Improving the Transduction Efficiency and Antitumor Effect of Conditionally Replicative Adenovirus by Application of 6-cyclohexyl Methyl-β-D-maltoside
Abstract
1. Introduction
2. Results and Discussion
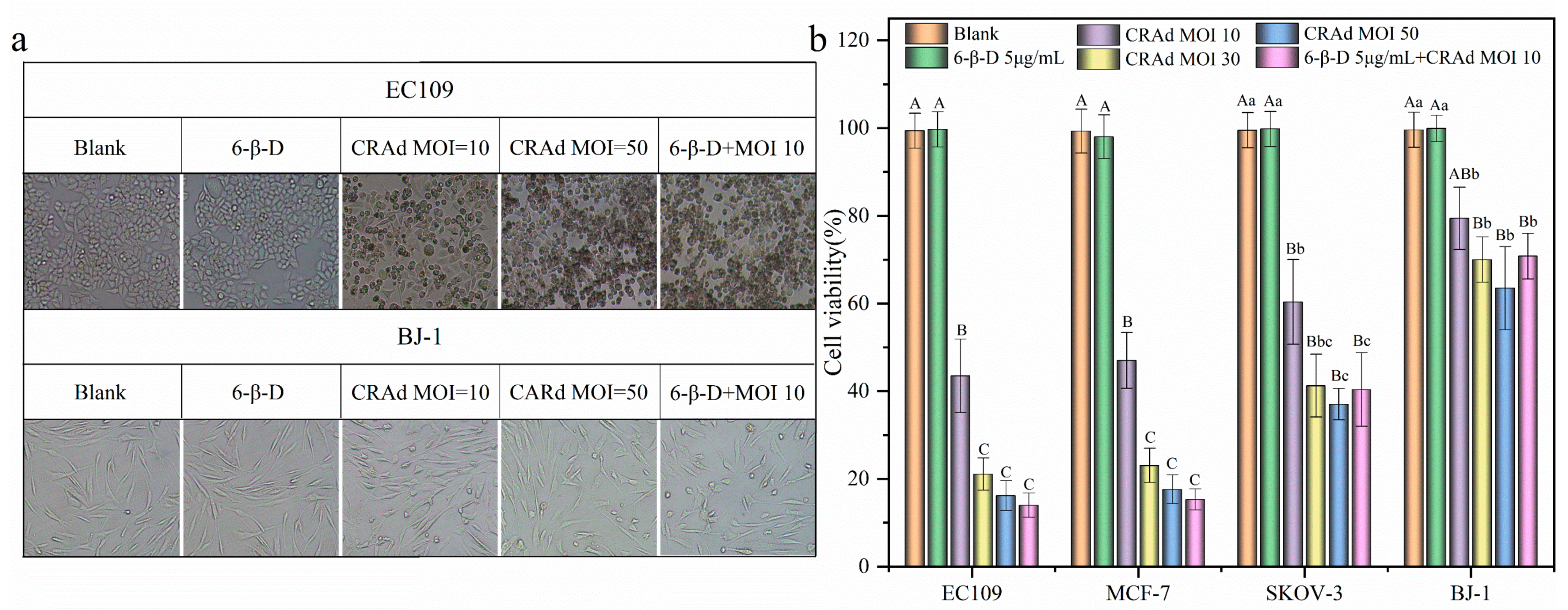
2.1. Conditional Replication Adenoviruses Can Kill Cancer Cells Specifically
2.2. High Transduction Efficiency and Low Cytotoxicity of 6-β-D
2.3. Application of 6-β-D Improved the Oncolytic Effect of CRAd
2.4. Combining 6-β-D with CRAd Increased Virus Replication in Cancer Cells
3. Materials and Methods
3.1. Cell Lines and Drugs
3.2. Cytotoxicity Assay
3.3. Transduction Efficiency
3.4. Survival Inhibition Effect Assay
3.5. RNA Preparation and RT-PCR
3.6. Viral Titer Assay
3.7. Flow Cytometry (FCM)
3.8. Caspase-3 Activity Analysis
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pastor, R.; Goedegebuure, P.S.; Curiel, D.T. Understanding and addressing barriers to successful adenovirus-based virotherapy for ovarian cancer. Cancer Gene Ther. 2021, 28, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Allawadhi, P.; Khurana, A.; Singh, V.; Navik, U.; Pasumarthi, S.K.; Khurana, I.; Banothu, A.K.; Weiskirchen, R.; Bharani, K.K. Gene therapy: Comprehensive overview and therapeutic applications. Life Sci. 2022, 294, 120375. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Nishikawaji, Y.; Kawakami, H.; Kosai, K.-I. Adenovirus Biology, Recombinant Adenovirus, and Adenovirus Usage in Gene Therapy. Viruses 2021, 13, 2502. [Google Scholar] [CrossRef]
- Muhammad, T.; Sakhawat, A.; Khan, A.A.; Ma, L.; Gjerset, R.A.; Huang, Y. Mesenchymal stem cell-mediated delivery of therapeutic adenoviral vectors to prostate cancer. Stem Cell Res. Ther. 2019, 10, 190. [Google Scholar] [CrossRef]
- Sasso, E.; Froechlich, G.; Cotugno, G.; D’Alise, A.M.; Gentile, C.; Bignone, V.; De Lucia, M.; Petrovic, B.; Campadelli-Fiume, G.; Scarselli, E.; et al. Replicative conditioning of Herpes simplex type 1 virus by Survivin promoter, combined to ERBB2 retargeting, improves tumour cell-restricted oncolysis. Sci. Rep. 2020, 10, 4307. [Google Scholar] [CrossRef]
- Chen, J.-S.; Liu, J.-C.; Shen, L.; Rau, K.-M.; Kuo, H.-P.; Li, Y.M.; Shi, D.; Lee, Y.-C.; Chang, K.-J.; Hung, M.-C. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004, 11, 740–747. [Google Scholar] [CrossRef]
- Ryan, B.M.; O’Donovan, N.; Duffy, M.J. Survivin: A new target for anti-cancer therapy. Cancer Treat. Rev. 2009, 35, 553–562. [Google Scholar] [CrossRef]
- Mu, X.; Wang, X.; Wei, Y.; Wen, C.; Zhang, Q.; Xu, C.; Liu, C.; Zhang, C.; Meng, F.; Zhao, N.; et al. ApoE-modified liposomes mediate the antitumour effect of survivin promoter-driven HSVtk in hepatocellular carcinoma. Cancer Gene Ther. 2020, 27, 754–767. [Google Scholar] [CrossRef]
