Salivary Pellicle Formed on Dental Composites Evaluated by Mass Spectrometry—An In Situ Study
Abstract
:1. Introduction
2. Results
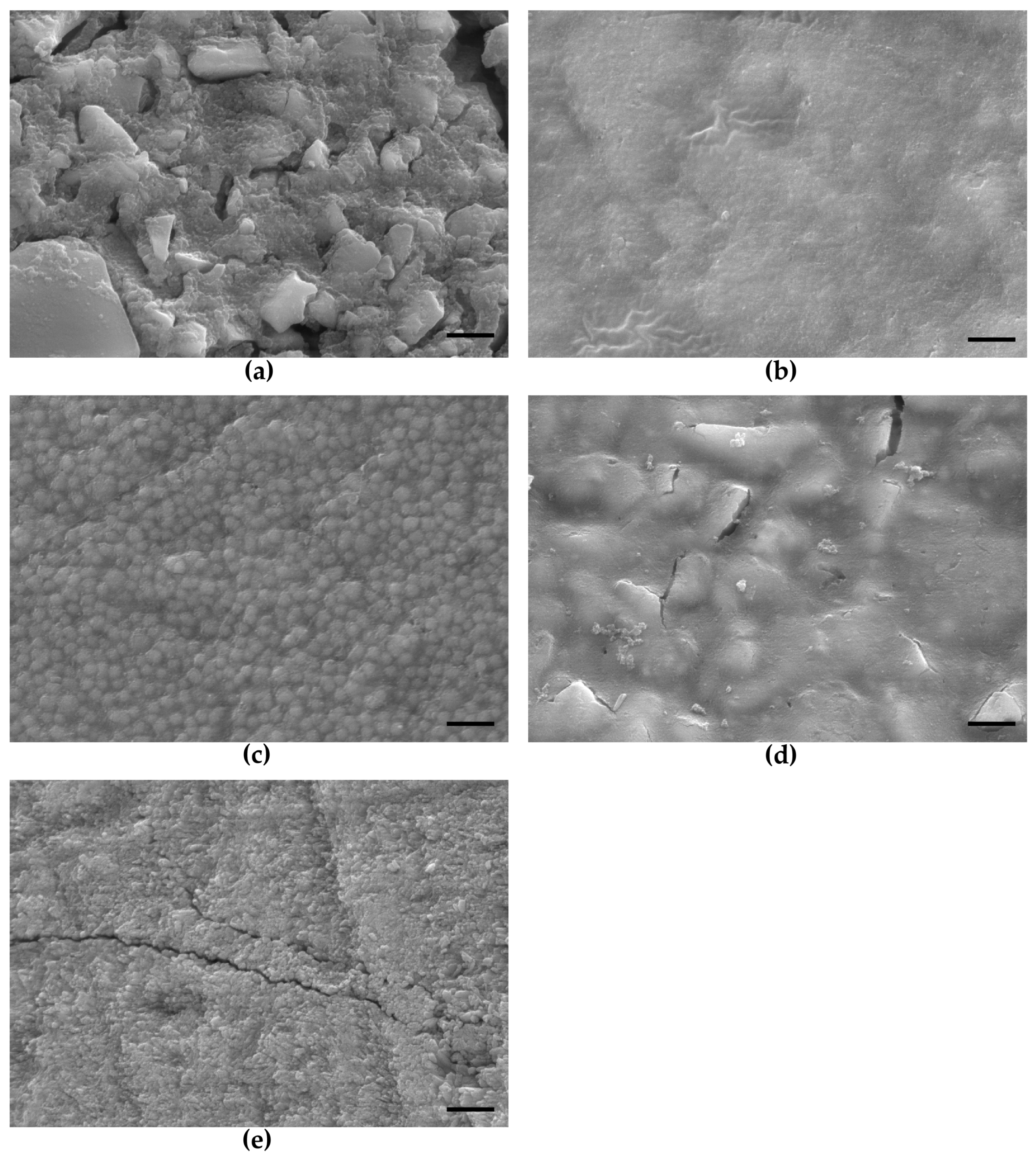
2.1. SEM Evaluation
2.2. Profilometry
2.3. SELDI–TOF–MS
3. Discussion
4. Materials and Methods
4.1. Recruitment of Participants
4.2. Sample Preparation
4.3. Scanning Electron Microscopic Examination
4.4. Mechanical and Optical Profilometry
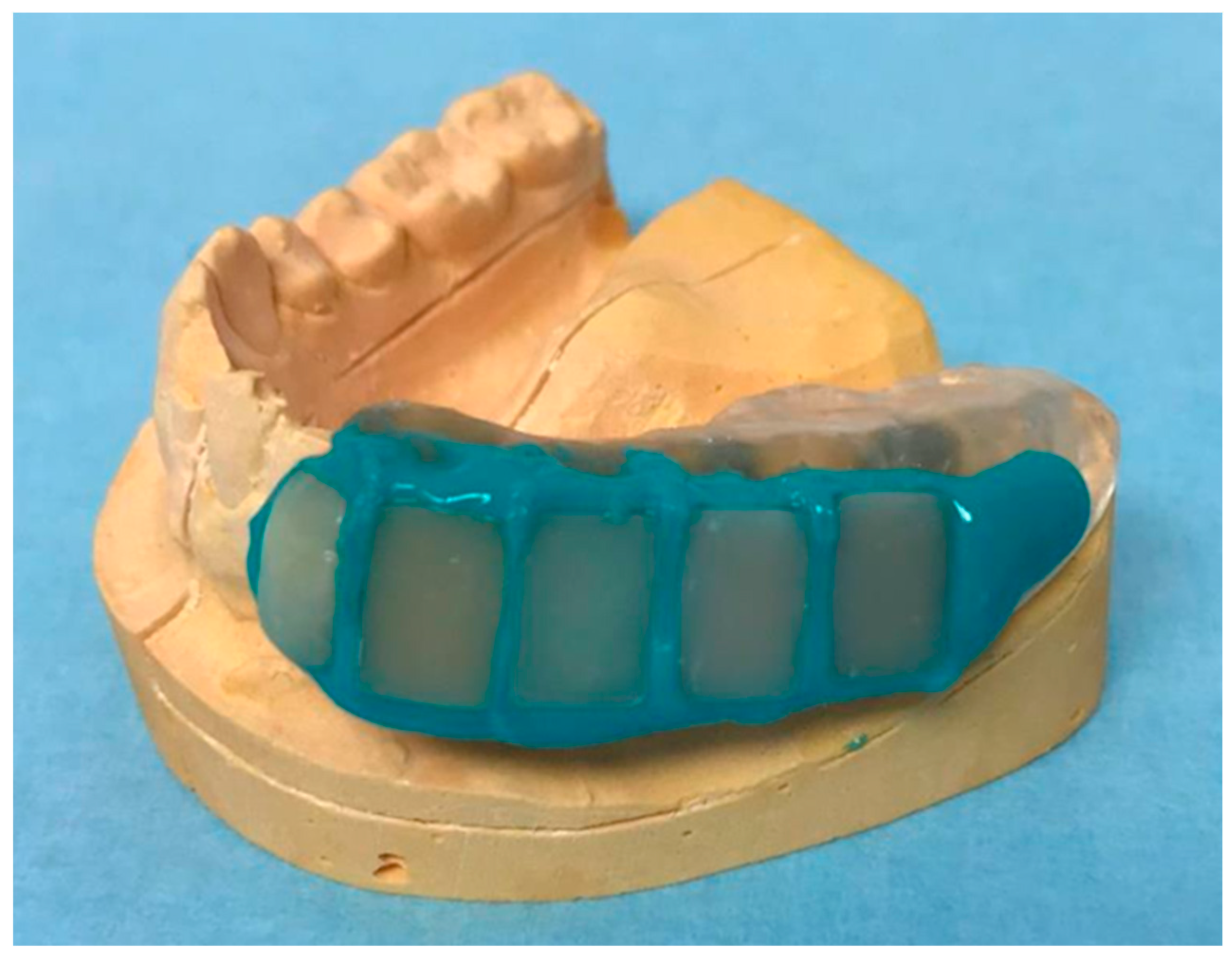
4.5. Fabrication of the Miniplast Splint
4.6. Oral Exposure of the Specimens
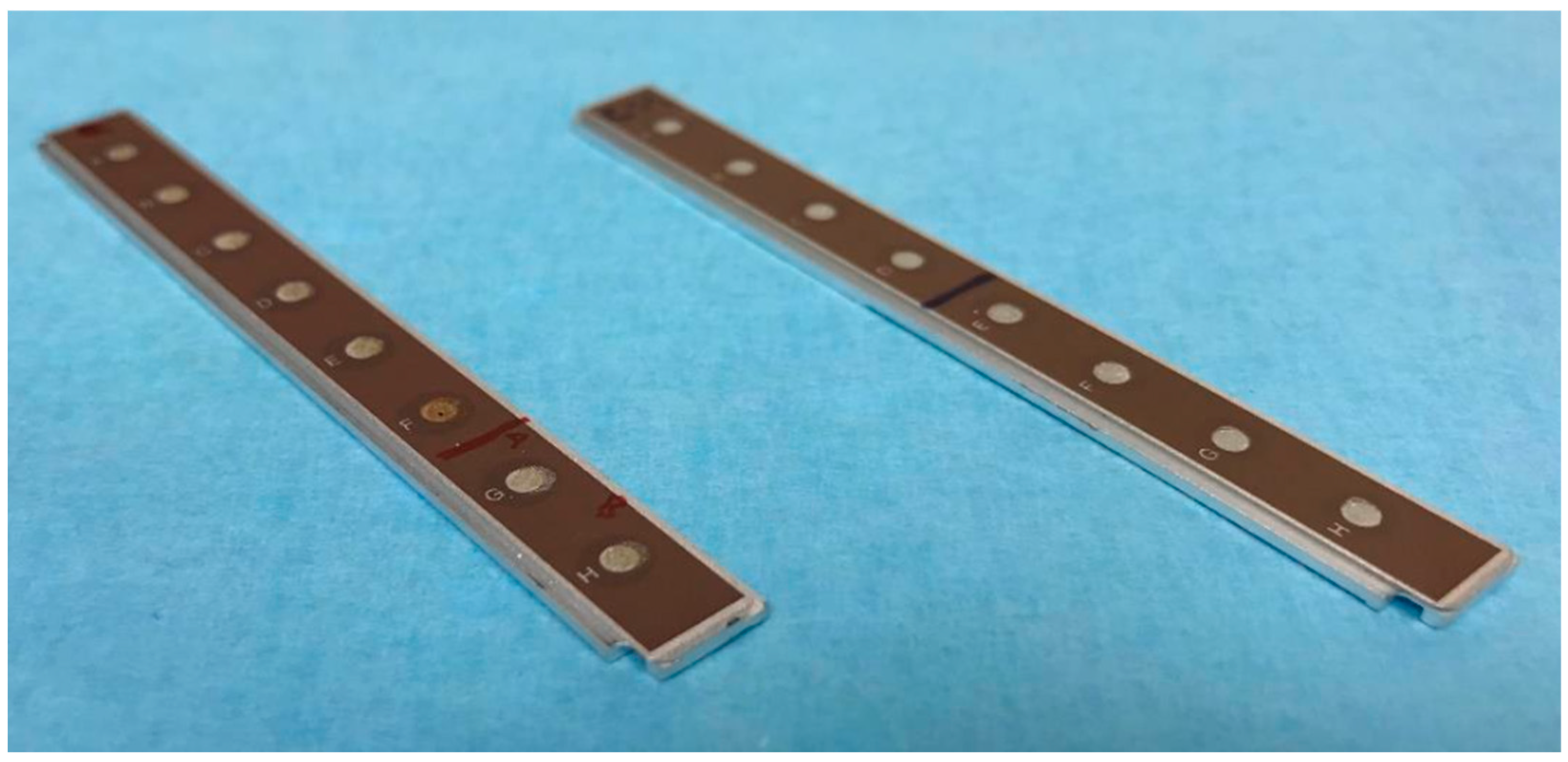
4.7. Surface-Enhanced Laser Desorption/Ionization Time of Flight Mass Spectrometry (SELDI–TOF–MS) Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chawhuaveang, D.D.; Yu, O.Y.; Yin, I.X.; Lam, W.Y.; Mei, M.L.; Chu, C.H. Acquired salivary pellicle and oral diseases: A literature review. J. Dent. Sci. 2021, 16, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Lindh, L.; Aroonsang, W.; Sotres, J.; Arnebrant, T. Salivary pellicles. Monogr. Oral Sci. 2014, 24, 30–39. [Google Scholar] [PubMed]
