Virtual Screening and Binding Analysis of Potential CD58 Inhibitors in Colorectal Cancer (CRC)
Abstract
:1. Introduction
2. Results and Discussion
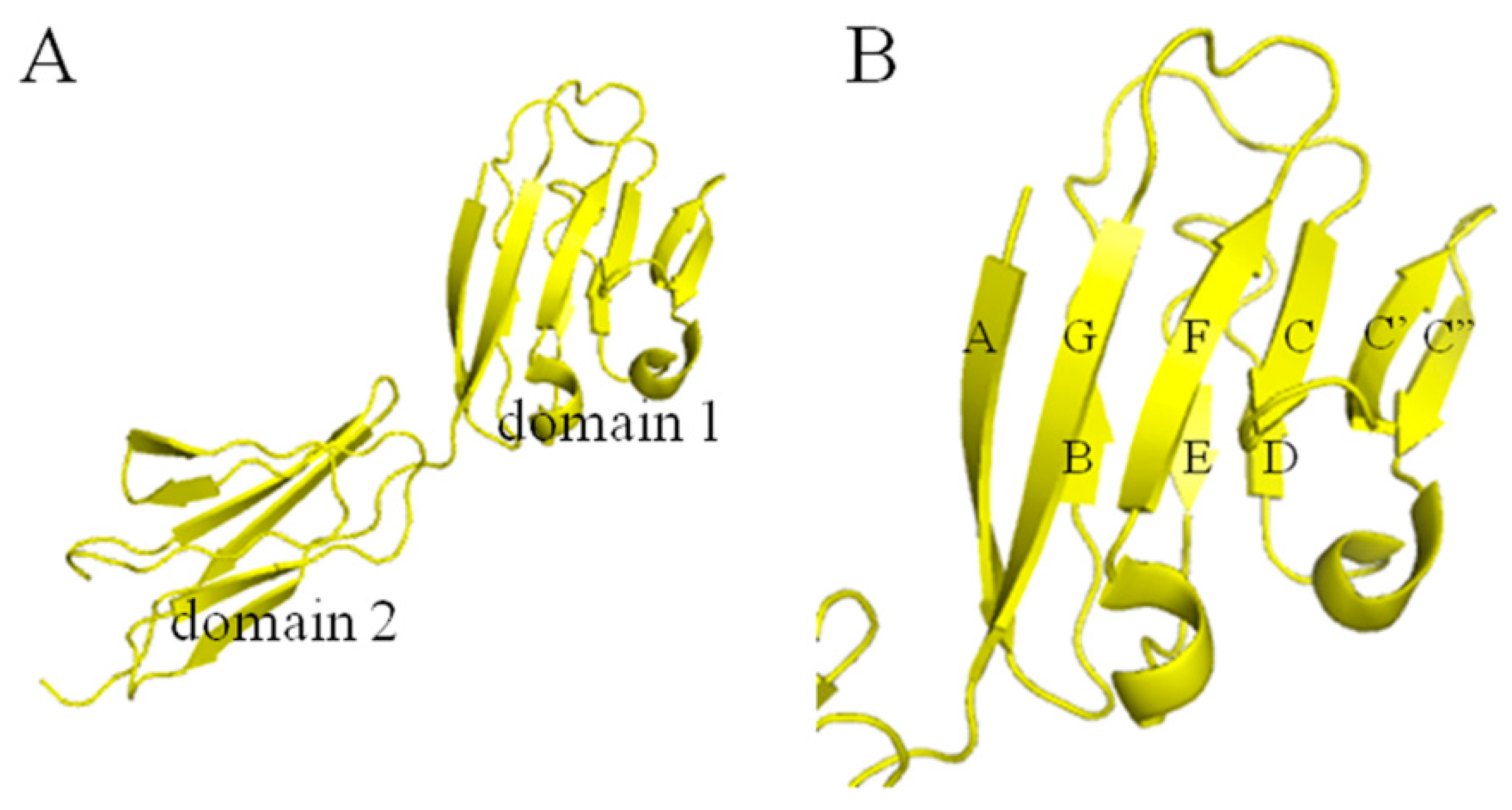
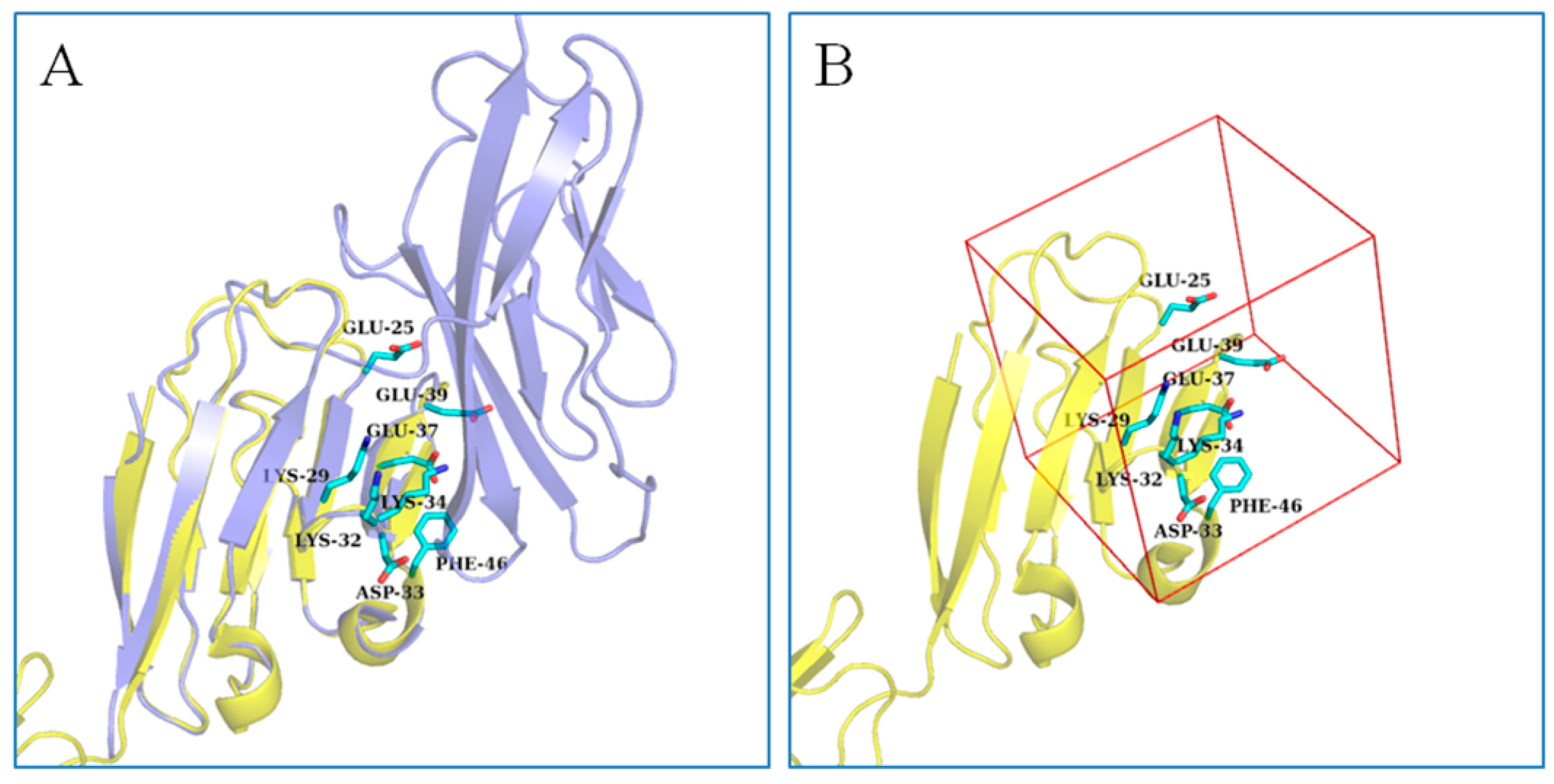
2.1. Active Site Analysis
2.2. Virtual Screening and Proliferation Inhibition of SW620 Cell Line
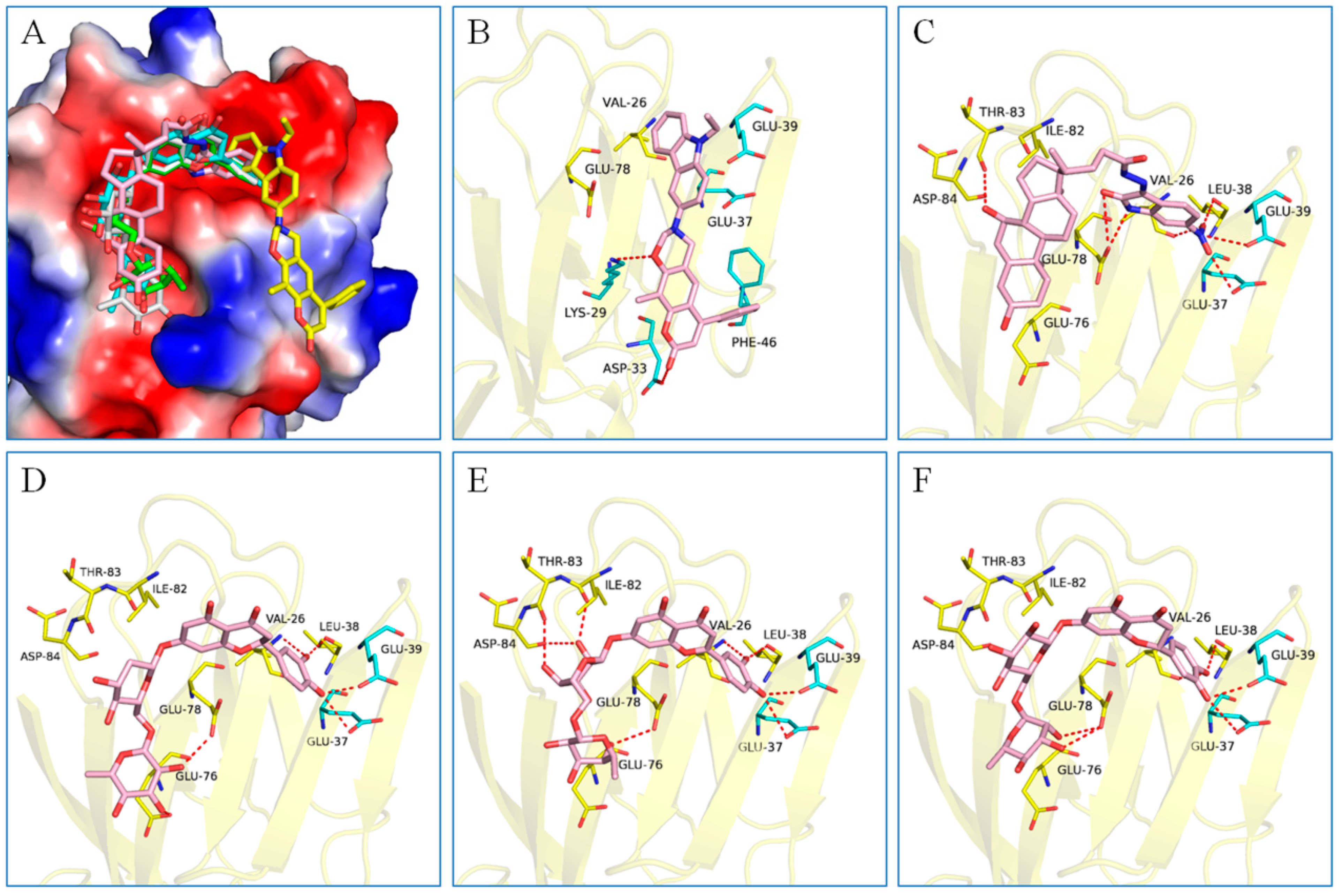
2.3. Binding Analysis of the Five Good Candidates
2.4. MD Simulations
2.5. ADMET Prediction
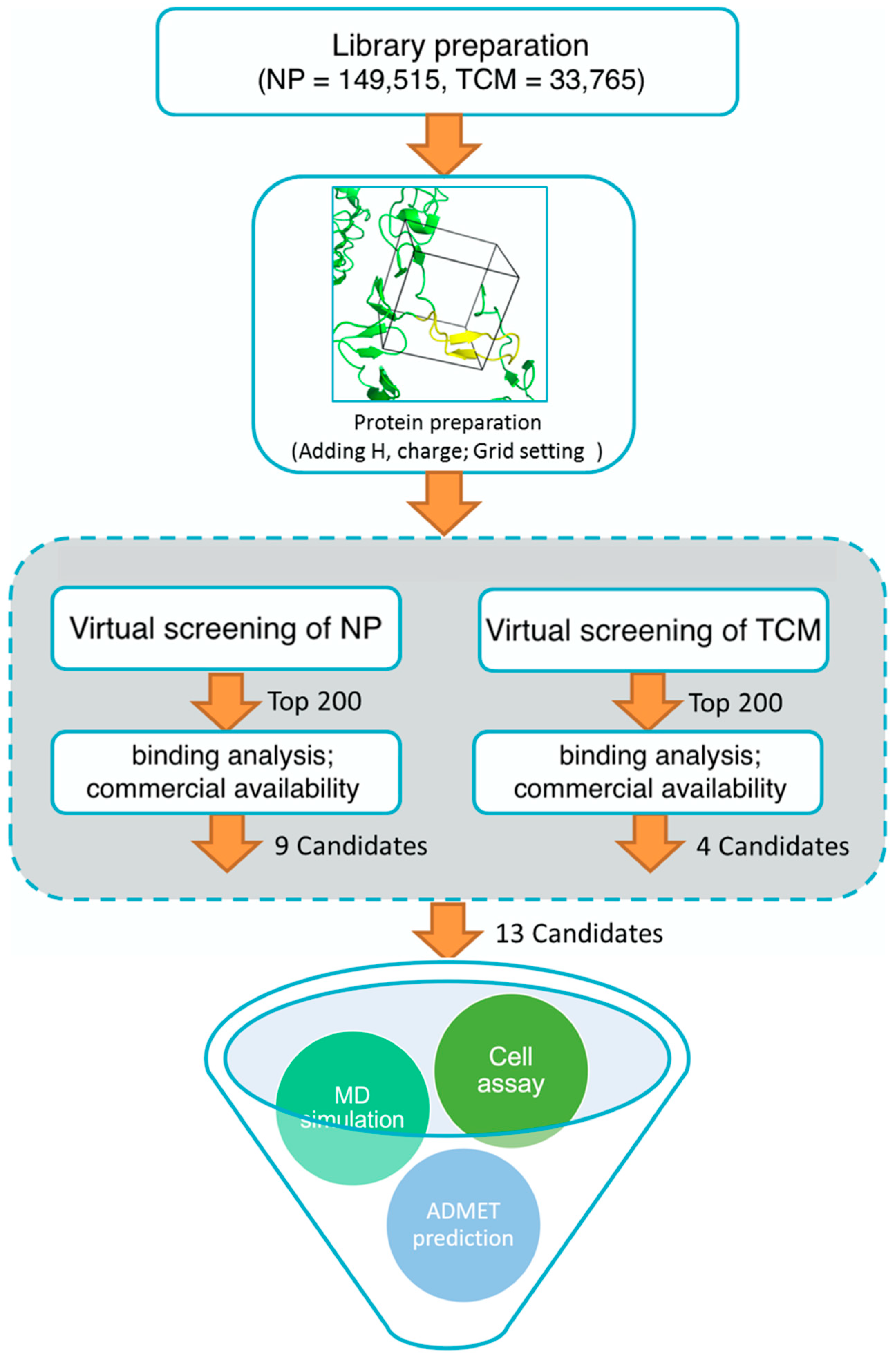
3. Experimental Methods
3.1. Library Download
3.2. Protein Preparation and High Throughput Virtual Screening
3.3. Chemistry
3.4. In Vitro Assay on SW620 Cell Lines
3.5. MD Simulations
3.6. ADMET Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yin, J.; Bai, Z.; Zhang, J.; Zheng, Z.; Yao, H.; Ye, P.; Li, J.; Gao, X.; Zhang, Z. Burden of colorectal cancer in China, 1990− 2017: Findings from the Global Burden of Disease Study 2017. Chin. J. Cancer Res. 2019, 31, 489. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A Tumorigenic Subpopulation with Stem Cell Properties in Melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective Identification of Tumorigenic Prostate Cancer Stem Cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Zhang, H.; Damelin, M.; Geles, K.G.; Grindley, J.C.; Dirks, P.B. Tumour-initiating cells: Challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 2009, 8, 806–823. [Google Scholar] [CrossRef]
- Wicha, M.S.; Liu, S.; Dontu, G. Cancer Stem Cells: An Old Idea—A Paradigm Shift. Cancer Res. 2006, 66, 1883–1890. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Ashkenazi, R.; Gentry, S.N.; Jackson, T.L. Pathways to Tumorigenesis—Modeling Mutation Acquisition in Stem Cells and Their Progeny. Neoplasia 2008, 10, 1170-IN6. [Google Scholar] [CrossRef]