- You, Z.; Fischer, D.C.; Tong, X.; Hasenburg, A.; Aguilar-Cordova, E.; Kieback, D.G. Coxsackievirus–adenovirus receptor expression in ovarian cancer cell lines is associated with increased adenovirus transduction efficiency and transgene expression. Cancer Gene Ther. 2001, 8, 168–175. [Google Scholar] [CrossRef]
- Kim, J.-S.; Lee, S.-H.; Cho, Y.-S.; Choi, J.-J.; Kim, Y.H.; Lee, J.-H. Enhancement of the Adenoviral Sensitivity of Human Ovarian Cancer Cells by Transient Expression of Coxsackievirus and Adenovirus Receptor (CAR). Gynecol. Oncol. 2002, 85, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, W.; Hamada, K.; Tagawa, M.; Morinaga, T.; Gotoh, A. Cancer Cell-specific Transfection of hCas9 Gene Using Ad5F35 Vector. Anticancer Res. 2021, 41, 3731–3740. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Li, Y.; Li, S.; Li, W.; Zhu, Y.; Wang, J.; Liu, X.; Yue, Y.; Jin, N.; Li, X. Anti-tumor effect of a dual cancer-specific recombinant adenovirus on ovarian cancer cells. Exp. Cell Res. 2020, 396, 112185. [Google Scholar] [CrossRef]
- Tian, Y.; Yao, W.; He, D.; Xu, Y.; Li, Y.; Zhu, Y.; Fang, J.; Bai, B.; Li, X.; Sun, L.; et al. A dual cancer-specific recombinant adenovirus suppresses the growth of liver cancer cells in vivo and in vitro. Anti-Cancer Drugs 2020, 31, 110–122. [Google Scholar] [CrossRef]
- Xiang, C.; Tenkumo, T.; Ogawa, T.; Kanda, Y.; Nakamura, K.; Shirato, M.; Sokolova, V.; Epple, M.; Kamano, Y.; Egusa, H.; et al. Gene transfection achieved by utilizing antibacterial calcium phosphate nanoparticles for enhanced regenerative therapy. Acta Biomater. 2021, 119, 375–389. [Google Scholar] [CrossRef]
- Sokolova, V.; Epple, M. Biological and Medical Applications of Calcium Phosphate Nanoparticles. Chemistry 2021, 27, 7471–7488. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.R.K.; Shou, Y.; Chan, B.; Logan, K.; Tay, A. Materials for Improving Immune Cell Transfection. Adv. Mater. 2021, 33, e2007421. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Patel, A.K.; Rhym, L.H.; Palmiero, U.C.; Bhat, B.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Systemic delivery of mRNA and DNA to the lung using polymer-lipid nanoparticles. Biomaterials 2021, 275, 120966. [Google Scholar] [CrossRef]
- Ravula, V.; Lo, Y.-L.; Wang, L.-F.; Patri, S.V. Gemini Lipopeptide Bearing an Ultrashort Peptide for Enhanced Transfection Efficiency and Cancer-Cell-Specific Cytotoxicity. ACS Omega 2021, 6, 22955–22968. [Google Scholar] [CrossRef]
- Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. [Google Scholar] [CrossRef]
