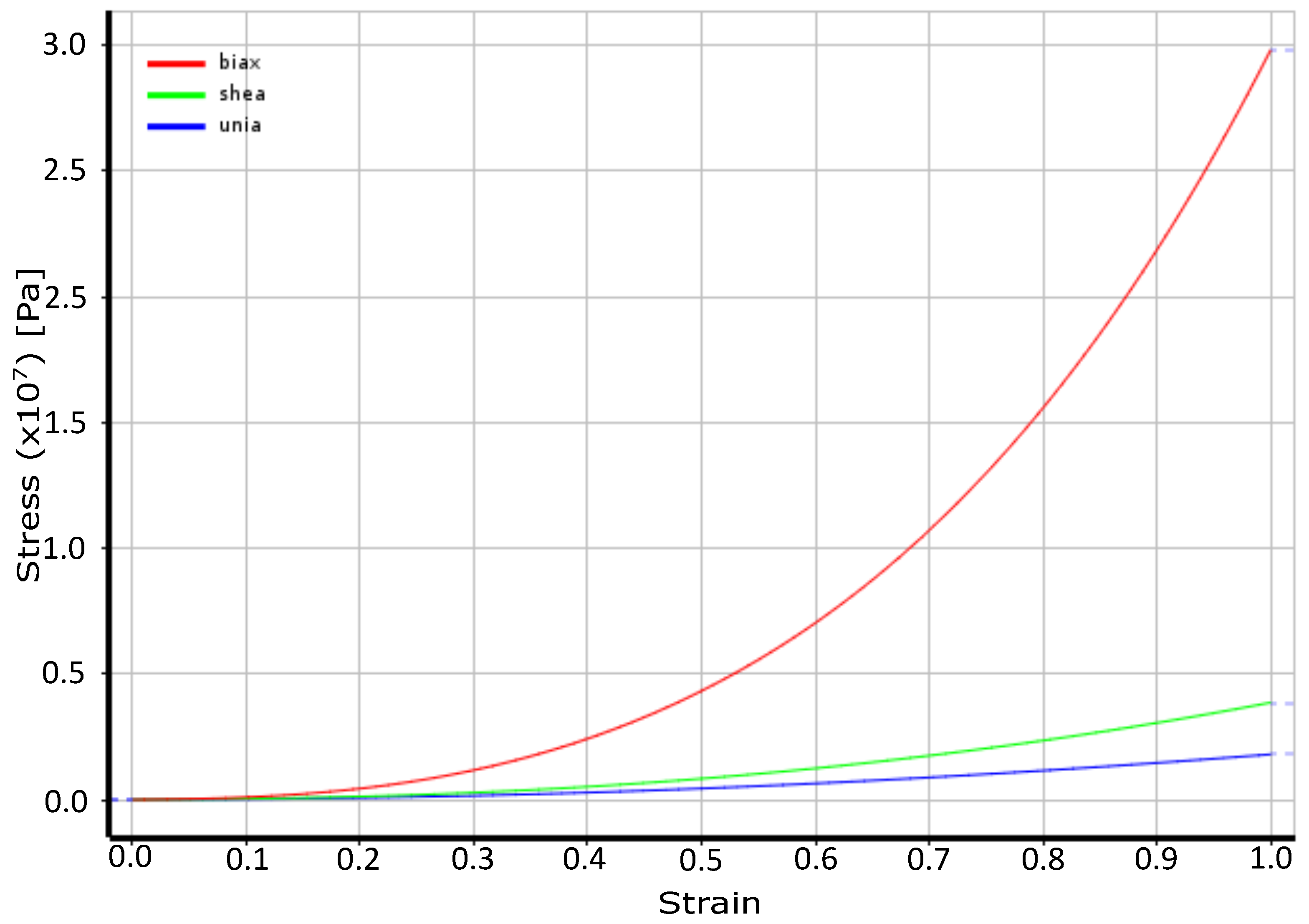
4.1. Material Properties: Rectum Wall
4.2. Peristaltic Movement
- Type I contractions: simple monophasic waves of low width and short duration. These contractions form holes on the surface creation pressures of 5 to 10 cm of (490.333–980.665 Pa), their duration varies from 5 to10 s, and their frequency is 8 to 12/min [34].
