Structure–Function Analysis of RBP7910: An Editosome Z-Binding Protein in Trypanosomatids
Abstract
:1. Introduction
2. Results and Discussion
2.1. Binding Affinity of RBP7910 with Potential Oligonucleotide Ligands
2.2. Three-Dimensional Structure Prediction of RBP7910
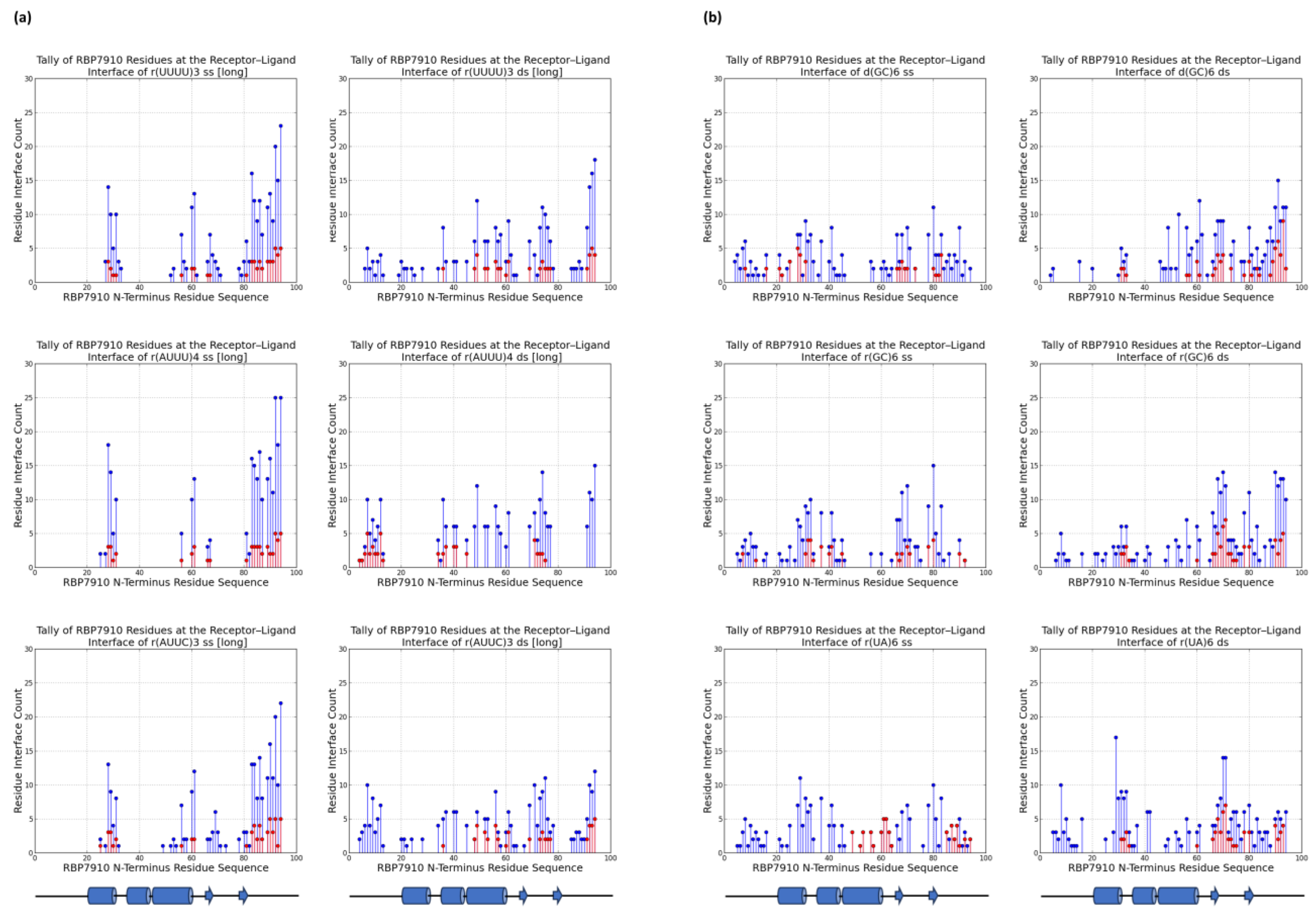
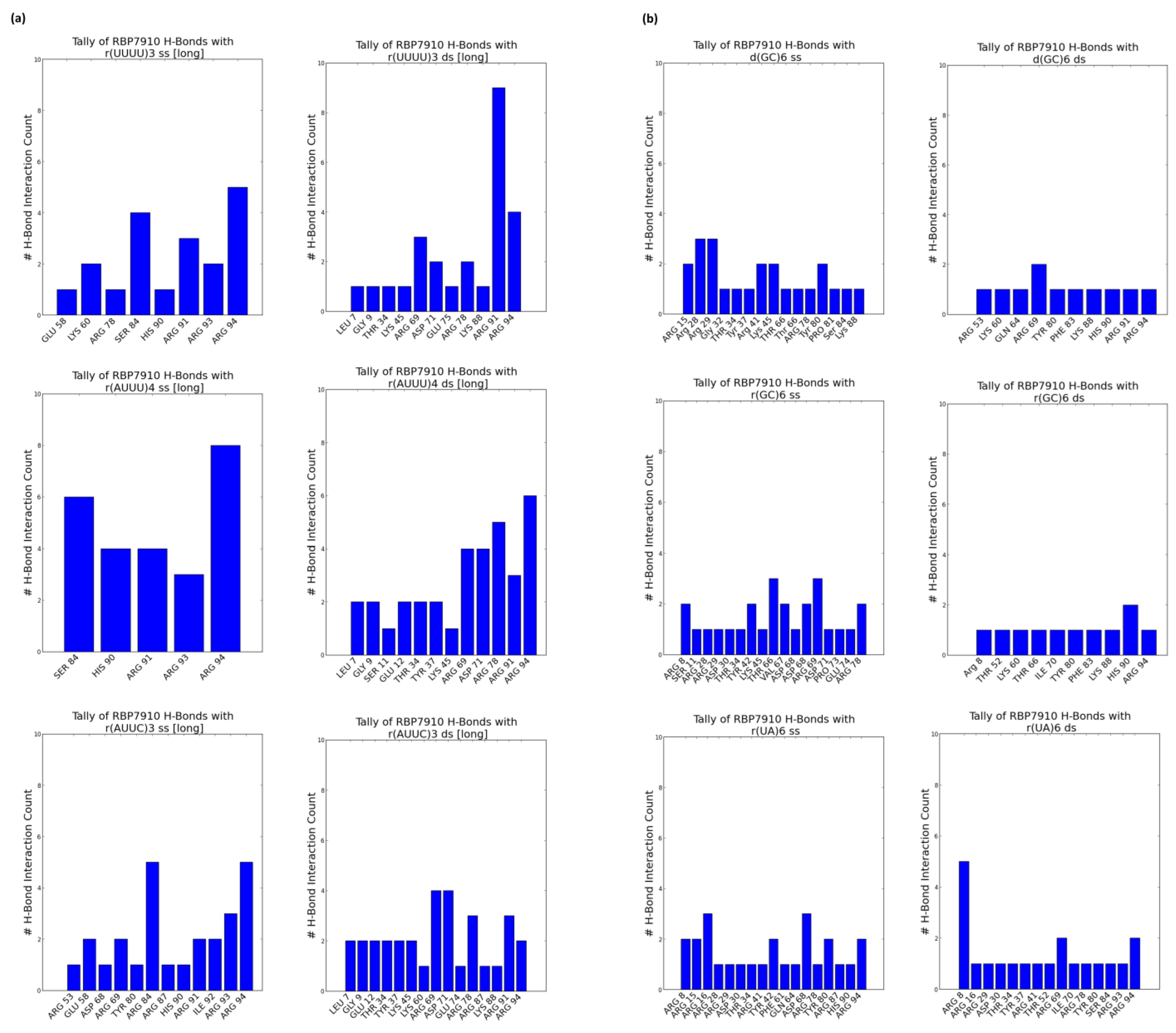
2.3. Prediction of the Binding Interface via Molecular Docking
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. Estimating the Binding Affinity of RBP7910 with Different Ligands
3.3. Modeling the 3D Structure of RBP7910
3.4. Molecular Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, V.; Madina, B.R.; Gulati, S.; Vashisht, A.A.; Kanyumbu, C.; Pieters, B.; Shakir, A.; Wohlschlegel, J.A.; Read, L.K.; Mooers, B.H.M. REH2C helicase and GRBC subcomplexes may base pair through mRNA and small guide RNA in kinetoplastid editosomes. J. Biol. Chem. 2016, 291, 5753–5764. [Google Scholar] [CrossRef] [PubMed]
- Aphasizheva, I.; Alfonzo, J.; Carnes, J.; Cestari, I.; Cruz-Reyes, J.; Göringer, H.U.; Hajduk, S.; Lukeš, J.; Madison-Antenucci, S.; Maslov, D.A. Lexis and grammar of mitochondrial RNA processing in trypanosomes. Trends Parasitol. 2020, 36, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Aphasizheva, I.; Aphasizhev, R. U-insertion/deletion mRNA-editing holoenzyme: Definition in sight. Trends Parasitol. 2016, 32, 144–156. [Google Scholar] [CrossRef]
- Nikpour, N.; Salavati, R. The RNA binding activity of the first identified trypanosome protein with Z-DNA-binding domains. Sci. Rep. 2019, 9, 5904. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. Z-DNA and Z-RNA in human disease. Commun. Biol. 2019, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Kim, D.; Kim, K.K.; Kim, Y.-G.; Hohng, S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011, 133, 668–671. [Google Scholar] [CrossRef]
