Exploring Natural Alkaloids from Brazilian Biodiversity as Potential Inhibitors of the Aedes aegypti Juvenile Hormone Enzyme: A Computational Approach for Vector Mosquito Control
Abstract
:1. Introduction
2. Results and Discussion
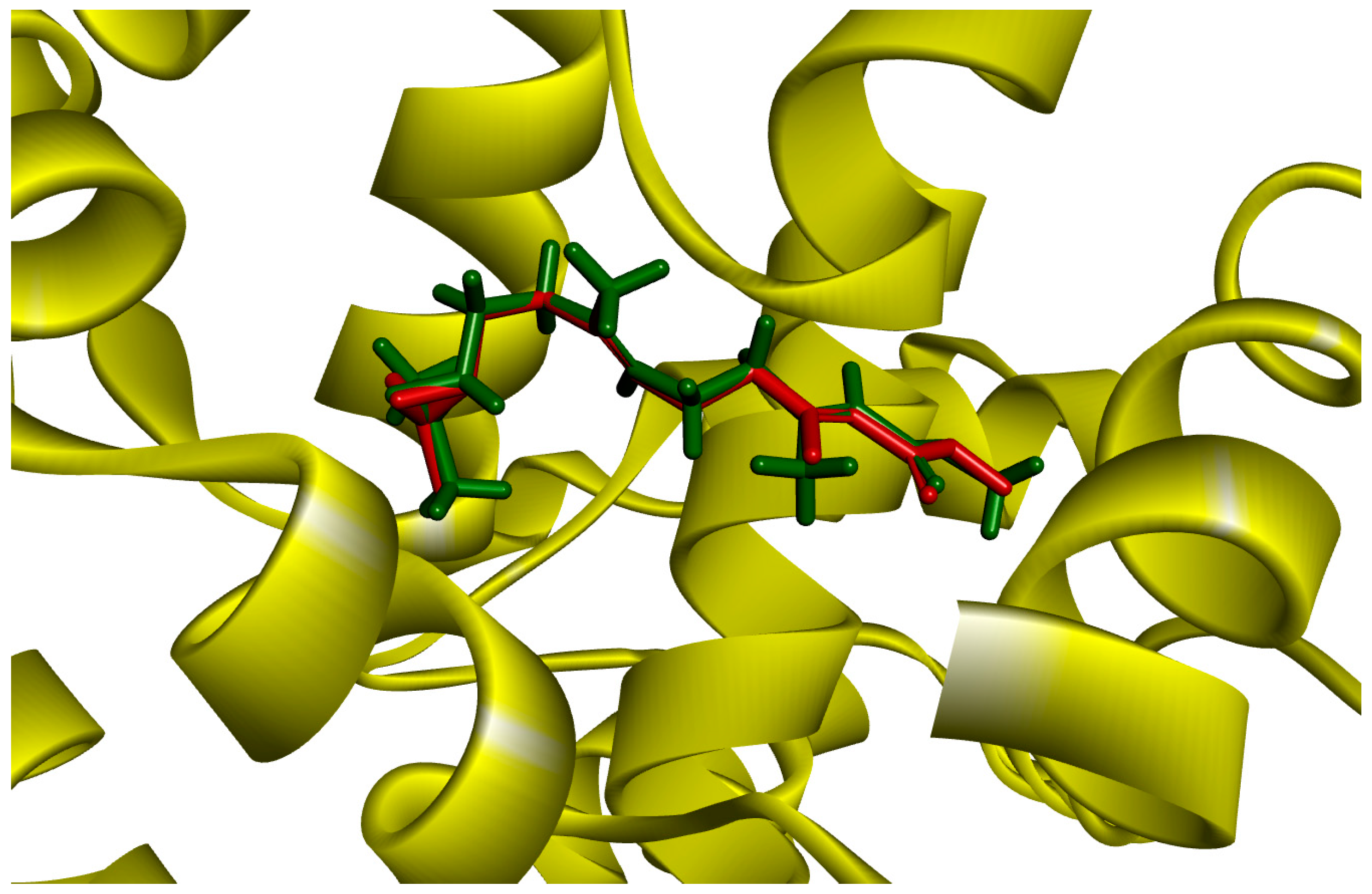
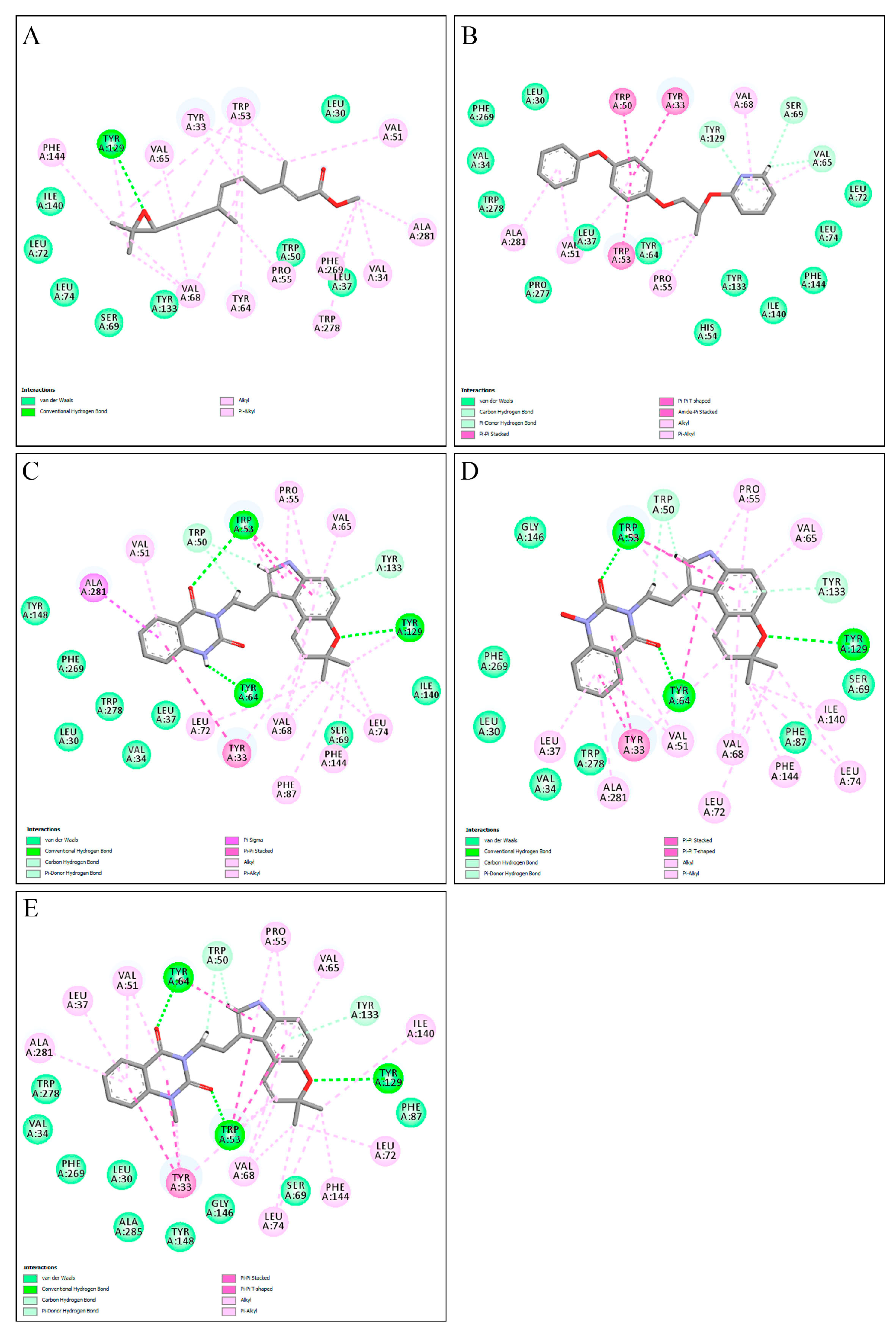
2.1. Molecular Docking
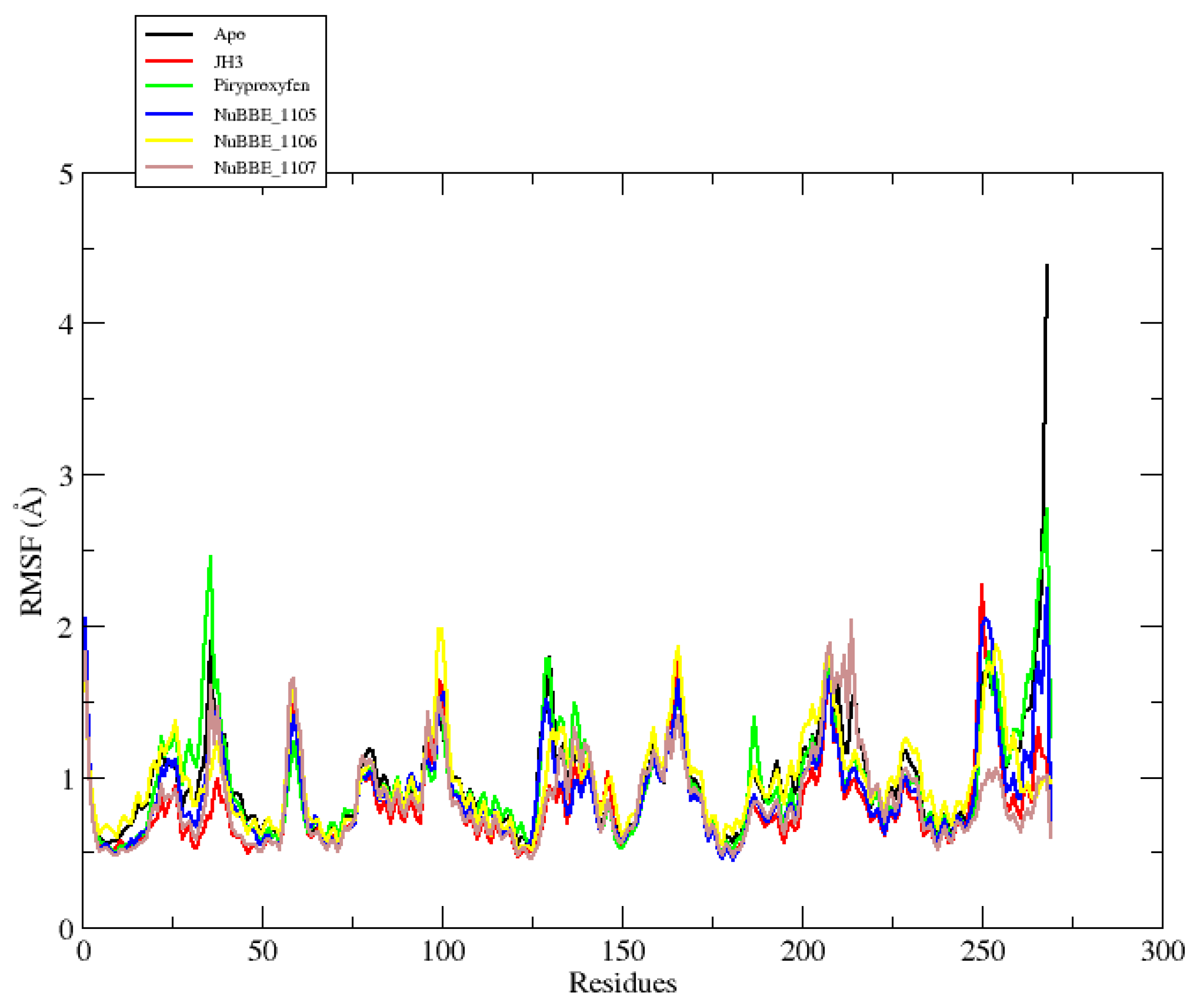
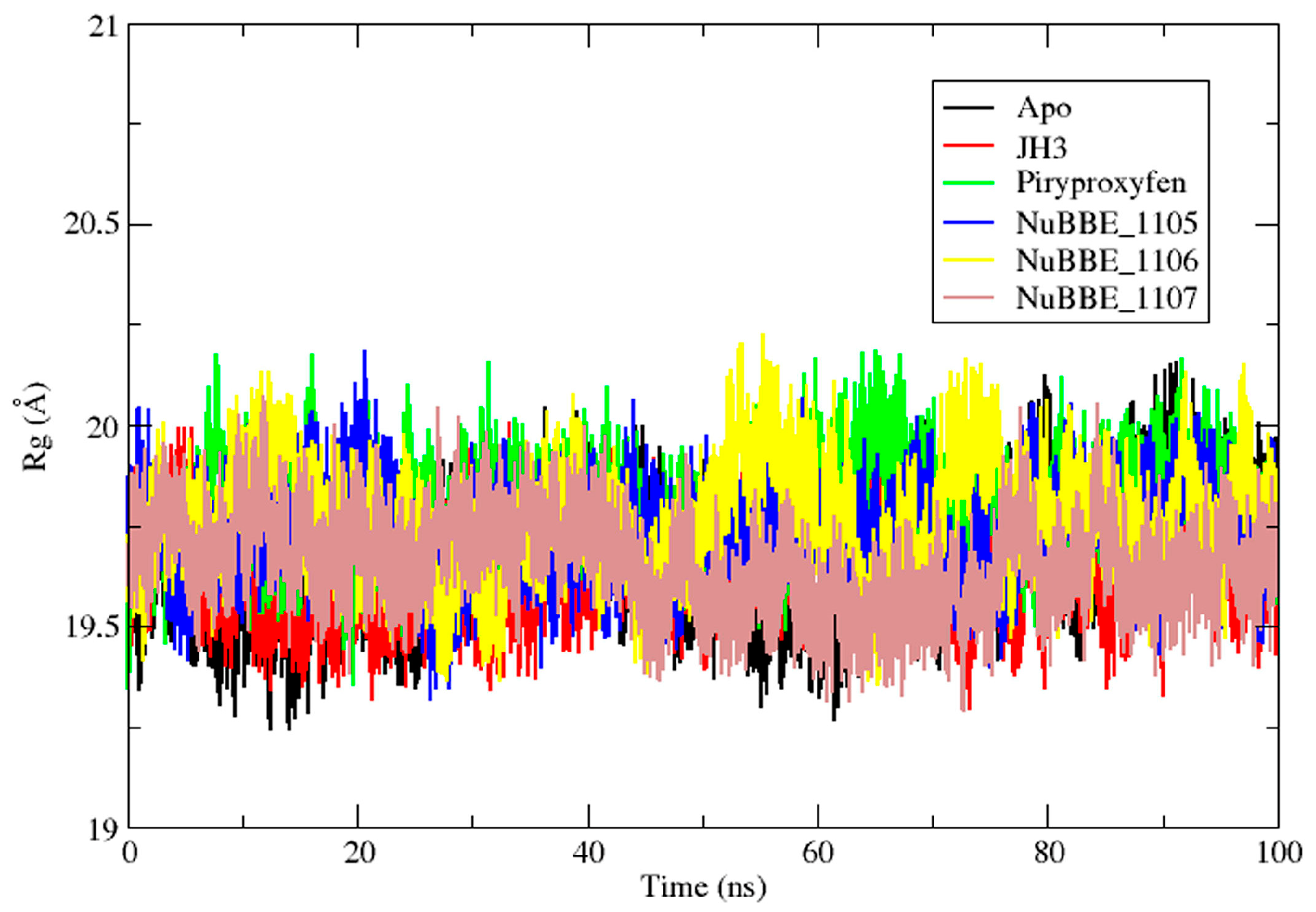
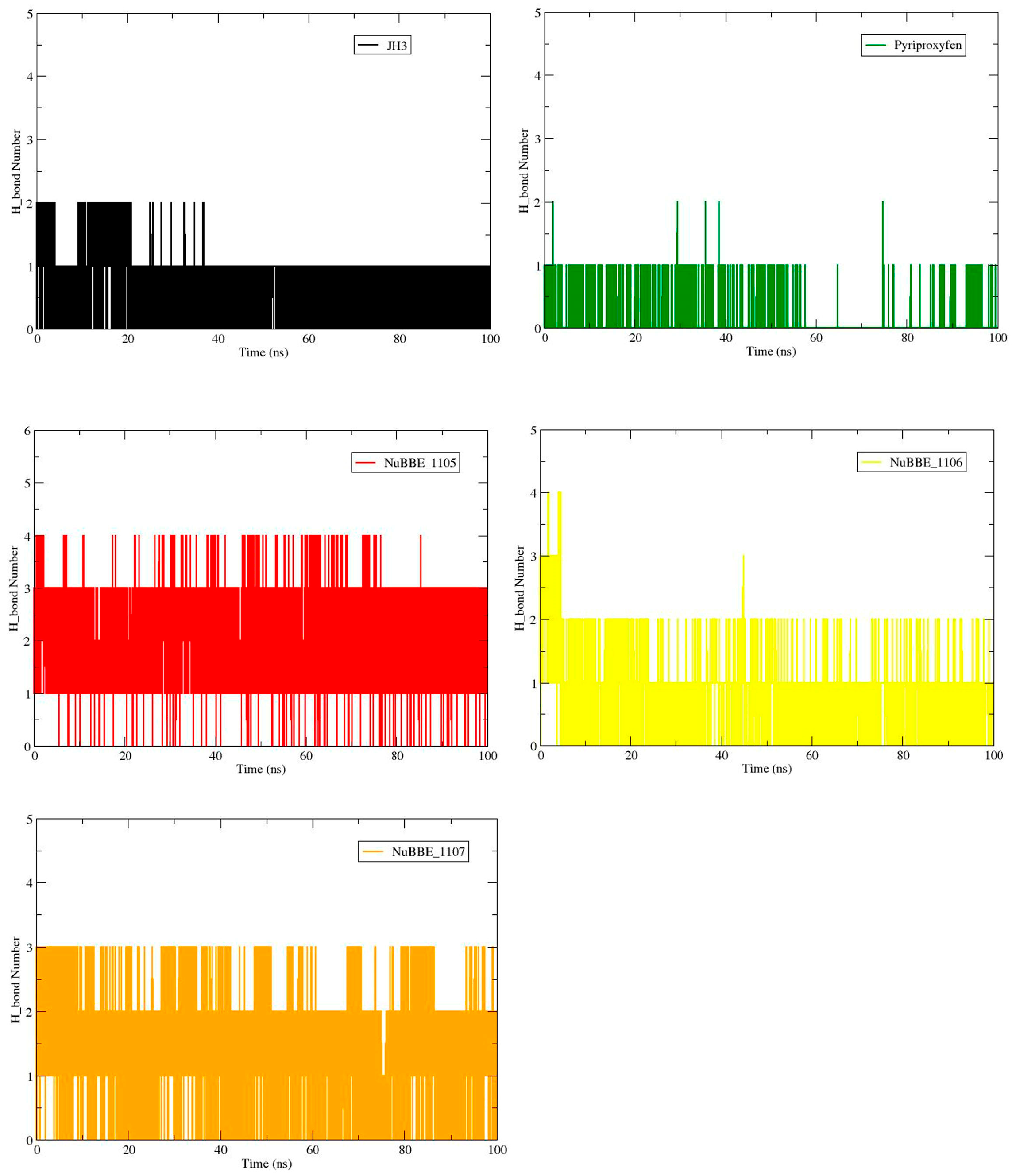
2.2. Molecular Dynamics
2.3. Binding Free Energy Calculations
3. Materials and Methods
3.1. Molecular Docking
3.2. Molecular Dynamics Simulations
3.3. Binding Free Energy and Residual Decomposition Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Vector-Borne Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases (accessed on 18 May 2023).
- Engdahl, C.S.; Tikhe, C.V.; Dimopoulos, G. Discovery of Novel Natural Products for Mosquito Control. Parasites Vectors 2022, 15, 481. [Google Scholar] [CrossRef]
- Paixão, E.S.; Teixeira, M.G.; Rodrigues, L.C. Zika, Chikungunya and Dengue: The Causes and Threats of New and Re-Emerging Arboviral Diseases. BMJ Glob. Health 2018, 3, e000530. [Google Scholar] [CrossRef]
- Kraemer, M.U.G.; Reiner, R.C.; Brady, O.J.; Messina, J.P.; Gilbert, M.; Pigott, D.M.; Yi, D.; Johnson, K.; Earl, L.; Marczak, L.B.; et al. Past and Future Spread of the Arbovirus Vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol. 2019, 4, 854–863. [Google Scholar] [CrossRef]
- Ferreira, Q.R.; Lemos, F.F.; Moura, M.N.; Nascimento, J.O.; Novaes, A.F.; Barcelos, I.S.; Fernandes, L.A.; Amaral, L.S.; Barreto, F.K.; Melo, F.F. Role of the Microbiome in Aedes spp. Vector Competence: What Do We Know? Viruses 2023, 15, 779. [Google Scholar] [CrossRef]
- Godoy, R.S.M.; dos Santos Felix, L.; da Silva Orfanó, A.; Chaves, B.A.; Nogueira, P.M.; dos Anjos Costa, B.; Soares, A.S.; Oliveira, C.C.A.; Nacif-Pimenta, R.; Silva, B.M.; et al. Dengue and Zika Virus Infection Patterns Vary among Aedes aegypti Field Populations from Belo Horizonte, a Brazilian Endemic City. PLoS Negl. Trop. Dis. 2021, 15, e0009839. [Google Scholar] [CrossRef]
- Wilke, A.B.B.; Vasquez, C.; Carvajal, A.; Medina, J.; Chase, C.; Cardenas, G.; Mutebi, J.-P.; Petrie, W.D.; Beier, J.C. Proliferation of Aedes aegypti in Urban Environments Mediated by the Availability of Key Aquatic Habitats. Sci. Rep. 2020, 10, 12925. [Google Scholar] [CrossRef]
- Morrison, A.C.; Zielinski-Gutierrez, E.; Scott, T.W.; Rosenberg, R. Defining Challenges and Proposing Solutions for Control of the Virus Vector Aedes aegypti. PLoS Med. 2008, 5, e68. [Google Scholar] [CrossRef]
- Löwy, I. Leaking Containers: Success and Failure in Controlling the Mosquito Aedes aegypti in Brazil. Am. J. Public Health 2017, 107, 517–524. [Google Scholar] [CrossRef]