- Type II contractions: These have a greater width 8, 15 to 30 (784.532, 1471, 2942 Pa), and last longer (25 to 30 s), their frequency is 2/min; both contractions act to mix the stool [34].
- Type III contractions: These represent a change in the base pressure, generally lower than 10 (980.665 Pa), with superposition of type I and II waves [34].
4.3. Stool
- A Dirichlet-type condition for the fluid velocity at the entry border given by
- A Neumann condition given by
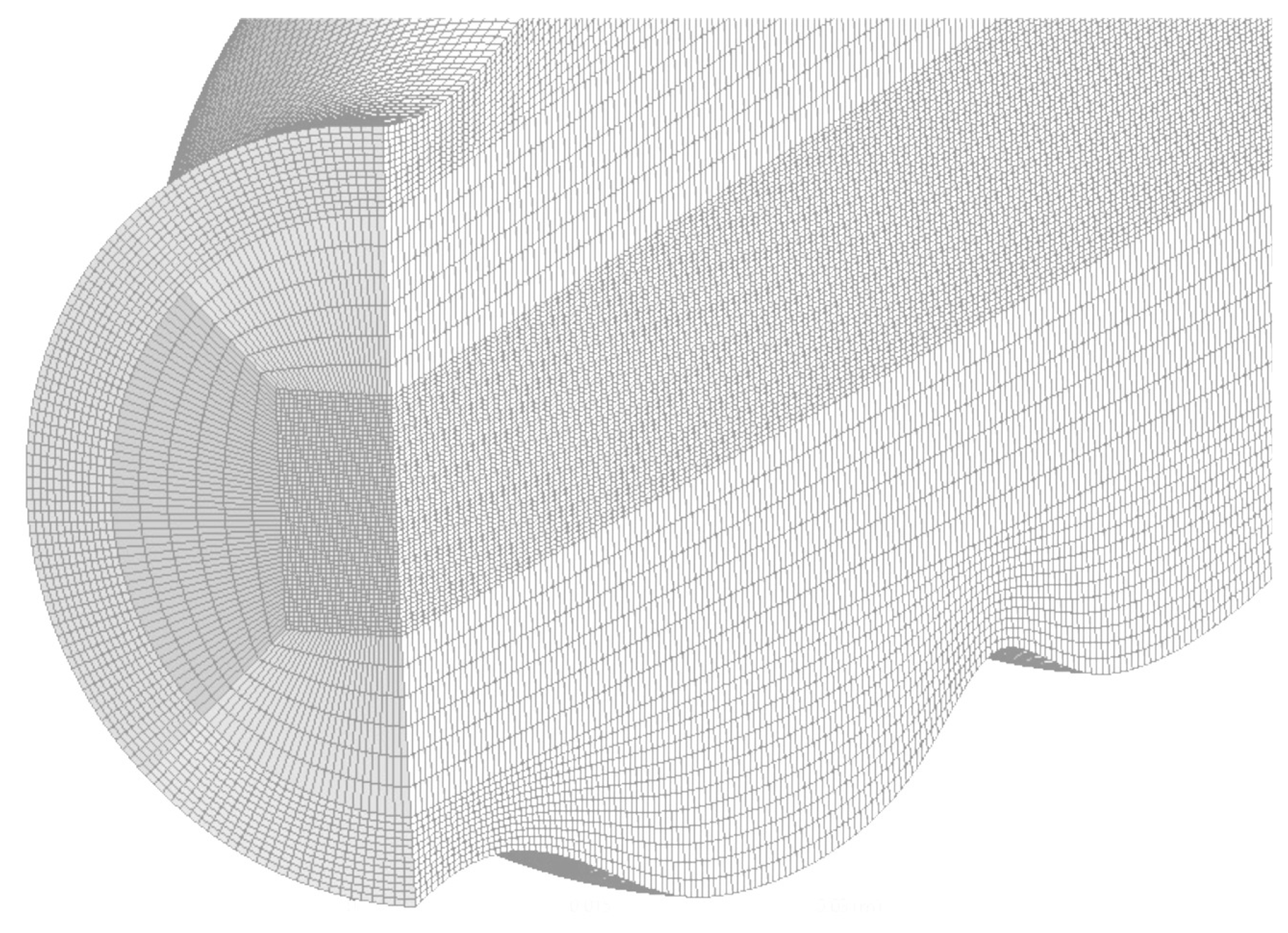
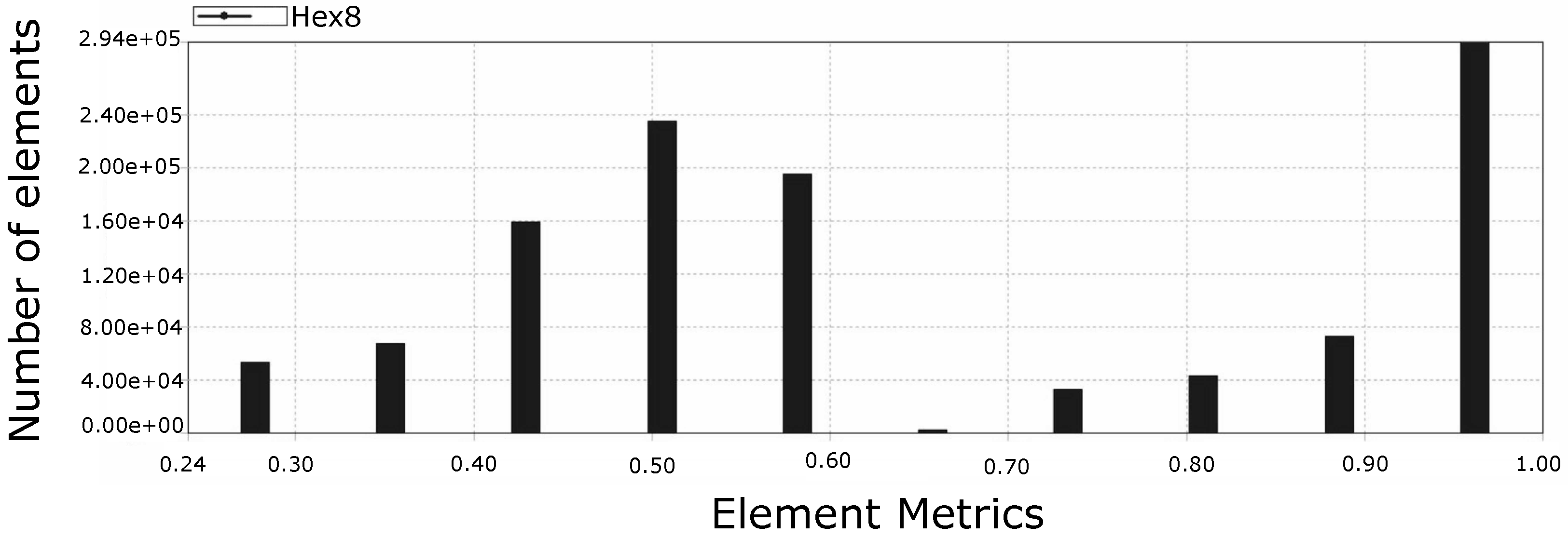
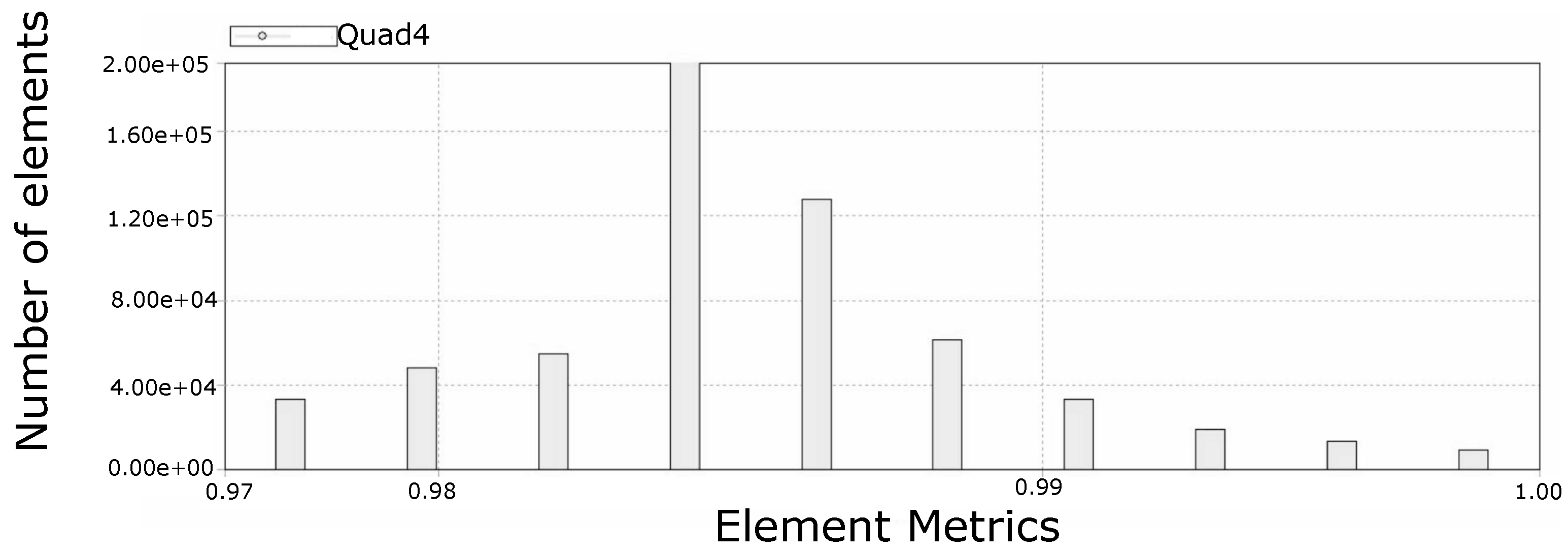
4.4. Mesh
4.5. Description of the Biomarker Particles
4.5.1. Colon Epithelial Cells
4.5.2. Exfoliation Processes
4.5.3. Calculation of Parameters for the Injection of Particles through the Rectum Wall
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Estimated Number of Incident Cases, Both Sexes, Worldwide (Top 10 Cancer Sites) in 2018; World Health Organization: Geneva, Switzerland, 2018.
- Metkar, S.K.; Girigoswami, K. Diagnostic biosensors in medicine—A review. Biocatal. Agric. Biotechnol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, H.; Zhou, Q.; Song, Q.; Rui, J.; Zou, B.; Zhou, G. Digital quantification of gene methylation in stool DNA by emulsion-PCR coupled with hydrogel immobilized bead-array. Biosens. Bioelectron. 2017, 92, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Oussalah, A.; Rischer, S.; Bensenane, M.; Conroy, G.; Filhine-Tresarrieu, P.; Debard, R.; Forest-Tramoy, D.; Josse, T.; Reinicke, D.; Garcia, M.; et al. Plasma mSEPT9: A Novel Circulating Cell-free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. EBioMedicine 2018, 30, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Jia, J.; Yu, H.; Peng, X.; Xiao, W.; Gong, Y.; Zhou, G.; Han, X.; Li, Y. The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J. Cancer Res. Clin. Oncol. 2017, 143, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Hwang, S.M.; Kim, T.S.; Kim, D.W.; Park, D.J.; Kang, S.B.; Kim, H.H.; Park, K.U. Circulating Methylated Septin 9 Nucleic Acid in the Plasma of Patients with Gastrointestinal Cancer in the Stomach and Colon. Transl. Oncol. 2013, 6, 290–296. [Google Scholar] [CrossRef]