- De Rosa, M.; de Sanctis, D.; Rosario, A.L.; Archer, M.; Rich, A.; Athanasiadis, A.; Carrondo, M.A. Crystal structure of a junction between two Z-DNA helices. Proc. Natl. Acad. Sci. USA 2010, 107, 9088–9092. [Google Scholar] [CrossRef]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar] [CrossRef]
- Ha, S.C.; Kim, D.; Hwang, H.-Y.; Rich, A.; Kim, Y.-G.; Kim, K.K. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc. Natl. Acad. Sci. USA 2008, 105, 20671–20676. [Google Scholar] [CrossRef]
- Kang, H.-J.; Le, T.V.T.; Kim, K.; Hur, J.; Kim, K.K.; Park, H.-J. Novel interaction of the Z-DNA binding domain of human ADAR1 with the oncogenic c-Myc promoter G-quadruplex. J. Mol. Biol. 2014, 426, 2594–2604. [Google Scholar] [CrossRef]
- Leeder, W.-M.; Hummel, N.F.C.; Göringer, H.U. Multiple G-quartet structures in pre-edited mRNAs suggest evolutionary driving force for RNA editing in trypanosomes. Sci. Rep. 2016, 6, 29810. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011, 43, 1090–1103. [Google Scholar] [CrossRef]
- Herbert, A. Z-DNA and Z-RNA: Methods—Past and Future. In Z-DNA: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2023; pp. 295–329. [Google Scholar]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, R.; Lee, Y. RUPEE: A fast and accurate purely geometric protein structure search. PLoS ONE 2019, 14, e0213712. [Google Scholar] [CrossRef]
- Ayoub, R.; Lee, Y. Protein structure search to support the development of protein structure prediction methods. Proteins Struct. Funct. Bioinform. 2021, 89, 648–658. [Google Scholar] [CrossRef]