- Gan, S.J.; Leong, Y.Q.; bin Barhanuddin, M.F.H.; Wong, S.T.; Wong, S.F.; Mak, J.W.; Ahmad, R.B. Dengue Fever and Insecticide Resistance in Aedes Mosquitoes in Southeast Asia: A Review. Parasites Vectors 2021, 14, 315. [Google Scholar] [CrossRef]
- Alomar, A.A.; Eastmond, B.H.; Alto, B.W. Juvenile Hormone Analog Enhances Zika Virus Infection in Aedes aegypti. Sci. Rep. 2021, 11, 21062. [Google Scholar] [CrossRef]
- Benelli, G.; Jeffries, C.L.; Walker, T. Biological Control of Mosquito Vectors: Past, Present, and Future. Insects 2016, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Melo-Santos, M.A.V.; Varjal-Melo, J.J.M.; Araújo, A.P.; Gomes, T.C.S.; Paiva, M.H.S.; Regis, L.N.; Furtado, A.F.; Magalhaes, T.; Macoris, M.L.G.; Andrighetti, M.T.M.; et al. Resistance to the Organophosphate Temephos: Mechanisms, Evolution and Reversion in an Aedes aegypti Laboratory Strain from Brazil. Acta Trop. 2010, 113, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Parker-Crockett, C.; Connelly, C.R.; Siegfried, B.; Alto, B. Influence of Pyrethroid Resistance on Vector Competency for Zika Virus by Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 2021, 58, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Shafie, F.A.; Tahir, M.P.M.; Sabri, N.M. Aedes Mosquitoes Resistance in Urban Community Setting. Procedia-Soc. Behav. Sci. 2012, 36, 70–76. [Google Scholar] [CrossRef]
- Walia, S.; Saha, S.; Tripathi, V.; Sharma, K.K. Phytochemical Biopesticides: Some Recent Developments. Phytochem. Rev. 2017, 16, 989–1007. [Google Scholar] [CrossRef]
- Wan-Norafikah, O.; Nazni, W.A.; Lee, H.L.; Zainol-Ariffin, P.; Sofian-Azirun, M. Permethrin Resistance in Aedes aegypti (Linnaeus) Collected from Kuala Lumpur, Malaysia. J. Asia-Pac. Entomol. 2010, 13, 175–182. [Google Scholar] [CrossRef]
- de Souza Wuillda, A.C.J.; Campos Martins, R.C.; das Neves Costa, F. Larvicidal Activity of Secondary Plant Metabolites in Aedes aegypti Control: An Overview of the Previous 6 Years. Nat. Product. Commun. 2019, 14, 1934578X19862893. [Google Scholar] [CrossRef]
- Valli, M.; Atanázio, L.C.V.; Monteiro, G.C.; Coelho, R.R.; Demarque, D.P.; Andricopulo, A.D.; Espindola, L.S.; da Silva Bolzani, V. The Potential of Biologically Active Brazilian Plant Species as a Strategy to Search for Molecular Models for Mosquito Control. Planta Med. 2021, 87, 6–23. [Google Scholar] [CrossRef]
- Chaudhary, S.; Kanwar, R.K.; Sehgal, A.; Cahill, D.M.; Barrow, C.J.; Sehgal, R.; Kanwar, J.R. Progress on Azadirachta Indica Based Biopesticides in Replacing Synthetic Toxic Pesticides. Front. Plant Sci. 2017, 8, 610. [Google Scholar] [CrossRef]