- Deeb, D.; Gao, X.; Jiang, H.; Arbab, A.S.; Dulchavsky, S.A.; Gautam, S.C. Growth inhibitory and apoptosis-inducing effects of xanthohumol, a prenylated chalone present in hops, in human prostate cancer cells. Anticancer Res. 2010, 30, 3333–3339. [Google Scholar]
- Ahlquist, D.A.; Taylor, W.R.; Mahoney, D.W.; Zou, H.; Domanico, M.; Thibodeau, S.N.; Boardman, L.A.; Berger, B.M.; Lidgard, G.P. The Stool DNA Test Is More Accurate Than the Plasma Septin 9 Test in Detecting Colorectal Neoplasia. Clin. Gastroenterol. Hepatol. 2012, 10, 272–277. [Google Scholar] [CrossRef]
- De Maio, G.; Rengucci, C.; Zoli, W.; Calistri, D. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World J. Gastroenterol. 2014, 20, 957–967. [Google Scholar] [CrossRef]
- Vatandoost, N.; Ghanbari, J.; Mojaver, M.; Avan, A.; Ghayour-Mobarhan, M.; Nedaeinia, R.; Salehi, R. Early detection of colorectal cancer: From conventional methods to novel biomarkers. J. Cancer Res. Clin. Oncol. 2016, 142, 341–351. [Google Scholar] [CrossRef]
- Saeed, M.; Shoaib, A.; Kandimalla, R.; Javed, S.; Almatroudi, A.; Gupta, R.; Aqil, F. Microbe-based therapies for colorectal cancer: Advantages and limitations. Semin. Cancer Biol. 2022, 86, 652–665. [Google Scholar] [CrossRef]
- Hlquist, D.A.A.A. Stool DNA screening for colorectal neoplasia: Biological and technical basis for high detection rates colorectal cancer screening. Pathology 2012, 44, 80–88. [Google Scholar] [CrossRef]
- Yousefi, A.T.; Ikeda, S.; Rusop Mahmood, M.; Yousefi, H.T. Simulation of Nano Sensor Based on Carbon Nanostructures in Order to Form Multifunctional Delivery Platforms. Adv. Mater. Res. 2013, 832, 778–782. [Google Scholar] [CrossRef]
