Differences of Atomic-Level Interactions between Midazolam and Two CYP Isoforms 3A4 and 3A5
Abstract
:1. Introduction
2. Simulation Methods
2.1. Model Preparation
2.2. Molecular Dynamics Simulation
2.3. MM-PBSA Calculation
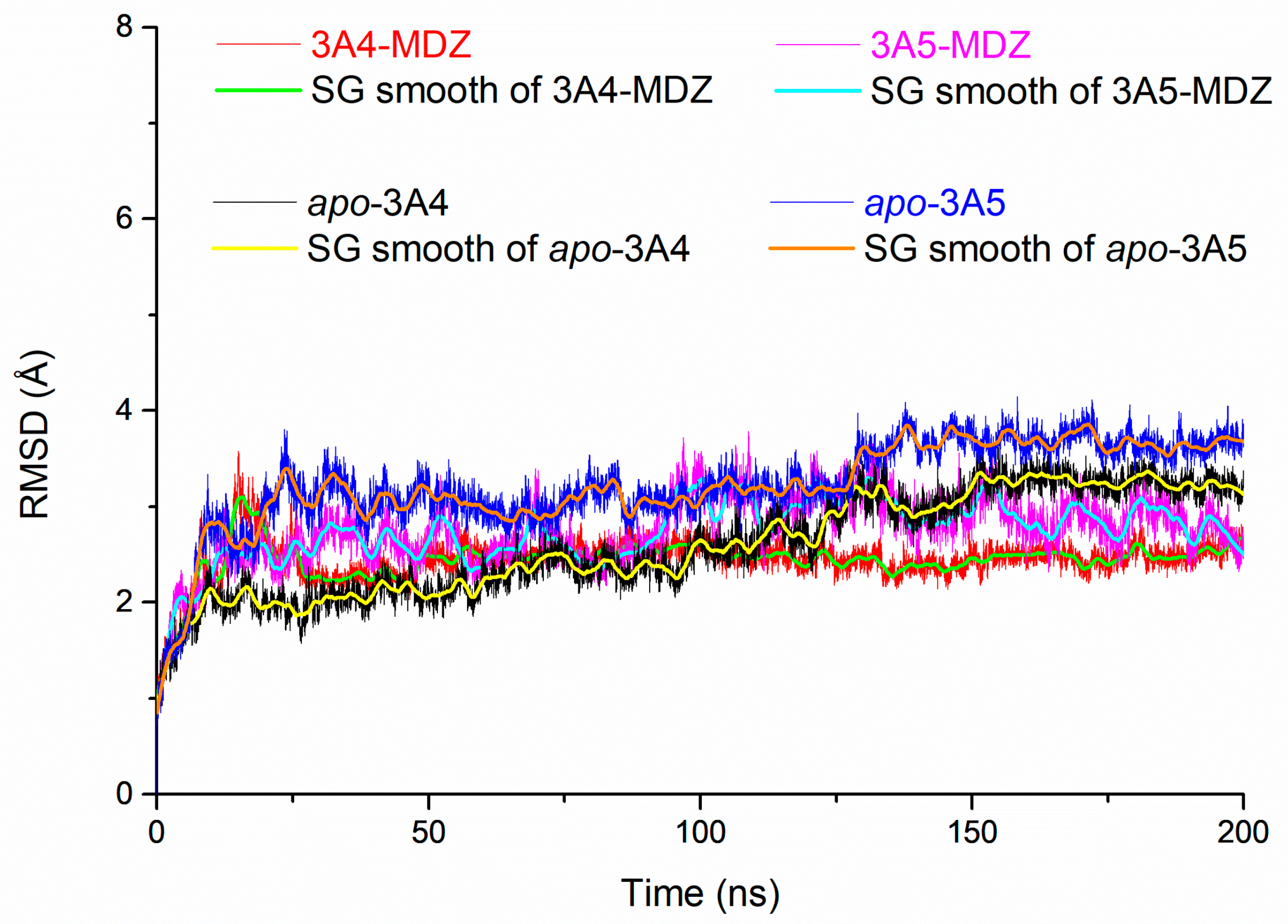
2.4. Analysis
3. Result and Discussion
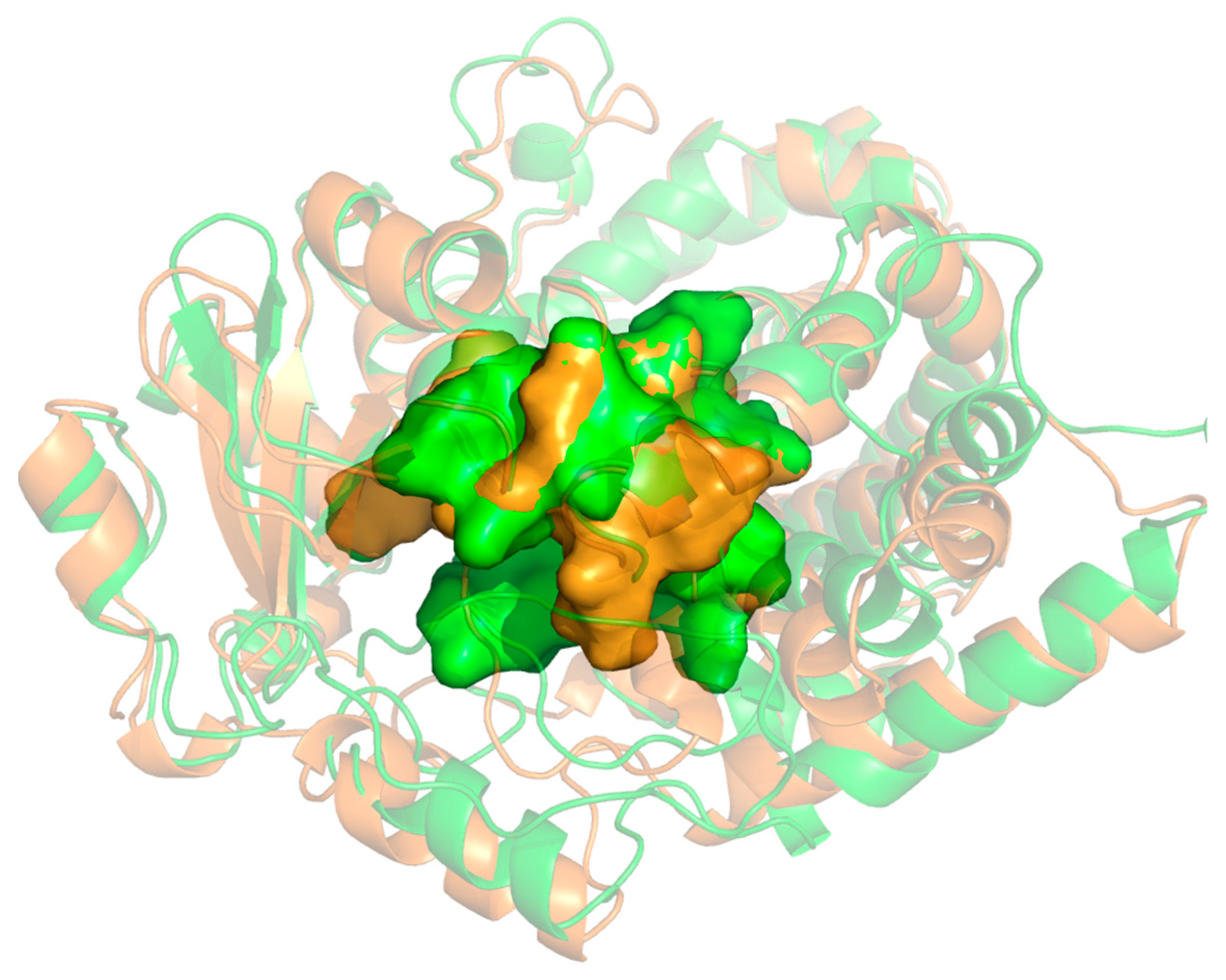
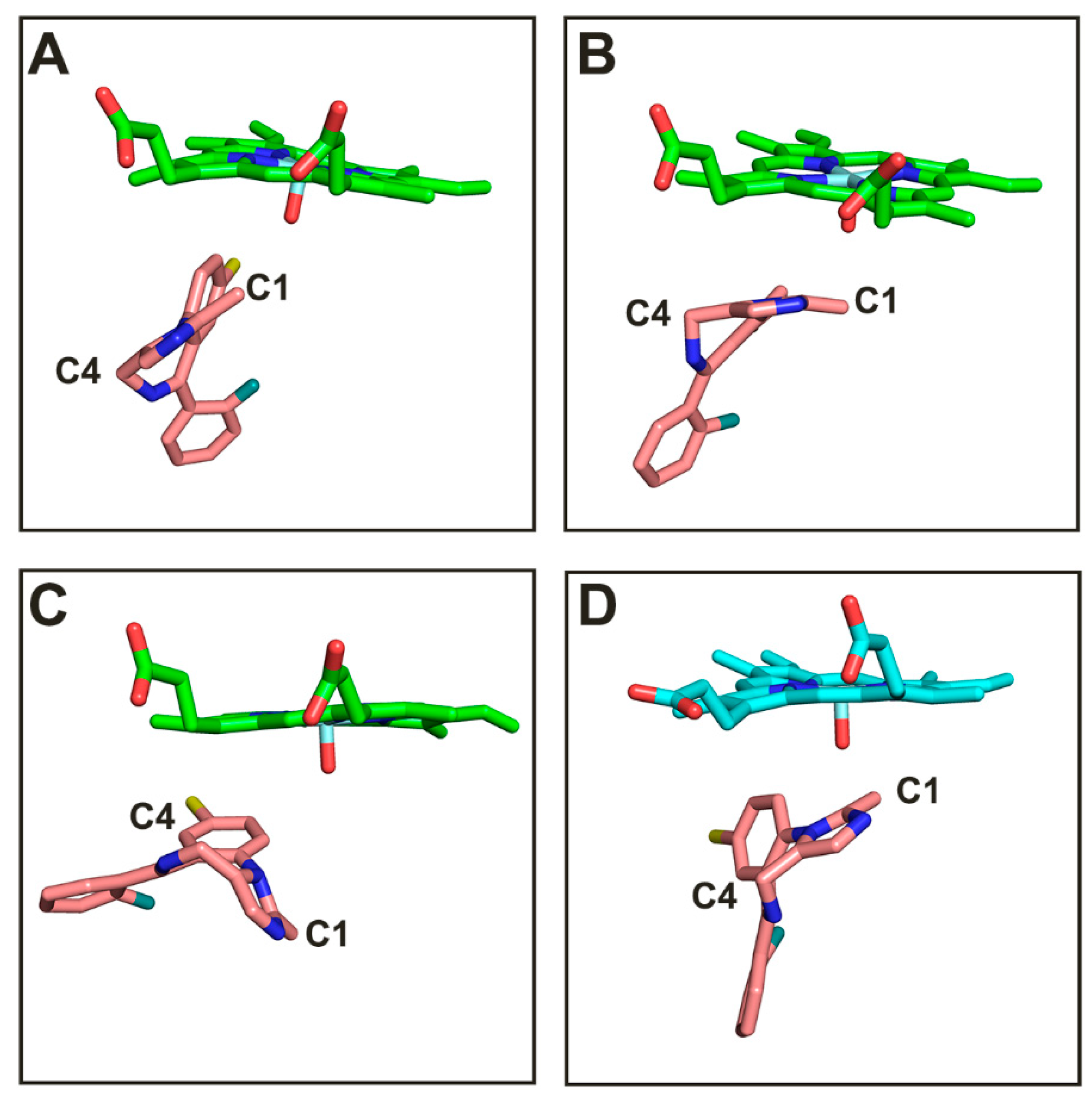
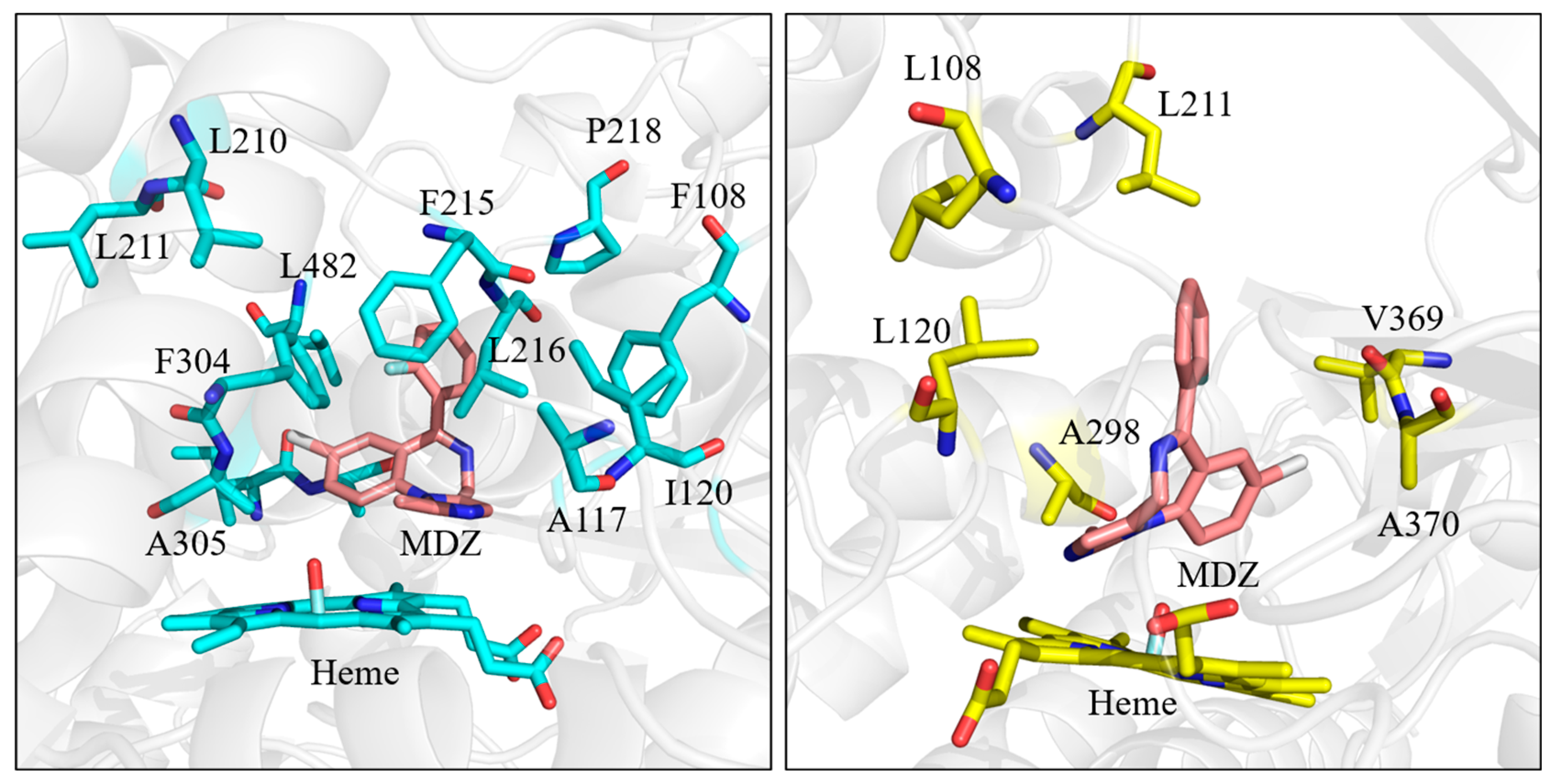
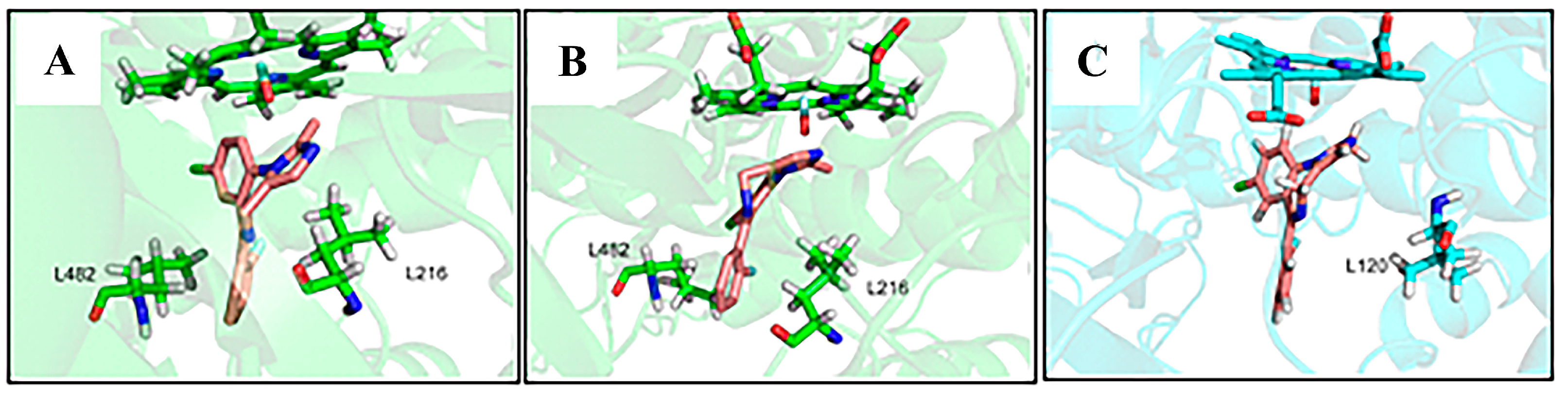
3.1. Analysis of Enzyme-Substrate Binding Pattern
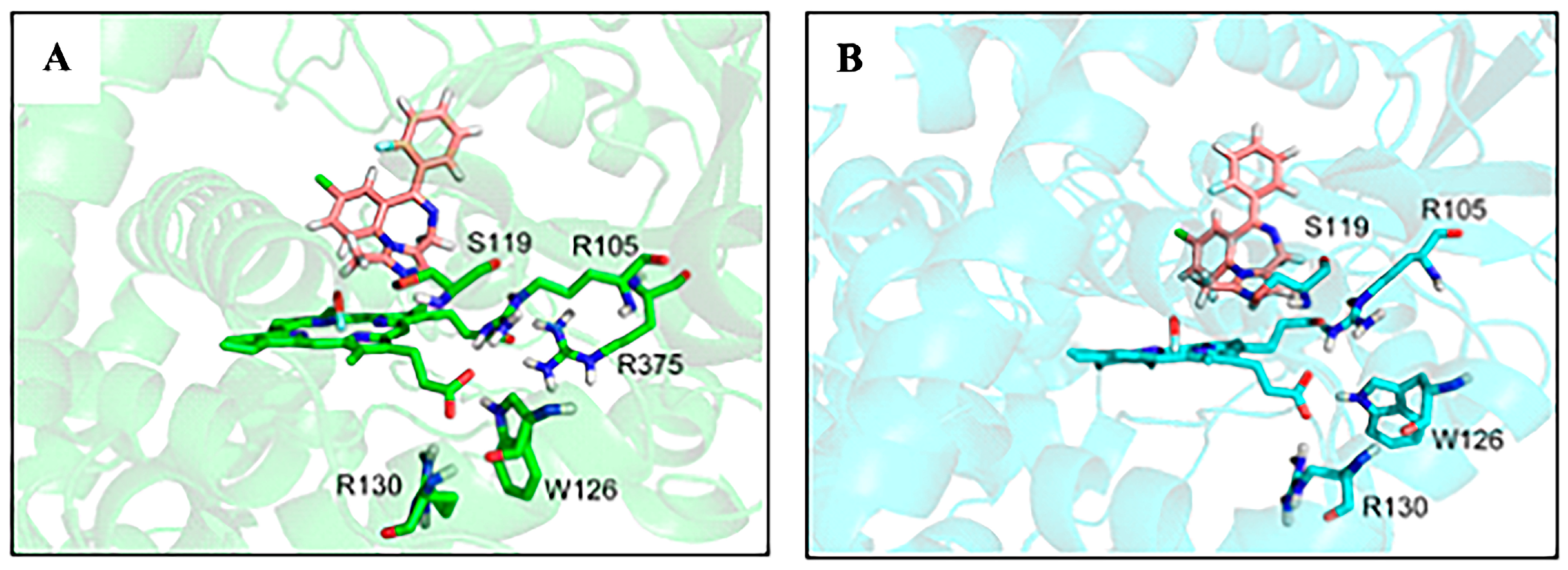
3.2. Factors That Stabilize Substrate Binding
3.3. Binding Free Energy Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.C. New perspectives on the impact of cytochrome P450 3A expression for pediatric pharmacology. Drug. Discov. Today 2006, 11, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Ince, I.; Knibbe, C.A.J.; Danhof, M.; de Wildt, S.N. Developmental changes in the expression and function of cytochrome P450 3A isoforms: Evidence from in vitro and in vivo investigations. Clin. Pharmacokinet. 2013, 52, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Koudriakova, T.; Iatsimirskaia, E.; Utkin, I.; Gangl, E.; Vouros, P.; Storozhuk, E.; Orza, D.; Marinina, J.; Gerber, N. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: Mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug. Metab. Dispos. 1998, 26, 552–561. [Google Scholar] [PubMed]
- Patki, K.C.; Von Moltke, L.L.; Greenblatt, D.J. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: Role of cyp3a4 and cyp3a5. Drug. Metab. Dispos. 2003, 31, 938–944. [Google Scholar] [CrossRef]
- Yamazaki, H.; Nakamoto, M.; Shimizu, M.; Murayama, N.; Niwa, T. Potential impact of cytochrome P450 3A5 in human liver on drug interactions with triazoles. Br. J. Clin. Pharmacol. 2010, 69, 593–597. [Google Scholar] [CrossRef]