- Vermeulen, L.; Felipe De Sousa, E.M.; Van Der Heijden, M.; Cameron, K.; De Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef]
- Xu, S.; Wen, Z.; Jiang, Q.; Zhu, L.; Feng, S.; Zhao, Y.; Wu, J.; Dong, Q.; Mao, J.; Zhu, Y. CD58, a novel surface marker, promotes self-renewal of tumor-initiating cells in colorectal cancer. Oncogene 2015, 34, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.A.; Mentzer, S.J.; Kamarck, M.E.; Hart, J.; Biro, P.A.; Strominger, J.L.; Burakoff, S.J. Gene mapping and somatic cell hybrid analysis of the role of human lymphocyte function-associated an-tigen-3 (LFA-3) in CTL-target cell interactions. J. Immunol. 1986, 136, 3085–3091. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L.; Sanders, M.E.; Shaw, S.; Springer, T.A. Purified lymphocyte function-associated antigen 3 binds to CD2 and mediates T lymphocyte adhesion. J. Exp. Med. 1987, 165, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, M.L.; Sanders, M.E.; Selvaraj, P.; Dustin, M.L.; Springer, T.A. Rosetting of activated human T lymphocytes with autologous erythrocytes. Definition of the re-ceptor and ligand molecules as CD2 and lymphocyte function-associated antigen 3 (LFA-3). J. Exp. Med. 1987, 165, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Moingeon, P.; Chang, H.-C.; Wallner, B.P.; Stebbins, C.; Frey, A.Z.; Reinherz, E.L. CD2-mediated adhesion facilitates T lymphocyte antigen recognition function. Nature 1989, 339, 312–314. [Google Scholar] [CrossRef] [PubMed]
- Ebert, E.C.; Panja, A.; Praveen, R. Human intestinal intraepithelial lymphocytes and epithelial cells coinduce interleukin-8 production through the CD2-CD58 interaction. Am. J. Physiol. Liver Physiol. 2009, 296, G671–G677. [Google Scholar] [CrossRef]