- Khursheed, A.; Rather, M.A.; Jain, V.; Wani, A.R.; Rasool, S.; Nazir, R.; Malik, N.A.; Majid, S.A. Plant Based Natural Products as Potential Ecofriendly and Safer Biopesticides: A Comprehensive Overview of Their Advantages over Conventional Pesticides, Limitations and Regulatory Aspects. Microb. Pathog. 2022, 173, 105854. [Google Scholar] [CrossRef]
- Şengül Demirak, M.Ş.; Canpolat, E. Plant-Based Bioinsecticides for Mosquito Control: Impact on Insecticide Resistance and Disease Transmission. Insects 2022, 13, 162. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.H.; Roberts, J.M.; Pope, T.W. Terpene Based Biopesticides as Potential Alternatives to Synthetic Insecticides for Control of Aphid Pests on Protected Ornamentals. Crop Prot. 2018, 110, 125–130. [Google Scholar] [CrossRef]
- Chapagain, B.P.; Saharan, V.; Wiesman, Z. Larvicidal Activity of Saponins from Balanites Aegyptiaca Callus against Aedes aegypti Mosquito. Bioresour. Technol. 2008, 99, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- de Luna, J.S.; dos Santos, A.F.; de Lima, M.R.F.; de Omena, M.C.; de Mendonça, F.A.C.; Bieber, L.W.; Sant’Ana, A.E.G. A Study of the Larvicidal and Molluscicidal Activities of Some Medicinal Plants from Northeast Brazil. J. Ethnopharmacol. 2005, 97, 199–206. [Google Scholar] [CrossRef]
- Falkowski, M.; Jahn-Oyac, A.; Odonne, G.; Flora, C.; Estevez, Y.; Touré, S.; Boulogne, I.; Robinson, J.-C.; Béreau, D.; Petit, P.; et al. Towards the Optimization of Botanical Insecticides Research: Aedes aegypti Larvicidal Natural Products in French Guiana. Acta Trop. 2020, 201, 105179. [Google Scholar] [CrossRef] [PubMed]
- Pontual, E.V.; Napoleão, T.H.; Dias de Assis, C.R.; de Souza Bezerra, R.; Xavier, H.S.; do Amaral Ferraz Navarro, D.M.; Coelho, L.C.B.B.; Paiva, P.M.G. Effect of Moringa Oleifera Flower Extract on Larval Trypsin and Acetylcholinesterase Activities in Aedes aegypti. Arch. Insect Biochem. Physiol. 2012, 79, 135–152. [Google Scholar] [CrossRef]
- Sá, R.A.; de Lima Santos, N.D.; da Silva, C.S.; Napoleão, T.H.; Gomes, F.S.; Cavada, B.S.; Coelho, L.C.B.B.; do Am-Aral Ferraz Navarro, D.M.; Bieber, L.W.; Paiva, P.M.G. Larvicidal Activity of Lectins from Myracrodruon Urundeuva on Aedes aegypti. Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2009, 149, 300–306. [Google Scholar] [CrossRef]
- Silvério, M.R.S.; Espindola, L.S.; Lopes, N.P.; Vieira, P.C. Plant Natural Products for the Control of Aedes aegypti: The Main Vector of Important Arboviruses. Molecules 2020, 25, 3484. [Google Scholar] [CrossRef]
- Balachandran, C.; Anbalagan, S.; Kandeepan, C.; Arun Nagendran, N.; Jayakumar, M.; Fathi Abd-Allah, E.; Alqarawi, A.A.; Hashem, A.; Baskar, K. Molecular Docking Studies of Natural Alkaloids as Acetylcholinesterase (AChE1) Inhibitors in Aedes aegypti. J. Asia-Pac. Entomol. 2021, 24, 645–652. [Google Scholar] [CrossRef]