- Tseng, E.; Walsky, R.L.; Luzietti, R.A., Jr.; Harris, J.J.; Kosa, R.E.; Goosen, T.C.; Zientek, M.A.; Obach, R.S. Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). J. Drug. Metab. Dispos. 2014, 42, 1163–1173. [Google Scholar] [CrossRef]
- Walsky, R.L.; Obach, R.S.; Hyland, R.; Kang, P.; Zhou, S.; West, M.; Geoghegan, K.F.; Helal, C.J.; Walker, G.S.; Goosen, T.C. Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for de lineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug. Metab. Dispos. 2012, 40, 1686–1697. [Google Scholar] [CrossRef]
- Williams, J.A.; Ring, B.J.; Cantrell, V.E.; Jones, D.R.; Eckstein, J.; Ruterbories, K.; Hamman, M.A.; Hall, S.D.; Wrighton, S.A. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug. Metab. Dispos. 2002, 30, 883–891. [Google Scholar] [CrossRef]
- Daly, A.K. Significance of the Minor Cytochrome P450 3A Isoforms. Clin. Pharmacokinet. 2006, 45, 13–31. [Google Scholar] [CrossRef]
- Johnsson, A.W.; Malmebo, S.; Cotter, A.J.; Andersson, T.B.; Johansson, I.; Edwards, R.J.; Boobis, A.R.; Sundberg, M.I. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. J. Drug. Metab. Dispos. 2003, 31, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, Y. MD Investigation on the Interaction between Carbamazepine and Two CYP Isoforms, CYP3A4 and CYP3A5. Int. J. Mol. Sci. 2023, 24, 2188. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.H.; Savas, U.; Johnson, E.F. The X-Ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5. Mole. Pharm. 2018, 93, 14–24. [Google Scholar] [CrossRef]
- Li, X.H.; Jeso, V.; Heyward, S.; Walker, G.S.; Sharma, R.; Micalizio, G.C.; Cameron, M.D. Characterization of T-5 N-oxide formation as the first highly selective measure of CYP3A5 activity. J. Drug. Metab. Dispos. 2014, 42, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.H.; Hendrix, C.W.; Bumpus, N.N. Cytochrome P450 3A5 plays a prominent role in the oxidative metabolism of the anti-human immunodeficiency virus drug maraviroc. J. Drug. Metab. Dispos. 2012, 40, 2221–2230. [Google Scholar] [CrossRef] [PubMed]