- Ravi, A.; Krishna, R.M.A.; Christen, J.B. Modeling and Simulation of Dual Application Capacitive MEMS Sensor. In Proceedings of the 2014 COMSOL Conference, Arizona State University, Tempe, AZ, USA; 2014; pp. 1–4. [Google Scholar]
- Lacatus, E.; Alecu, G.C.; Tudor, A. Models for Simulation Based Selection of 3D Multilayered Graphene Biosensors. In Proceedings of the 2015 COMSOL Conference, Grenoble, France, 14–16 October 2015; pp. 3–8. [Google Scholar] [CrossRef]
- Kang, G.; Márquez, C.; Barat, A.; Byrne, A.T.; Prehn, J.H.M.; Sorribes, J.; César, E. Colorectal tumour simulation using agent based modelling and high performance computing. Future Gener. Comput. Syst. 2017, 67, 397–408. [Google Scholar] [CrossRef]
- Mustapha, K.; Gilli, Q.; Frayret, J.M.; Lahrichi, N.; Karimi, E. Agent-based Simulation Patient Model for Colon and Colorectal Cancer Care Trajectory. Procedia Comput. Sci. 2016, 100, 188–197. [Google Scholar] [CrossRef]
- Dzwinel, W.; Kłusek, A.; Vasilyev, O.V. Supermodeling in simulation of melanoma progression. Procedia Comput. Sci. 2016, 80, 999–1010. [Google Scholar] [CrossRef]
- Bethge, A.; Schumacher, U.; Wedemann, G. Simulation of metastatic progression using a computer model including chemotherapy and radiation therapy. J. Biomed. Inform. 2015, 57, 74–87. [Google Scholar] [CrossRef]
- Wedemann, G.; Bethge, A.; Haustein, V.; Schumacher, U. Computer simulation of the metastatic progression. In Methods in Molecular Biology, 2nd ed.; Dwek, M., Schumacher, U., Brooks, S.A., Eds.; Springer: New York, NY, USA, 2014; Chapter 8; Volume 1070, pp. 107–116. [Google Scholar] [CrossRef]
- Liu, Y.; Shah, S.; Tan, J. Computational Modeling of Nanoparticle Targeted Drug Delivery. Rev. Nanosci. Nanotechnol. 2012, 1, 66–83. [Google Scholar] [CrossRef]