- Itoh, Y.; Joh, T.; Tanida, S.; Sasaki, M.; Kataoka, H.; Itoh, K.; Oshima, T.; Ogasawara, N.; Togawa, S.; Wada, T.; et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine 2005, 29, 275–282. [Google Scholar] [CrossRef]
- Čačev, T.; Radošević, S.; Križanac, Š.; Kapitanović, S. Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis 2008, 29, 1572–1580. [Google Scholar] [CrossRef]
- Wallich, R.; Brenner, C.; Brand, Y.; Roux, M.; Reister, M.; Meuer, S. Gene structure, promoter characterization, and basis for alternative mRNA splicing of the human CD58 gene. J. Immunol. 1998, 160, 2862–2871. [Google Scholar] [CrossRef]
- Withka, J.M.; Wyss, D.F.; Wagner, G.; Arulanandam, A.R.; Reinherz, E.L.; Recny, M.A. Structure of the glycosylated adhesion domain of human T lymphocyte glycoprotein CD2. Structure 1993, 1, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Bodian, D.L.; Jones, E.; Harlos, K.; Stuart, D.I.; Davis, S.J. Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 Å resolution. Structure 1994, 2, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Wyss, D.F.; Choi, J.S.; Li, J.; Knoppers, M.H.; Willis, K.J.; Arulanandam, A.R.N.; Smolyar, A.; Reinherz, E.L.; Wagner, G. Conformation and Function of the N-Linked Glycan in the Adhesion Domain of Human CD2. Science 1995, 269, 1273–1278. [Google Scholar] [CrossRef]
- Ikemizu, S.; Sparks, L.M.; van der Merwe, P.A.; Harlos, K.; Stuart, D.I.; Jones, E.Y.; Davis, S.J. Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-Å resolution. Proc. Natl. Acad. Sci. USA 1999, 96, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chow, V.T.; Jois, S.D. A novel, rapid and sensitive heterotypic cell adhesion assay for CD2–CD58 interaction, and its application for testing inhibitory peptides. J. Immunol. Methods 2004, 291, 39–49. [Google Scholar] [CrossRef]