- Gorski, J.C.; Hall, S.D.; Jones, D.R.; Branden, M.V.; Wrighton, S.A. Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem. Pharmacol. 1994, 47, 1643–1653. [Google Scholar] [CrossRef]
- Lin, Y.S.; Lockwood, G.F.; Graham, M.A.; Brian, W.R.; Loi, C.M.; Dobrinska, M.R.; Shen, D.D.; Watkins, P.B.; Wilkinson, G.R.; Kharasch, E.D.; et al. In-vivo phenotyping for CYP3A by a single-point determination of midazolam plasma concentration. Pharmacogenetics 2001, 11, 781–791. [Google Scholar] [CrossRef]
- Thummel, K.E.; Wilkinson, G.R. In vitro and in vivo drug interactions involving human CYP3A. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 389–430. [Google Scholar] [CrossRef]
- Shaik, S.; Cohen, S.; Wang, Y.; Chen, H.; Kumar, D.; Thiel, W. P450 enzymes: Their structure, reactivity, and selectivity-modeled by QM/MM calculations. Chem. Rev. 2010, 110, 949–1017. [Google Scholar] [CrossRef]
- Lonsdale, R.; Harvey, J.N.; Mulholland, A.J. A practical guide to modelling enzyme-catalysed reactions. Chem. Soc. Rev. 2012, 41, 3025–3038. [Google Scholar] [CrossRef]
- Meunier, B.; de Visser, S.P.; Shaik, S. Mechanism of Oxidation Reactions Catalyzed by Cytochrome P450 Enzymes. Chem. Rev. 2004, 104, 3947–3980. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xie, Z.; Liu, Z.; Zhou, S.; Ma, L.; Liu, W.; Huang, J.W.; Ko, T.P.; Li, X.; Hu, Y.; et al. Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase. Nat. Commun. 2020, 11, 2676. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, K. Theoretical study on kinetic isotope effects in the C-H bond activation of alkanes by iron-oxo complexes. Coordin. Chem. Rev. 2002, 226, 251–259. [Google Scholar] [CrossRef]