- Wehbie, M.; Onyia, K.K.; Mahler, F.; Le Roy, A.; Deletraz, A.; Bouchemal, I.; Vargas, C.; Babalola, J.O.; Breyton, C.; Ebel, C.; et al. Maltose-Based Fluorinated Surfactants for Membrane-Protein Extraction and Stabilization. Langmuir 2021, 37, 2111–2122. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Liu, L.; Reinhart, C.; Michel, H. Heterologous expression of human Neuromedin U receptor 1 and its subsequent solubilization and purification. Biochim. Biophys. Acta 2008, 1778, 2203–2209. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sakurai, F.; Nishimae, F.; Takayama, K.; Mizuguchi, H. Optimization of an E1A Gene Expression Cassette in an Oncolytic Adenovirus for Efficient Tumor Cell Killing Activity. Anticancer Res. 2021, 41, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Brachtlova, T.; Van Beusechem, V.W. Unleashing the Full Potential of Oncolytic Adenoviruses against Cancer by Applying RNA Interference: The Force Awakens. Cells 2018, 7, 228. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, C.; Liu, W.; Chen, W.; Yin, Y.; Li, C.-W.; Hsu, J.L.; Sun, J.; Zhou, Q.; Li, H.; et al. SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics 2020, 10, 4627–4643. [Google Scholar] [CrossRef]
- He, R.; Cui, M.; Lin, H.; Zhao, L.; Wang, J.; Chen, S.; Shao, Z. Melatonin resists oxidative stress-induced apoptosis in nucleus pulposus cells. Life Sci. 2018, 199, 122–130. [Google Scholar] [CrossRef]




| Time | Treatment | Fold (Titer/PFU/mL) | |
|---|---|---|---|
| SKOV-3 | EC109 | ||
| 3 h | CRAd | 1 (105.3) | 1 (105.5) |
| 6-β-D + CRAd | 1.50 (105.5) | 1.58 (105.7) | |
| 96 h | CRAd | 1 (109.5) | 1 (1010.4) |
| 6-β-D + CRAd | 1.86 (1010) | 4.90 (1011.2) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Fang, Y.; Meng, X.; Wang, X.; Liu, W.; Liu, M.; Zhang, P. Improving the Transduction Efficiency and Antitumor Effect of Conditionally Replicative Adenovirus by Application of 6-cyclohexyl Methyl-β-D-maltoside. Molecules 2023, 28, 528. https://doi.org/10.3390/molecules28020528
Lu W, Fang Y, Meng X, Wang X, Liu W, Liu M, Zhang P. Improving the Transduction Efficiency and Antitumor Effect of Conditionally Replicative Adenovirus by Application of 6-cyclohexyl Methyl-β-D-maltoside. Molecules. 2023; 28(2):528. https://doi.org/10.3390/molecules28020528
Chicago/Turabian StyleLu, Wenjing, Yaping Fang, Xue Meng, Xiaoli Wang, Wenbo Liu, Mengdong Liu, and Ping Zhang. 2023. "Improving the Transduction Efficiency and Antitumor Effect of Conditionally Replicative Adenovirus by Application of 6-cyclohexyl Methyl-β-D-maltoside" Molecules 28, no. 2: 528. https://doi.org/10.3390/molecules28020528
APA StyleLu, W., Fang, Y., Meng, X., Wang, X., Liu, W., Liu, M., & Zhang, P. (2023). Improving the Transduction Efficiency and Antitumor Effect of Conditionally Replicative Adenovirus by Application of 6-cyclohexyl Methyl-β-D-maltoside. Molecules, 28(2), 528. https://doi.org/10.3390/molecules28020528