- Bartas, M.; Slychko, K.; Brázda, V.; Červeň, J.; Beaudoin, C.A.; Blundell, T.L.; Pečinka, P. Searching for new Z-DNA/Z-RNA binding proteins based on structural similarity to experimentally validated Zα domain. Int. J. Mol. Sci. 2022, 23, 768. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Pawar, S.S.; Rohane, S.H. Review on discovery studio: An important tool for molecular docking. Asian J. Res. Chem. 2021, 14, 86–88. [Google Scholar] [CrossRef]
- Li, H.; Huang, E.; Zhang, Y.; Huang, S.; Xiao, Y. HDOCK update for modeling protein-RNA/DNA complex structures. Protein Sci. 2022, 31, e4441. [Google Scholar] [CrossRef] [PubMed]
- Dejung, M.; Subota, I.; Bucerius, F.; Dindar, G.; Freiwald, A.; Engstler, M.; Boshart, M.; Butter, F.; Janzen, C.J. Quantitative proteomics uncovers novel factors involved in developmental differentiation of Trypanosoma brucei. PLoS Pathog. 2016, 12, e1005439. [Google Scholar] [CrossRef] [PubMed]

| Ligands * | Kd (nM) | Kd Confidence ** |
|---|---|---|
| r(UUUU)3 ss | 147.31 | 19.65 |
| r(UUUU)3 ds | 1714.05 | 203.31 |
| r(AUUU)4 ss | 179.74 | 15 |
| r(AUUU)4 ds | 1884.56 | 419.35 |
| r(AUUC)3 ss | 272.46 | 52.51 |
| r(AUUC)3 ds | 1231.75 | 230.59 |
| r(UA)6 ss | 880.23 | 199.4 |
| r(UA)6 ds | 1340.73 | 231.92 |
| d(GC)6 ds | 444.22 | 297.65 |
| d(GC)6 ss | 21.99 | 24.13 |
| r(GC)6 ds | 3217.29 | 752.31 |
| r(GC)6 ss | 2031.26 | 270.45 |
| Ligand | Ligand Sequence | SD * |
|---|---|---|
| r(UUUU)3 ss | 5′-GGUGGGUUAUUUUUUUUUUUUUAUCAACUGGG-3′ | 6.899 |
| r(UUUU)3 ds | /Antisense | 5.530 |
| r(AUUU)4 ss | 5′GGUGGGUUAUAUUUAUUUAUUUAUUUAUCAACUGGG-3′ | 7.470 |
| r(AUUU)4 ds | /Antisense | 7.107 |
| r(AUUC)3 ss | 5′-GGUGGGUUAUAUUCAUUCAUUCAUCAACUGGG-3′ | 6.530 |
| r(AUUC)3 ds | /Antisense | 4.917 |
| d(GC)6 ss | 5′-CGCGCGCGCGCGATCAACTGGG-3′ | 6.249 |
| d(GC)6 ds | /Antisense | 16.677 |
| r(GC)6 ss | 5′-CGCGCGCGCGCGAUCAACUGGG-3′ | 12.336 |
| r(GC)6 ds | /Antisense | 3.523 |
| r(UA)6 ss | 5′-UAUAUAUAUAUAAUCAACUGGG-3′ | 10.156 |
| r(UA)6 ds | /Antisense | 3.392 |
| Type of Oligos | Sequence |