- Muthusamy, R.; Bharatam, P.V. Molecular Modeling Studies on Cytochrome P450-mediated Drug Metabolism. Curr. Drug Metab. 2021, 22, 683–697. [Google Scholar]
- Chai, L.; Zhang, H.; Song, R.; Yang, H.; Yu, H.; Paneth, P.; Kepp, K.P.; Akamatsu, M.; Ji, L. Precision Biotransformation of Emerging Pollutants by Human Cytochrome P450 Using Computational-Experimental Synergy: A Case Study of Tris(1,3-dichloro-2-propyl) Phosphate. Environ. Sci. Technol. 2021, 55, 14037–14050. [Google Scholar] [CrossRef]
- Dixit, V.; Hariparsad, N.; Li, F.; Desai, P.; Thummel, K.E.; Unadkat, J.D. Cytochrome P450 enzymes and transporters induced by anti-human immunodeficiency virus protease inhibitors in human hepatocytes: Implications for predicting clinical drug interactions. Drug Metab. Dispos. 2007, 35, 1853–1859. [Google Scholar] [CrossRef]
- Kalathiya, U.; Padariya, M. Structure-based design and evaluation of novel N-phenyl-1H-indol-2-amine derivatives for fat mass and obesity-associated (FTO) protein inhibition. Comput. Biol. Chem. J. 2016, 64, 414–425. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W. Comparison of simple potential functial functions for simulating liquid water. Chem. Phys. J. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Case, D.A.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.A.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; Kaus, J.; Kovalenko, A.; Lee, T.S.; et al. AMBER 2016 Reference Manual; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C.J. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory. Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Shahrokh, K.; Orendt, A.; Yost, G.S.; Cheatham, T.E., 3rd. Quantum mechanically derived AMBER-compatible heme parameters for various states of the cytochrome P450 catalytic cycle. J. Comput. Chem. 2012, 33, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L.J. Particle Mesh Ewald: An N• Log (N) Method for Ewald Sums in Large Systems. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Sitkoff, D.; Sharp, K.A.; Honig, B. Accurate calculation of hydration free energies using macroscopic solvent models. Phys. Chem. J. 1994, 98, 1978–1988. [Google Scholar] [CrossRef]
- Hou, T.; Zhang, W.; Case, D.A.; Wang, W. Characterization of Domain–Peptide Interaction Interface: A Case Study on the Amphiphysin-1 SH3 Domain. Mol. Biol. J. 2008, 376, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.C.; McKinnon, R.A.; Miners, J.O. Cytochrome P450 structutr-function: Insights from molecular dynamics simulations. Drug Metab. Rev. 2016, 48, 434–452. [Google Scholar] [CrossRef]
- Stoll, A.; Loke, S.; Joseph, J.F.; Machalz, D.; Torre, X.D.L.; Botre, F.; Wolber, G.; Bureik, M.; Parr, M.K. Fine-mapping of the substrate specificity of human steroid 21-hydroxylase (CYP21A2). Steroid Biochem. Mol. Biol. 2019, 194, 105446. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Structural basis for regiospecific midazolam oxidation by human cytochrome P450 3A4. Proc. Natl. Acad. Sci. USA 2017, 114, 486–491. [Google Scholar] [CrossRef]
- Khan, K.K.; He, Y.Q.; Domanski, T.L.; Halpert, J.R. Midazolam oxidation by cytochrome P450 3A4 and active-site mutants: An evaluation of multiple binding sites and of the metabolic pathway that leads to enzyme inactivation. J. Mol. Pharmacol. 2002, 61, 495–506. [Google Scholar] [CrossRef]
- Roussel, F.; Khan, K.K.; Halpert, J.R. The importance of SRS-1 residues in catalytic specificity of human cytochrome P450 3A4. J. Arch. Biochem. Biophys. 2000, 374, 269–278. [Google Scholar] [CrossRef]