- Jining, L.; Makagiansar, I.; Yusuf-Makagiansar, H.; Chow, V.T.K.; Siahaan, T.J.; Jois, S.D.S. Design, structure and biological activity of β-turn peptides of CD2 protein for inhibition of T-cell adhesion. JBIC J. Biol. Inorg. Chem. 2004, 271, 2873–2886. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, C.; Ke, S.; Satyanarayanajois, S.D. Structure-Based Rational Design of β-Hairpin Peptides from Discontinuous Epitopes of Cluster of Differentiation 2 (CD2) Protein To Modulate Cell Adhesion Interaction. J. Med. Chem. 2007, 50, 4038–4047. [Google Scholar] [CrossRef]
- Giddu, S.; Subramanian, V.; Yoon, H.S.; Satyanarayanajois, S.D. Design of β-Hairpin Peptides for Modulation of Cell Adhesion by β-Turn Constraint. J. Med. Chem. 2009, 52, 726–736. [Google Scholar] [CrossRef]
- Satyanarayanajois, S.D.; Büyüktimkin, B.; Gokhale, A.; Ronald, S.; Siahaan, T.J.; Latendresse, J.R. A Peptide from the Beta-strand Region of CD2 Protein that Inhibits Cell Adhesion and Suppresses Arthritis in a Mouse Model. Chem. Biol. Drug Des. 2010, 76, 234–244. [Google Scholar] [CrossRef]
- Gokhale, A.; Kanthala, S.; Latendresse, J.; Taneja, V.; Satyanarayanajois, S. Immunosuppression by Co-stimulatory Molecules: Inhibition of CD 2-CD 48/CD 58 Interaction by Peptides from CD2 to Suppress Progression of Collagen-induced Arthritis in Mice. Chem. Biol. Drug Des. 2013, 82, 106–118. [Google Scholar] [CrossRef]
- Sable, R.; Durek, T.; Taneja, V.; Craik, D.J.; Pallerla, S.; Gauthier, T.; Jois, S. Constrained cyclic peptides as immunomodulatory inhibitors of the CD2: CD58 protein–protein in-teraction. ACS Chem. Biol. 2016, 11, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