- Tóth, K.; Wasserkort, R.; Sipos, F.; Kalmár, A.; Wichmann, B.; Leiszter, K.; Valcz, G.; Juhász, M.; Miheller, P.; Patai, Á.V.; et al. Detection of methylated Septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS ONE 2014, 9, e115415. [Google Scholar] [CrossRef]
- ANSYS Inc. ANSYS Help; ANSYS Inc.: Canonsburg, PA, USA, 2019. [Google Scholar]
- He, X. Modeling of the Interaction between Colon and Colonoscope during a Colonoscopy. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2018. [Google Scholar]
- Sokolis, D.P.; Sassani, S.G. Microstructure-based constitutive modeling for the large intestine validated by histological observations. J. Mech. Behav. Biomed. Mater. 2013, 21, 149–166. [Google Scholar] [CrossRef]
- Yazdanpanh-Ardakani, K.; Niroomand-Oscuii, H. New Approach in Modeling Peristaltic Transport of Non-Newtonian Fluid. J. Mech. Med. Biol. 2013, 13, 1350052. [Google Scholar] [CrossRef]
- Yu, Y. Numerical Methods for Fluid-Structure Interaction: Analysis and Simulations. Ph.D. Thesis, Brown University, Providence, RI, USA, 2014. [Google Scholar]
- TermehYousefi, A.; Bagheri, S.; Shahnazar, S.; Rahman, M.H.; Kadri, N.A. Computational local stiffness analysis of biological cell: High aspect ratio single wall carbon nanotube tip. Mater. Sci. Eng. C 2016, 59, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, G.S.; Maday, Y.; Osorno, J.B. Solución del Problema de Dominios Acoplados con Interaccion Fluido-Estructura (F-E) en un Dispositivo de Asistencia Ventricular Cardiaca. Ph.D. Thesis, Universidad Pontificia Bolivariana, Medellin, Colombia, 2011. [Google Scholar]
- Mcintosh, R.L.; Anderson, V. Erratum: “A Comprehensive Tissue Properties Database Provided for the Thermal Assessment of a Human At Rest”. Biophys. Rev. Lett. 2013, 8, 99–100. [Google Scholar] [CrossRef]
- Christensen, M.B.; Oberg, K.; Wolchok, J.C. Tensile Properties of the Rectal and Sigmoid Colon: A Comparative Analysis of Human and Porcine Tissue; Springer: Berlin/Heidelberg, Germany, 2015; Volume 4. [Google Scholar] [CrossRef]