- Wang, X.F.; Sun, J.; Wang, X.L.; Tian, J.K.; Tian, Z.W.; Zhang, J.L.; Jia, R.J. MD investigation on the binding of microphthalmia-associated transcription factor with DNA. Saudi Chem. Soc. 2022, 26, 101420. [Google Scholar] [CrossRef]


| Complex | The Recipient and the Donor | Ratio (%) |
|---|---|---|
| CYP3A4-MDZ | heme@O1A-105R@HH21-NH2 | 65.1 |
| heme@O2D-105R@HH22-NH2 | 83.9 | |
| heme@O2D-105R@HH12-NH1 | 77.4 | |
| heme@O1D-126W@HE1-NE1 | 92.5 | |
| heme@O2D-126W@HE1-NE1 | 75.5 | |
| heme@O1D-130R@NH11-NH1 | 94.0 | |
| heme@O1A-375R@NH12-NH1 | 76.5 | |
| heme@O1A-375R@NH22-NH2 | 86.6 | |
| heme@O2A-375R@NH22-NH2 | 89.5 | |
| heme@O2A-375R@NH12-NH1 | 52.9 | |
| MDZ@NAN-119S@HG-OG | 93.8 | |
| CYP3A5-MDZ | heme@O1D-105R@HH22-NH2 | 68.4 |
| heme@O2A-105R@HH21-NH2 | 52.1 | |
| heme@O1A-105R@HH21-NH2 | 43.4 | |
| heme@O2A-105R@HE-NE | 34.8 | |
| heme@O1A-105R@HE-NE | 31.4 | |
| heme@O2D-105R@HH22-NH2 | 33.5 | |
| heme@O1D-126W@HE1-NE1 | 90.7 | |
| heme@O2D-126W@HE1-NE1 | 85.5 | |
| heme@O2D-130R@NH11-NH1 | 50.7 | |
| MDZ@NAN-119S@HG-OG | 77.9 |
| Complex | Salt Bridge Action | Ratio (%) |
|---|---|---|
| CYP3A4-MDZ | 374E@OE1-106R@HH12-NH1 | 67.30 |
| 374E@OE1-106R@HH11-NH1 | 67.30 | |
| 374E@OE2-106R@HH12-NH1 | 61.57 | |
| 374E@OE2-106R@HH11-NH1 | 61.57 | |
| 374E@OE2-106R@HE-NE | 43.03 | |
| 374E@OE1-106R@HE-NE | 54.53 | |
| 441N@OD1-130R@HH22-NH2 | 96.40 | |
| 441N@OD1-130R@HH21-NH2 | 96.40 | |
| 441N@OD1-130R@HH12-NH1 | 89.17 | |
| 441N@OD1-130R@HH11-NH1 | 89.17 | |
| 441N@ND2-130R@HH22-NH2 | 37.70 | |
| 441N@ND2-130R@HH21-NH2 | 37.70 | |
| CYP3A5-MDZ | 76E@OE1-106R@HH12-NH1 | 87.37 |
| 76E@OE1-106R@HH11-NH1 | 87.37 | |
| 76E@OE1-106R@HH22-NH2 | 88.67 | |
| 76E@OE1-106R@HH21-NH2 | 88.67 | |
| 76E@OE2-106R@HH12-NH1 | 79.87 | |
| 76E@OE2-106R@HH11-NH1 | 79.87 | |
| 76E@OE2-106R@HH22-NH2 | 64.03 | |
| 76E@OE2-106R@HH21-NH2 | 64.03 | |
| 374E@OE2-106R@HH12-NH1 | 33.97 | |
| 374E@OE2-106R@HH11-NH1 | 33.97 | |