- Leherte, L.; Petit, A.; Jacquemin, D.; Vercauteren, D.P.; Laurent, A.D. Investigating cyclic peptides inhibiting CD2–CD58 interactions through molecular dynamics and molecular docking methods. J. Comput. Mol. Des. 2018, 32, 1295–1313. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Sable, R.; Shrestha, L.; Dahal, A.; Gauthier, T.; Taneja, V.; Jois, S. Modulation of co-stimulatory signal from CD2–CD58 proteins by a grafted peptide. Chem. Biol. Drug Des. 2021, 97, 607–627. [Google Scholar] [CrossRef] [PubMed]
- Shoichet, B.K. Virtual screening of chemical libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Omar, H.; del Cueto, M.; Nematiaram, T.; Troisi, A. High-throughput virtual screening for organic electronics: A comparative study of alternative strategies. J. Mater. Chem. C 2021, 9, 13557–13583. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Bombarelli, R.; Aguilera-Iparraguirre, J.; Hirzel, T.D.; Duvenaud, D.; Maclaurin, D.; Blood-Forsythe, M.A.; Chae, H.S.; Einzinger, M.; Ha, D.-G.; Wu, T.; et al. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat. Mater. 2016, 15, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.H.S.; Ferreira, R.S.; Caffarena, E.R. Integrating Molecular Docking and Molecular Dynamics Simulations. In Docking Screens for Drug Discovery; Springer: New York, NY, USA, 2019; pp. 13–34. [Google Scholar]
- Al-Karmalawy, A.A.; Dahab, M.A.; Metwaly, A.M.; Elhady, S.S.; Elkaeed, E.B.; Eissa, I.H.; Darwish, K.M. Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the h ACE2 receptor. Front. Chem. 2021, 9, 661230. [Google Scholar] [CrossRef]
- Vittorio, S.; Gitto, R.; Adornato, I.; Russo, E.; De Luca, L. In Silico Strategy for Targeting the mTOR Kinase at Rapamycin Binding Site by Small Molecules. Molecules 2021, 26, 1103. [Google Scholar] [CrossRef]