- Garcez, W.S.; Garcez, F.R.; da Silva, L.M.G.E.; Hamerski, L. Larvicidal Activity against Aedes aegypti of Some Plants Native to the West-Central Region of Brazil. Bioresour. Technol. 2009, 100, 6647–6650. [Google Scholar] [CrossRef]
- Kim, S.-I.; Ahn, Y.-J. Larvicidal Activity of Lignans and Alkaloid Identified in Zanthoxylum Piperitum Bark toward Insecticide-Susceptible and Wild Culex Pipiens Pallens and Aedes aegypti. Parasites Vectors 2017, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Liu, Q.Z.; Du, S.S.; Deng, Z.W. Mosquito Larvicidal Activity of Alkaloids and Limonoids Derived from Evodia Rutaecarpa Unripe Fruits against Aedes albopictus (Diptera: Culicidae). Parasitol. Res. 2012, 111, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, R.; Baskar, K.; Ignacimuthu, S.; Maria Packiam, S.; Rajapandiyan, K. Bioactivity of Couroupita guianensis Aubl. against Filarial and Dengue Vectors and Non-Target Fish. S. Afr. J. Bot. 2019, 125, 46–53. [Google Scholar] [CrossRef]
- Wachira, S.W.; Omar, S.; Jacob, J.W.; Wahome, M.; Alborn, H.T.; Spring, D.R.; Masiga, D.K.; Torto, B. Toxicity of Six Plant Extracts and Two Pyridone Alkaloids from Ricinus communis against the Malaria Vector Anopheles gambiae. Parasites Vectors 2014, 7, 312. [Google Scholar] [CrossRef]
- Wu, Y.; Ren, D.; Gao, C.; Li, J.; Du, B.; Wang, Z.; Qian, S. Recent Advances for Alkaloids as Botanical Pesticides for Use in Organic Agriculture. Int. J. Pest. Manag. 2021, 69, 288–298. [Google Scholar] [CrossRef]
- Xia, J.; Liu, Y.; Zhou, Y.; Zhang, J.; Li, C.; Yin, X.; Tian, X.; Zhang, X. Two Novel Alkaloids from Corydalis curviflora Maxim. and Their Insecticidal Activity. Pest. Manag. Sci. 2020, 76, 2360–2367. [Google Scholar] [CrossRef]
- Yan, H.; Xie, N.; Zhong, C.; Su, A.; Hui, X.; Zhang, X.; Jin, Z.; Li, Z.; Feng, J.; He, J. Aphicidal Activities of Amaryllidaceae Alkaloids from Bulbs of Lycoris radiata against Aphis citricola. Ind. Crops Prod. 2018, 123, 372–378. [Google Scholar] [CrossRef]
- Chowański, S.; Adamski, Z.; Marciniak, P.; Rosiński, G.; Büyükgüzel, E.; Büyükgüzel, K.; Falabella, P.; Scrano, L.; Ventrella, E.; Lelario, F.; et al. A Review of Bioinsecticidal Activity of Solanaceae Alkaloids. Toxins 2016, 8, 60. [Google Scholar] [CrossRef]
- Zifruddin, A.N.; Mohamad Yusoff, M.A.; Abd Ghani, N.S.; Nor Muhammad, N.A.; Lam, K.W.; Hassan, M. Ensemble-Based, High-Throughput Virtual Screening of Potential Inhibitor Targeting Putative Farnesol Dehydrogenase of Metisa plana (Lepidoptera: Psychidae). Comput. Biol. Chem. 2023, 103, 107811. [Google Scholar] [CrossRef]