- Omari, T.I.; Rudolph, C.D. Chapter 112—Gastrointestinal Motility. In Fetal and Neonatal Physiology, 3rd ed.; Polin, R.A., Fox, W.W., Abman, S.H., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2004; pp. 1125–1138. [Google Scholar] [CrossRef]
- Baker, M.L.; Tune, J.M.; Hightower, D. Intraluminal Pressure Measurements During Barium Enema. Am. J. Roentgenol. 1981, 137, 217–221. [Google Scholar]
- Arróniz, M.A.C.; Palacios, A.C.; Zebadúa, Ó.A. Anatomía y fisiología de colon. In Gastroenterología; Torres, E.P., Francis, J.M.A., Sahagún, F.B., Stalnikowitz, D.K., Eds.; McGraw-Hill Education: New York, NY, USA, 2015. [Google Scholar]
- Toklu, E. A new mathematical model of peristaltic flow on esophageal bolus transport. Sci. Res. Essays 2011, 6, 6606–6614. [Google Scholar] [CrossRef]
- Sarna, S.K. Giant migrating contractions and their myoelectric correlates in the small intestine. Am. J. Physiol.-Gastrointest. Liver Physiol. 1987, 253, G697–G705. [Google Scholar] [CrossRef]
- Painter, N.S.; Truelove, S.C. The intraluminal pressure patterns in diverticulosis of the colon Part I Resting patterns of pressure. Gut 1964, 5, 201–207. [Google Scholar] [CrossRef]
- Chen, J.H.; Yu, Y.; Yang, Z.; Yu, W.Z.; Chen, W.L.; Yu, H.; Kim, M.J.M.; Huang, M.; Tan, S.; Luo, H.; et al. Intraluminal pressure patterns in the human colon assessed by high-resolution manometry. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Penn, R.; Ward, B.J.; Strande, L.; Maurer, M. Review of synthetic human faeces and faecal sludge for sanitation and wastewater research. Water Res. 2018, 132, 222–240. [Google Scholar] [CrossRef]
- Sinnott, M.D.; Cleary, P.W.; Arkwright, J.W.; Dinning, P.G. Investigating the relationships between peristaltic contraction and fluid transport in the human colon using Smoothed Particle Hydrodynamics. Comput. Biol. Med. 2012, 42, 492–503. [Google Scholar] [CrossRef]