|---|---|
| d(GC)6-Sense Cy5 | 5′-GGTGGGTTATCGCGCGCGCGCGATCAACTGGG-3′ |
| d(GC)6-Antisense | 5′-CCCAGTTGATCGCGCGCGCGCGATAACCCACC-3′ |
| r(GC)6-Sense Cy5 | 5′-GGUGGGUUAUCGCGCGCGCGCGAUCAACUGGG-3′ |
| r(GC)6-Antisense RNA | 5′-CCCAGUUGAUCGCGCGCGCGCGAUAACCCACC-3′ |
| r(UA)6-Sense Cy5 | 5′-GGUGGGUUAUUAUAUAUAUAUAAUCAACUGGG-3′ |
| r(UA)6-Antisense | 5′-CCCAGUUGAUUAUAUAUAUAUAAUAACCCACC-3′ |
| r(AUUU)4-Sense Cy5 | 5′-GGUGGGUUAUAUUUAUUUAUUUAUUUAUCAACUGGG/3Cy5Sp/-3′ |
| r(AUUU)4-Antisense | 5′-CCCAGUUGAUAAAUAAAUAAAUAAAUAUAACCCACC-3′ |
| r(AUUC)3-Sense Cy5 | 5′-GGUGGGUUAUAUUCAUUCAUUCAUCAACUGGG/3Cy5Sp/-3′ |
| r(AUUC)3-Antisense | 5′-CCCAGUUGAUGAAUGAAUGAAUAUAACCCACC-3′ |
| r(UUUU)3-Sense Cy5 | 5′-GGUGGGUUAUUUUUUUUUUUUUAUCAACUGGG/3Cy5Sp/-3′ |
| r(UUUU)3-Antisense | 5′-CCCAGUUGAUAAAAAAAAAAAAAUAACCCACC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehlert, C.; Poorinmohammad, N.; Mohammaei, S.; Zhang, L.; Salavati, R. Structure–Function Analysis of RBP7910: An Editosome Z-Binding Protein in Trypanosomatids. Molecules 2023, 28, 6963. https://doi.org/10.3390/molecules28196963
Ehlert C, Poorinmohammad N, Mohammaei S, Zhang L, Salavati R. Structure–Function Analysis of RBP7910: An Editosome Z-Binding Protein in Trypanosomatids. Molecules. 2023; 28(19):6963. https://doi.org/10.3390/molecules28196963
Chicago/Turabian StyleEhlert, Curtis, Naghmeh Poorinmohammad, Saba Mohammaei, Linhua Zhang, and Reza Salavati. 2023. "Structure–Function Analysis of RBP7910: An Editosome Z-Binding Protein in Trypanosomatids" Molecules 28, no. 19: 6963. https://doi.org/10.3390/molecules28196963
APA StyleEhlert, C., Poorinmohammad, N., Mohammaei, S., Zhang, L., & Salavati, R. (2023). Structure–Function Analysis of RBP7910: An Editosome Z-Binding Protein in Trypanosomatids. Molecules, 28(19), 6963. https://doi.org/10.3390/molecules28196963