- Goodman, W.G.; Cusson, M. 8—The Juvenile Hormones. In Insect Endocrinology; Gilbert, L.I., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 310–365. ISBN 978-0-12-384749-2. [Google Scholar]
- Jindra, M.; Bittova, L. The Juvenile Hormone Receptor as a Target of Juvenoid “Insect Growth Regulators”. Arch. Insect Biochem. Physiol. 2020, 103, e21615. [Google Scholar] [CrossRef]
- Smykal, V.; Dolezel, D. Evolution of Proteins Involved in the Final Steps of Juvenile Hormone Synthesis. J. Insect Physiol. 2023, 145, 104487. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Pham, V.; Jablonka, W.; Goodman, W.G.; Ribeiro, J.M.C.; Andersen, J.F. A Mosquito Hemolymph Odorant-Binding Protein Family Member Specifically Binds Juvenile Hormone. J. Biol. Chem. 2017, 292, 15329–15339. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Furuta, K.; Nagamine, K.; Shinoda, T.; Yonesu, K.; Okabe, T. Identification of a Juvenile-Hormone Signaling Inhibitor via High-Throughput Screening of a Chemical Library. Sci. Rep. 2020, 10, 18413. [Google Scholar] [CrossRef] [PubMed]
- Tumova, S.; Milacek, M.; Šnajdr, I.; Muthu, M.; Tuma, R.; Reha, D.; Jedlicka, P.; Bittova, L.; Novotna, A.; Majer, P.; et al. Unique Peptidic Agonists of a Juvenile Hormone Receptor with Species-Specific Effects on Insect Development and Reproduction. Proc. Natl. Acad. Sci. USA 2022, 119, e2215541119. [Google Scholar] [CrossRef]
- Smith, R.D.; Dunbar, J.B., Jr.; Ung, P.M.-U.; Esposito, E.X.; Yang, C.-Y.; Wang, S.; Carlson, H.A. CSAR Benchmark Exercise of 2010: Combined Evaluation Across All Submitted Scoring Functions. J. Chem. Inf. Model. 2011, 51, 2115–2131. [Google Scholar] [CrossRef]
- Liu, J.; Su, M.; Liu, Z.; Li, J.; Li, Y.; Wang, R. Enhance the Performance of Current Scoring Functions with the Aid of 3D Protein-Ligand Interaction Fingerprints. BMC Bioinform. 2017, 18, 343. [Google Scholar] [CrossRef]
- Akhoon, B.A.; Tiwari, H.; Nargotra, A. Chapter 3—In Silico Drug Design Methods for Drug Repurposing. In Silico Drug Design; Roy, K., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 47–84. ISBN 978-0-12-816125-8. [Google Scholar]
- Sacan, A.; Ekins, S.; Kortagere, S. Applications and Limitations of in Silico Models in Drug Discovery. Methods Mol. Biol. 2012, 910, 87–124. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive Evaluation of Ten Docking Programs on a Diverse Set of Protein–Ligand Complexes: The Prediction Accuracy of Sampling Power and Scoring Power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Lima, L.R.; Bastos, R.S.; Ferreira, E.F.B.; Leão, R.P.; Araújo, P.H.F.; Pita, S.S.D.R.; De Freitas, H.F.; Espejo-Román, J.M.; Dos Santos, E.L.V.S.; Ramos, R.D.S.; et al. Identification of Potential New Aedes aegypti Juvenile Hormone Inhibitors from N-Acyl Piperidine Derivatives: A Bioinformatics Approach. Int. J. Mol. Sci. 2022, 23, 9927. [Google Scholar] [CrossRef]