- Bourgault, Y.; Ethier, M.; Leblanc, V.G. ESAIM: Mathematical Modelling and Numerical Analysis Simulation of Electrophysiological Waves with an Unstructured Finite Element Method. ESAIM Math. Model. Numer. Anal. 2003, 37, 649–661. [Google Scholar] [CrossRef]
- Heil, M. An efficient solver for the fully coupled solution of large-displacement fluid-structure interaction problems. Comput. Methods Appl. Mech. Eng. 2004, 193, 1–23. [Google Scholar] [CrossRef]
- Olivella, X.; Agelet de Saracíbar, C. Mecánica de Medios Continuos Para Ingenieros; Universitat Politecnica de Catalunya: Barcelona, Spain, 2002; p. 329. [Google Scholar]
- Salvi, S.; Gurioli, G.; De Giorgi, U.; Conteduca, V.; Tedaldi, G.; Calistri, D.; Casadio, V. Cell-free DNA as a diagnostic marker for cancer: Current insights. OncoTargets Ther. 2016, 9, 6549–6559. [Google Scholar] [CrossRef]
- Jung, K.; Fleischhacker, M.; Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker-A critical appraisal of the literature. Clin. Chim. Acta 2010, 411, 1611–1624. [Google Scholar] [CrossRef]
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br. J. Cancer 2014, 111, 1482–1489. [Google Scholar] [CrossRef]
- Tanić, M.; Beck, S. Epigenome-wide association studies for cancer biomarker discovery in circulating cell-free DNA: Technical advances and challenges. Curr. Opin. Genet. Dev. 2017, 42, 48–55. [Google Scholar] [CrossRef]
- Salomo, M.; Kegler, K.; Gutsche, C.; Reinmuth, J.; Skokow, W.; Kremer, F.; Hahn, U.; Struhalla, M. The elastic properties of single double-stranded DNA chains of different lengths as measured with optical tweezers. Colloid Polym. Sci. 2006, 284, 1325–1331. [Google Scholar] [CrossRef]
- Venema, K. The TNO In Vitro Model of the Colon (TIM-2). In The Impact of Food Bioactives on Health; Springer International Publishing: Cham, Switzerland, 2015; pp. 293–304. [Google Scholar]
- Wiśniewski, J.R.; Ostasiewicz, P.; Duś, K.; Zielińska, D.F.; Gnad, F.; Mann, M. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 2012, 8, 611. [Google Scholar] [CrossRef]