- Li, X.; Miao, F.; Xin, R.; Tai, Z.; Pan, H.; Huang, H.; Yu, J.; Chen, Z.; Zhu, Q. Combining network pharmacology, molecular docking, molecular dynamics simulation, and experimental verification to examine the efficacy and immunoregulation mechanism of FHB granules on vitiligo. Front. Immunol. 2023, 14, 1194823. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.-C. TCM Database@Taiwan: The World’s Largest Traditional Chinese Medicine Database for Drug Screening In Silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Sun, Z.-Y.J.; Byron, O.; Campbell, G.; Wagner, G.; Wang, J.-H.; Reinherz, E.L. Molecular dissection of the CD2-C58 counter-receptor interface identifies CD2 Tyr86 and CD58 Lys34 residues as the functional “hot spot”. J. Mol. Biol. 2001, 312, 711–720. [Google Scholar] [CrossRef]
- Wang, X.; Ji, C.G.; Zhang, J.Z.H. Glycosylation Modulates Human CD2-CD58 Adhesion via Conformational Adjustment. J. Phys. Chem. B 2015, 119, 6493–6501. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Lobanov, M.I.; Bogatyreva, N.; Galzitskaia, O.V. Radius of gyration is indicator of compactness of protein structure. Mol. Biol. 2008, 42, 701–706. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Miyamoto, T.; Min, W.; Lillehoj, H.S. Lymphocyte Proliferation Response During Eimeria tenella Infection Assessed by a New, Reliable, Nonradioactive Colorimetric Assay. Avian Dis. 2002, 46, 10–16. [Google Scholar] [CrossRef]
- Tominaga, H.; Ishiyama, M.; Ohseto, F.; Sasamoto, K.; Hamamoto, T.; Suzuki, K.; Watanabe, M. A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal. Commun. 1999, 36, 47–50. [Google Scholar] [CrossRef]