- Ononamadu, C.J.; Abdalla, M.; Ihegboro, G.O.; Li, J.; Owolarafe, T.A.; John, T.D.; Tian, Q. In Silico Identification and Study of Potential Anti-Mosquito Juvenile Hormone Binding Protein (MJHBP) Compounds as Candidates for Dengue Virus—Vector Insecticides. Biochem. Biophys. Rep. 2021, 28, 101178. [Google Scholar] [CrossRef]
- da Silva Ramos, R.; da Silva Costa, J.; Campos Silva, R.; Vilhena da Costa, G.; Bruno Lobato Rodrigues, A.; de Menezes Rabelo, É.; Nonato Picanço Souto, R.; Anthony Taft, C.; Tomich de Paula da Silva, C.H.; Campos Rosa, J.M.; et al. Identification of Potential Inhibitors from Pyriproxyfen with Insecticidal Activity by Virtual Screening. Pharmaceuticals 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.S.; Macêdo, W.J.; Costa, J.S.; da Silva, C.H.; Rosa, J.M.; da Cruz, J.N.; de Oliveira, M.S.; de Aguiar Andrade, E.H.; e Silva, R.B.; Souto, R.N.P.; et al. Potential Inhibitors of the Enzyme Acetylcholinesterase and Juvenile Hormone with Insecticidal Activity: Study of the Binding Mode via Docking and Molecular Dynamics Simulations. J. Biomol. Struct. Dyn. 2020, 38, 4687–4709. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Igarashi, R.; Ushiku, Y.; Mitsutake, A. Analysis of Protein Folding Simulation with Moving Root Mean Square Deviation. J. Chem. Inf. Model. 2023, 63, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- da Costa, R.A.; da Rocha, J.A.P.; Pinheiro, A.S.; da Costa, A.D.S.S.; da Rocha, E.C.M.; Silva, R.C.; Gonçalves, A.D.S.; Santos, C.B.R.; Brasil, D.D.S.B. A Computational Approach Applied to the Study of Potential Allosteric Inhibitors Protease NS2B/NS3 from Dengue Virus. Molecules 2022, 27, 4118. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of Gyration as an Indicator of Protein Structure Compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Srivastava, H.K.; Sastry, G.N. Molecular Dynamics Investigation on a Series of HIV Protease Inhibitors: Assessing the Performance of MM-PBSA and MM-GBSA Approaches. J. Chem. Inf. Model. 2012, 52, 3088–3098. [Google Scholar] [CrossRef]
- Costa, R.A.; Cruz, J.N.; Nascimento, F.C.A.; Silva, S.G.; Silva, S.O.; Martelli, M.C.; Carvalho, S.M.L.; Santos, C.B.R.; Neto, A.M.J.C.; Brasil, D.S.B. Studies of NMR, Molecular Docking, and Molecular Dynamics Simulation of New Promising Inhibitors of Cruzaine from the Parasite Trypanosoma Cruzi. Med. Chem. Res. 2019, 28, 246–259. [Google Scholar] [CrossRef]