| 374E@OE1-106R@HH12-NH1 | 32.73 | |
| 374E@OE1-106R@HH11-NH1 | 32.73 |
| Complex | Residue | Score |
|---|---|---|
| CYP3A4-MDZ | Phe108 | −1.056 |
| Ala117 | 1.311 | |
| Ile120 | 0.056 | |
| Phe215 | 0.322 | |
| Leu216 | 0.211 | |
| Pro218 | 0.733 | |
| Phe304 | 1.744 | |
| Ala305 | 1.167 | |
| Ile369 | 0.889 | |
| Ala370 | 1.000 | |
| Leu482 | 0.133 | |
| CYP3A5-MDZ | Leu120 | −0.022 |
| Leu211 | −0.067 | |
| Ala297 | 1.167 | |
| Val369 | 1.144 | |
| Ala370 | 1.256 |
| 3A4-MDZ | 3A5-MDZ | |
|---|---|---|
| ∆Eele | −15.5 ± 2.5 | −12.0 ± 2.7 |
| ∆Evdw | −38.3 ± 2.7 | −30.5 ± 2.2 |
| ∆GPB | 23.5 ± 1.5 | 19.9 ± 1.8 |
| ∆GSA | −4.3 ± 0.2 | −3.3+0.2 |
| ∆Ggas | −53.8 ± 3.5 | −42.5 ± 3.6 |
| ∆Gsolv | 19.2 ± 1.5 | 16.6 ± 1.7 |
| a ∆Gpol | 8.00 | 7.95 |
| b ∆Gnonpol | −43.6 | −33.8 |
| c ∆GMMPB/SA | −34.6 ± 2.9 | −25.8 ± 2.5 |
| T∆S | −15.1 ± 7.4 | −14.2 ± 5.5 |
| d ∆Gbind | −19.5 | −11.6 |
| Residue | ∆Evdw | ∆Eele | ∆GPB | ∆GSA | ∆Gbind |
|---|---|---|---|---|---|
| Leu216 | −0.77 ± 0.57 | −0.15 ± 0.27 | 0.61 ± 0.30 | −0.36 ± 0.06 | −2.67 ± 0.64 |
| Ser119 | −0.28 ± 0.58 | −3.74 ± 0.98 | 1.90 ± 0.28 | −0.09 ± 0.03 | −2.20 ± 0.59 |
| Leu482 | −1.29 ± 0.31 | −0.12 ± 0.07 | 0.19 ± 0.05 | −0.23 ± 0.06 | −1.45 ± 0.34 |
| Ala370 | −1.17 ± 0.26 | −0.03 ± 0.05 | 0.19 ± 0.08 | −0.23 ± 0.05 | −1.23 ± 0.27 |
| Phe304 | −1.10 ± 0.38 | −0.07 ± 0.07 | 0.37 ± 0.14 | −0.16 ± 0.04 | −0.96 ± 0.32 |
| Ile369 | −0.70 ± 0.27 | −0.08 ± 0.07 | 0.05 ± 0.07 | −0.05 ± 0.02 | −0.78 ± 0.27 |
| Arg105 | −0.64 ± 0.31 | −1.43 ± 0.42 | 1.47 ± 0.40 | −0.07 ± 0.03 | −0.66 ± 0.41 |
| Asp217 | −0.83 ± 0.23 | −1.13 ± 0.34 | 1.54 ± 0.46 | −0.12 ± 0.03 | −0.54 ± 0.23 |
| Ile301 | −0.51 ± 0.13 | −0.17 ± 0.09 | 0.21 ± 0.09 | −0.03 ± 0.01 | −0.50 ± 0.13 |
| Thr309 | −0.43 ± 0.13 | −0.08 ± 0.10 | −0.08−0.08 | −0.05 ± 0.02 | −0.50 ± 0.13 |