- Kubota, T.; Hanatani, Y.; Tsuyuki, K.; Nakada, M.; Asanuma, F.; Okazaki, M.; Ishibiki, K.; Abe, O.; Nakajima, E.; Maruyama, K. Antitumor Effect of 3-[(4-Amino-2-Methyl-5-Pyrimidinyl) Methyl]-1-(2-Chloroethyl)-1-Nitrosourea (ACNU) on Human Colon Carcinomas Transplanted into Nude Mice. Jpn. J. Clin. Oncol. 1982, 12, 33–42. [Google Scholar]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Yu, W.; He, X.; Vanommeslaeghe, K.; MacKerell, A.D. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012, 33, 2451–2468. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]
- Cheng, F.; Li, W.; Zhou, Y.; Li, J.; Shen, J.; Lee, P.W.; Tang, Y. Prediction of human genes and diseases targeted by xenobiotics using predictive toxicogenomic-derived models (PTDMs). Mol. Biosyst. 2013, 9, 1316–1325. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef]
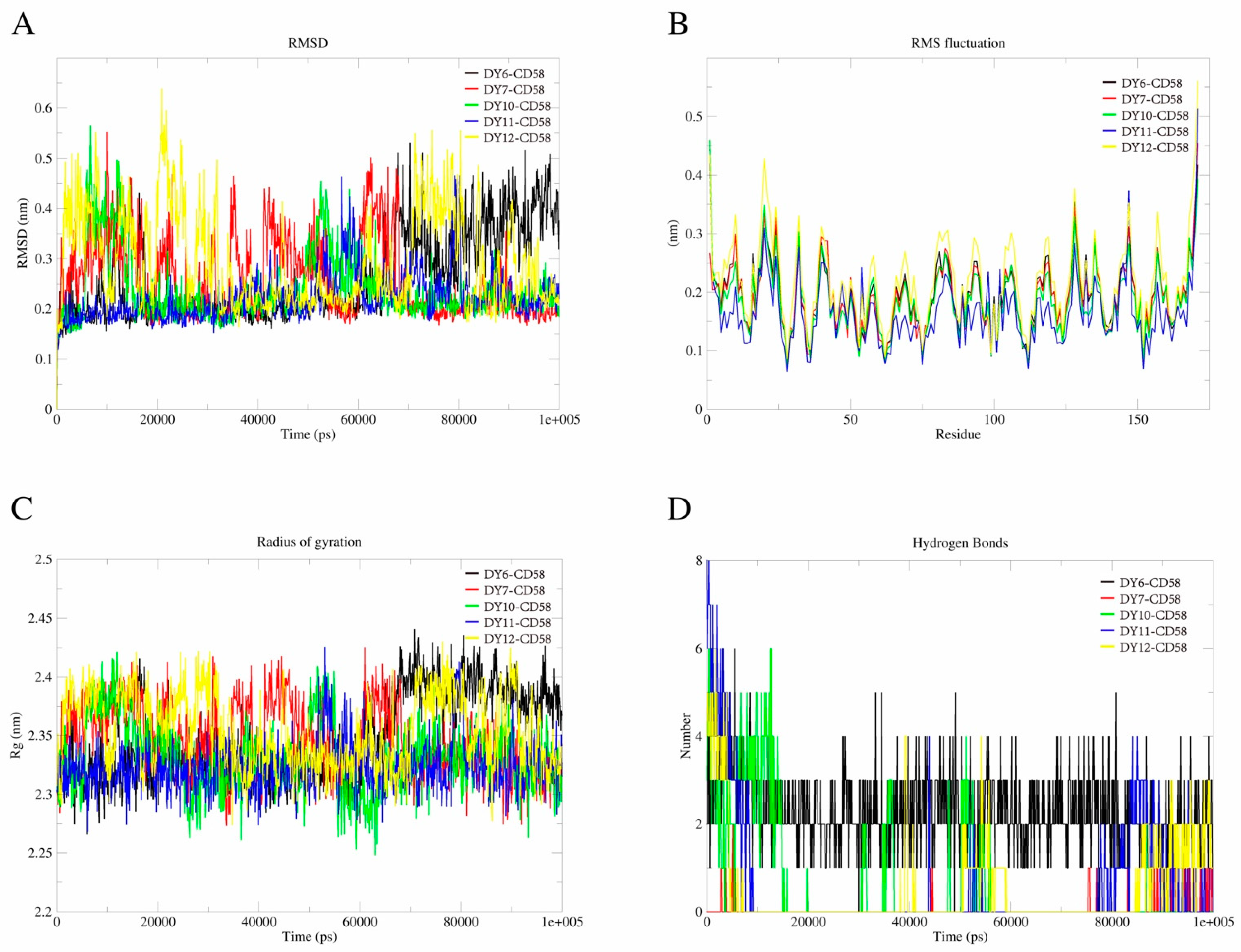
| Compound | MW a | Affinity (kcal/mol) | Structure | SW620 Cell Line b |
|---|---|---|---|---|
| IC50 (μM) | ||||
| DY1 | 496.5 | −9.0 | 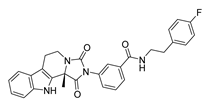 | >200 |
| DY2 | 575.7 | −9.2 | 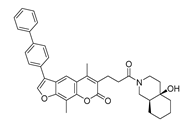 | >200 |
| DY3 | 412.4 | −8.8 | 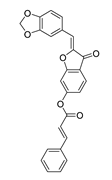 | >200 |
| DY4 | 482.5 | −8.7 | 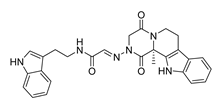 | >200 |
| DY5 | 630.7 | −8.8 | 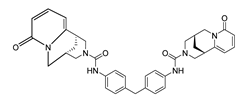 | >200 |
| DY6 | 580.7 | −9.0 | 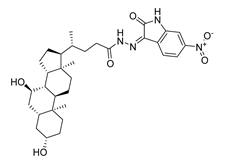 | 31.30 ± 0.88 |
| DY7 | 486.6 | −8.9 | 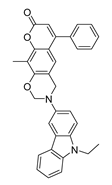 | 89.38 ± 15.28 |
| DY8 | 511.6 | −9.0 |  | >200 |
| DY9 | 722.0 | −8.8 |  | >200 |
| DY10 | 580.5 | −8.3 |  | 26.33 ± 0.23 |
| DY11 | 596.5 | −8.2 |  | 104.99 ± 7.86 |
| DY12 | 594.6 | −8.5 | 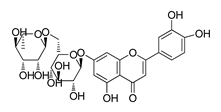 | 59.44 ± 3.40 |
| DY13 | 594.5 | −7.6 | 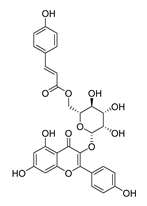 | >200 |