- Johnson, B.C.; Métifiot, M.; Pommier, Y.; Hughes, S.H. Molecular Dynamics Approaches Estimate the Binding Energy of HIV-1 Integrase Inhibitors and Correlate with In Vitro Activity. Antimicrob. Agents Chemother. 2012, 56, 411–419. [Google Scholar] [CrossRef]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein-Ligand Docking Using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Valli, M.; Dametto, A.C.; Pinto, M.E.F.; Freire, R.T.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. NuBBEDB: An Updated Database to Uncover Chemical and Biological Information from Brazilian Biodiversity. Sci. Rep. 2017, 7, 7215. [Google Scholar] [CrossRef] [PubMed]
- Valli, M.; dos Santos, R.N.; Figueira, L.D.; Nakajima, C.H.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. Development of a Natural Products Database from the Biodiversity of Brazil. J. Nat. Prod. 2013, 76, 439–444. [Google Scholar] [CrossRef]
- Lee, T.-S.; Cerutti, D.S.; Mermelstein, D.; Lin, C.; LeGrand, S.; Giese, T.J.; Roitberg, A.; Case, D.A.; Walker, R.C.; York, D.M. GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model. 2018, 58, 2043–2050. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L., III. A Modified TIP3P Water Potential for Simulation with Ewald Summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- da Costa, R.A.; da Rocha, J.A.; Pinheiro, A.S.; Da Costa, A.D.; da Rocha, E.C.; Josino, L.P.; da Silva Gonçalves, A.; e Lima, A.H.; Brasil, D.S.B. In Silico Identification of Novel Allosteric Inhibitors of Dengue Virus NS2B/NS3 Serine Protease: Scientific Paper. J. Serbian Chem. Soc. 2022, 87, 693–706. [Google Scholar] [CrossRef]
- Darden, T.; York, D.M.; Pedersen, N.L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.I.; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]


| Ligands | PLPChem | Structures |
|---|---|---|
| NuBBE_1107 (3-(2-(7,7-dimethyl-3,7-dihydropyrano [3,2-e]indol-1-yl)ethyl-1-methylquinazoline-2,4(1H,3H)-dione) | 113.05 | 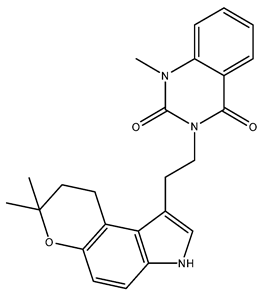 |
| NuBBE_1105 (3-(2-(7,7-dimethyl-3,7-dihydropyrano[3,2-e]indol-1-yl)ethyl)quinazoline-2,4(1H,3H)-dione) | 111.34 | 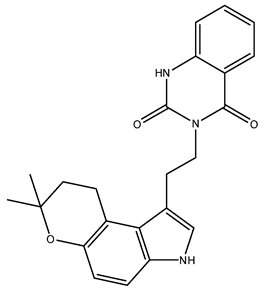 |
| NuBBE_1106 (3-(2-(7,7-dimethyl-3,7-dihydropyrano[3,2-e] indol-1-yl)ethyl)-1-hydroxyquinazoline-2,4(1H,3H)-dione) | 107.28 | 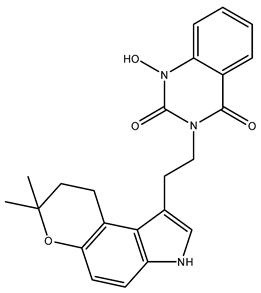 |
| JH3 (2E,6E)-9-[(2R)-3,3-dimetiloxiran-2-il]-3,7-dimetilnona-2,6-dienoato | 85.70 |  |
| Pyriproxyfen 2-[1-(4-phenoxyphenoxy)propan-2-yloxy]pyridine | 92.75 |  |
| Ligand ID | ΔEvdW | ΔEele | ΔGGB | ΔGSASA | ΔGbind |
|---|---|---|---|---|---|
| JH3 | −46.55 ± 0.07 | −8.96 ± 0.05 | 20.06 ± 0.01 | −6.15 ± 0.03 | −41.60 ± 0.07 |
| Pyriproxyfen | −45.99 ± 0.07 | −3.38 ± 0.07 | 19.98 ± 0.06 | −6.36 ± 0.01 | −35.75 ± 0.07 |