| Ala305 | −0.41 ± 0.18 | −0.09 ± 0.07 | −0.07−0.05 | −0.03 ± 0.02 | −0.42 ± 0.17 |
| HEM | −5.12 ± 0.63 | −0.88 ± 0.56 | 1.72 ± 0.42 | −0.35 ± 0.04 | −4.62 ± 0.76 |
| Residue | ∆Evdw | ∆Eele | ∆GPB | ∆GSA | ∆Gbind |
|---|---|---|---|---|---|
| Ser119 | −0.71 ± 0.65 | −3.78 ± 0.84 | 2.15 ± 0.30 | −0.15 ± 0.04 | −2.49 ± 0.52 |
| Thr302 | −1.68 ± 0.45 | −0.21 ± 0.22 | 0.45 ± 0.21 | −0.26 ± 0.04 | −1.70 ± 0.49 |
| Phe297 | −1.36 ± 0.47 | −0.36 ± 0.16 | 0.68 ± 0.21 | −0.19 ± 0.05 | −1.23 ± 0.43 |
| Arg105 | −0.68 ± 0.19 | −0.49 ± 0.44 | 0.48 ± 0.39 | −0.05 ± 0.02 | −0.74 ± 0.37 |
| Leu211 | −0.55 ± 0.21 | −0.08 ± 0.05 | 0.18 ± 0.07 | −0.18 ± 0.05 | −0.63 ± 0.23 |
| Ala370 | −0.50 ± 0.23 | −0.02 ± 0.07 | 0.16 ± 0.10 | −0.14 ± 0.05 | −0.50 ± 0.24 |
| Val369 | −0.51 ± 0.22 | −0.02 ± 0.18 | 0.14 ± 0.20 | 0.07 ± 0.03 | −0.47 ± 0.24 |
| Ala298 | −0.44 ± 0.14 | −0.08 ± 0.05 | 0.16 ± 0.08 | −0.02 ± 0.01 | −0.39 ± 0.15 |
| HEM | −5.12 ± 0.75 | −1.29 ± 0.69 | 2.48 ± 0.88 | −0.39 ± 0.05 | −4.32 ± 0.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zheng, Q.; Bai, F. Differences of Atomic-Level Interactions between Midazolam and Two CYP Isoforms 3A4 and 3A5. Molecules 2023, 28, 6900. https://doi.org/10.3390/molecules28196900
Liu S, Zheng Q, Bai F. Differences of Atomic-Level Interactions between Midazolam and Two CYP Isoforms 3A4 and 3A5. Molecules. 2023; 28(19):6900. https://doi.org/10.3390/molecules28196900
Chicago/Turabian StyleLiu, Shuhui, Qingchuan Zheng, and Fuquan Bai. 2023. "Differences of Atomic-Level Interactions between Midazolam and Two CYP Isoforms 3A4 and 3A5" Molecules 28, no. 19: 6900. https://doi.org/10.3390/molecules28196900
APA StyleLiu, S., Zheng, Q., & Bai, F. (2023). Differences of Atomic-Level Interactions between Midazolam and Two CYP Isoforms 3A4 and 3A5. Molecules, 28(19), 6900. https://doi.org/10.3390/molecules28196900