- Collins, F.S.; Lander, E.S.; Rogers, J. International human genome sequencing consortium, finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Martínez-Martín, D.; Fläschner, G.; Gaub, B.; Martin, S.; Newton, R.; Beerli, C.; Mercer, J.; Gerber, C.; Müller, D.J. Inertial picobalance reveals fast mass fluctuations in mammalian cells. Nature 2017, 550, 500–505. [Google Scholar] [CrossRef]
- Fonseca, J.C.; Marques, J.C.; Paiva, A.A.; Freitas, A.M.; Madeira, V.M.; Jørgensen, S.E. Nuclear DNA in the determination of weighing factors to estimate exergy from organisms biomass. Ecol. Model. 2000, 126, 179–189. [Google Scholar] [CrossRef]
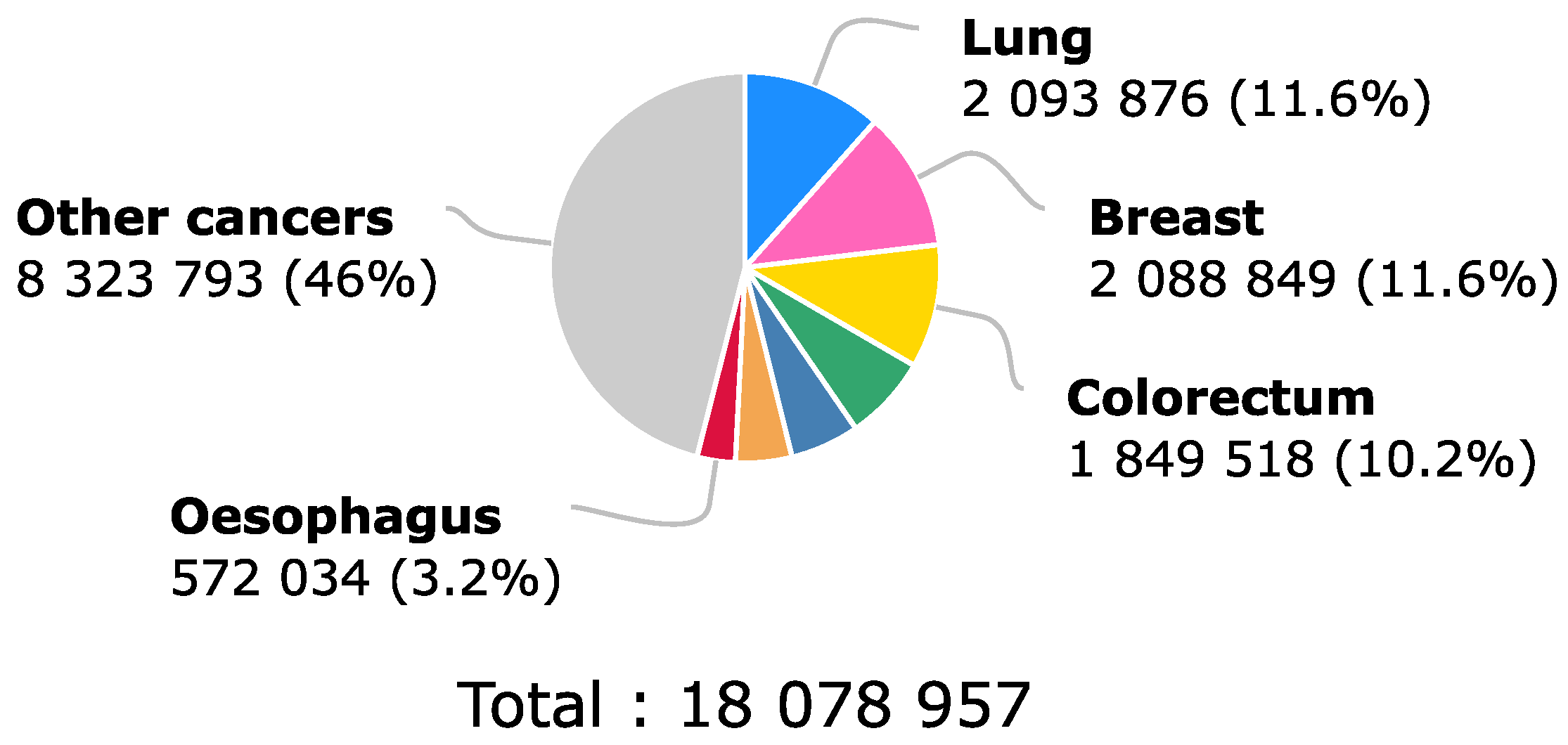




| Constant | Value (Pa) |
|---|---|
| 652.01 | |
| 42,835.25 | |
| 219,120.30 |
| Length (bp) | Length (nm) | Mass (kg) | Volume () | Density () |
|---|---|---|---|---|
| 200 | 68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallejo Morales, E.; Suárez Guerrero, G.; Hoyos Palacio, L.M. Computational Simulation of Colorectal Cancer Biomarker Particle Mobility in a 3D Model. Molecules 2023, 28, 589. https://doi.org/10.3390/molecules28020589
Vallejo Morales E, Suárez Guerrero G, Hoyos Palacio LM. Computational Simulation of Colorectal Cancer Biomarker Particle Mobility in a 3D Model. Molecules. 2023; 28(2):589. https://doi.org/10.3390/molecules28020589
Chicago/Turabian StyleVallejo Morales, Esteban, Gustavo Suárez Guerrero, and Lina M. Hoyos Palacio. 2023. "Computational Simulation of Colorectal Cancer Biomarker Particle Mobility in a 3D Model" Molecules 28, no. 2: 589. https://doi.org/10.3390/molecules28020589
APA StyleVallejo Morales, E., Suárez Guerrero, G., & Hoyos Palacio, L. M. (2023). Computational Simulation of Colorectal Cancer Biomarker Particle Mobility in a 3D Model. Molecules, 28(2), 589. https://doi.org/10.3390/molecules28020589