| NuBBE_1105 | −56.55 ± 0.08 | −20.50 ± 0.10 | 34.71 ± 0.05 | −6.49 ± 0.01 | −48.84 ± 0.09 |
| NuBBE_1106 | −39.90 ± 0.08 | −48.60 ± 0.19 | 81.33 ± 0.15 | −4.38 ± 0.01 | −11.55 ± 0.11 |
| NuBBE_1107 | −58.49 ± 0.07 | −17.44 ± 0.09 | 32.96 ± 0.05 | −6.73 ± 0.01 | −49.70 ± 0.08 |
| Complex | Hydrogen Bond Formation | Distance (Å) | Occupancy (%) |
|---|---|---|---|
| AagJHBP–H3 | Tyr129@HH-LIG@O3 | 2.77 | 83.47 |
| Tyr148@HH-LIG@O2 | 2.77 | 9.26 | |
| AagJHBP–Pyproxyfen | Tyr64@HH-LIG@O | 2.81 | 6.62 |
| Tyr133@HH-LIG@O1 | 2.81 | 2.41 | |
| AagJHBP–NuBBE_1105 | Tyr64@HH- LIG@O2 | 2.82 | 95.93 |
| Tyr129@HH -LIG@O1 | 2.83 | 65.50 | |
| Trp53@HE1-LIG@O | 2.88 | 38.26 | |
| AagJHBP–NuBBE_1106 | Tyr129@HH-LIG@O2 | 2.73 | 87.40 |
| Gly146@O-LIG@H4 | 2.59 | 5.39 | |
| AagJHBP–NuBBE_1107 | Tyr64@HH-LIG@O | 2.73 | 75.05 |
| Tyr129@HH-LIG@O1 | 2.83 | 46.13 | |
| Trp53@HE1-LIG@O2 | 2.82 | 20.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, R.A.d.; Costa, A.d.S.S.d.; Rocha, J.A.P.d.; Lima, M.R.d.C.; Rocha, E.C.M.d.; Nascimento, F.C.d.A.; Gomes, A.J.B.; Rego, J.d.A.R.d.; Brasil, D.d.S.B. Exploring Natural Alkaloids from Brazilian Biodiversity as Potential Inhibitors of the Aedes aegypti Juvenile Hormone Enzyme: A Computational Approach for Vector Mosquito Control. Molecules 2023, 28, 6871. https://doi.org/10.3390/molecules28196871
Costa RAd, Costa AdSSd, Rocha JAPd, Lima MRdC, Rocha ECMd, Nascimento FCdA, Gomes AJB, Rego JdARd, Brasil DdSB. Exploring Natural Alkaloids from Brazilian Biodiversity as Potential Inhibitors of the Aedes aegypti Juvenile Hormone Enzyme: A Computational Approach for Vector Mosquito Control. Molecules. 2023; 28(19):6871. https://doi.org/10.3390/molecules28196871
Chicago/Turabian StyleCosta, Renato Araújo da, Andréia do Socorro Silva da Costa, João Augusto Pereira da Rocha, Marlon Ramires da Costa Lima, Elaine Cristina Medeiros da Rocha, Fabiana Cristina de Araújo Nascimento, Anderson José Baia Gomes, José de Arimatéia Rodrigues do Rego, and Davi do Socorro Barros Brasil. 2023. "Exploring Natural Alkaloids from Brazilian Biodiversity as Potential Inhibitors of the Aedes aegypti Juvenile Hormone Enzyme: A Computational Approach for Vector Mosquito Control" Molecules 28, no. 19: 6871. https://doi.org/10.3390/molecules28196871
APA StyleCosta, R. A. d., Costa, A. d. S. S. d., Rocha, J. A. P. d., Lima, M. R. d. C., Rocha, E. C. M. d., Nascimento, F. C. d. A., Gomes, A. J. B., Rego, J. d. A. R. d., & Brasil, D. d. S. B. (2023). Exploring Natural Alkaloids from Brazilian Biodiversity as Potential Inhibitors of the Aedes aegypti Juvenile Hormone Enzyme: A Computational Approach for Vector Mosquito Control. Molecules, 28(19), 6871. https://doi.org/10.3390/molecules28196871