| Nimustine Hydrochloride | 309.2 |  | 59.44 ± 4.45 |
| ADMET Properties | DY6 | DY7 | DY10 | DY11 | DY12 |
|---|---|---|---|---|---|
| BBB | BBB− | BBB+ | BBB− | BBB− | BBB− |
| HIA | HIA+ | HIA+ | HIA+ | HIA+ | HIA+ |
| Pgp Substrate | Substrate | Non-substrate | Substrate | Substrate | Substrate |
| hERG Inhibition | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| AMES Toxicity | AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic |
| Carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens |
| Acute Oral Toxicity | low toxic | low toxic | low toxic | low toxic | low toxic |
| Molecular Weight | 580.73 | 486.57 | 580.54 | 596.54 | 594.52 |
| LogP | 5.25 | 7.25 | −0.43 | −1.46 | −1.39 |
| Rotatable Bonds | 6 | 3 | 6 | 6 | 6 |
| H Bond Acceptors | 10 | 5 | 14 | 15 | 15 |
| H Bond Donors | 4 | 0 | 8 | 9 | 9 |
| Surface Area | 161 | 213.67 | 233.03 | 237.82 | 236.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Yu, J.; Guo, Z. Virtual Screening and Binding Analysis of Potential CD58 Inhibitors in Colorectal Cancer (CRC). Molecules 2023, 28, 6819. https://doi.org/10.3390/molecules28196819
Guo R, Yu J, Guo Z. Virtual Screening and Binding Analysis of Potential CD58 Inhibitors in Colorectal Cancer (CRC). Molecules. 2023; 28(19):6819. https://doi.org/10.3390/molecules28196819
Chicago/Turabian StyleGuo, Rong, Jiangnan Yu, and Zhikun Guo. 2023. "Virtual Screening and Binding Analysis of Potential CD58 Inhibitors in Colorectal Cancer (CRC)" Molecules 28, no. 19: 6819. https://doi.org/10.3390/molecules28196819
APA StyleGuo, R., Yu, J., & Guo, Z. (2023). Virtual Screening and Binding Analysis of Potential CD58 Inhibitors in Colorectal Cancer (CRC). Molecules, 28(19), 6819. https://doi.org/10.3390/molecules28196819