- Siqueira, W.L.; Custodio, W.; McDonald, E.E. New insights into the composition and functions of the acquired enamel pellicle. J. Dent. Res. 2012, 91, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.A.; Ventura, T.M.S.; Cassiano, L.P.S.; Silva, C.M.S.; Martini, T.; Leite, A.L.; Rios, D.; Magalhães, A.C.; Buzalaf, M.A.R. Changes in the Proteomic Profile of Acquired Enamel Pellicles as a Function of Their Time of Formation and Hydrochloric Acid Exposure. Caries Res. 2018, 52, 367–377. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, Z.; Ding, C.; Li, J. Functional biomedical materials derived from proteins in the acquired salivary pellicle. J Mater. Chem. B 2021, 9, 6507–6520. [Google Scholar] [CrossRef]
- Hannig, M. Ultrastructural investigation of pellicle morphogenesis at two different intraoral sites during a 24-h period. Clin. Oral Investig. 1999, 3, 88–95. [Google Scholar] [CrossRef]
- Kreth, J.; Merritt, J.; Pfeifer, C.S.; Khajotia, S.; Ferracane, J.L. Interaction between the Oral Microbiome and Dental Composite Biomaterials: Where We Are and Where We Should Go. J. Dent. Res. 2020, 99, 1140–1149. [Google Scholar] [CrossRef]
- Hannig, M.; Joiner, A. The structure, function and properties of the acquired pellicle. Monogr. Oral Sci. 2006, 19, 29–64. [Google Scholar]
- Steiger-Ronay, V.; Kuster, I.M.; Wiedemeier, D.B.; Attin, T.; Wegehaupt, F.J. Erosive loss of tooth substance is dependent on enamel surface structure and presence of pellicle—An in vitro study. Arch. Oral Biol. 2020, 112, 104686. [Google Scholar] [CrossRef]
- Hannig, C.; Hannig, M.; Attin, T. Enzymes in the acquired enamel pellicle. Eur. J. Oral Sci. 2005, 113, 2–13. [Google Scholar] [CrossRef]
- Luo, J.; Wang, Y.; Wang, K.; Jiang, W.; Li, X.; Zhang, L. Comparative proteomic analysis on acquired enamel pellicle at two time points in caries-susceptible and caries-free subjects. J. Dent. 2020, 94, 103301. [Google Scholar] [CrossRef] [PubMed]
- Lynge Pedersen, A.M.; Belstrøm, D. The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 2019, 80 (Suppl. 1), S3–S12. [Google Scholar] [CrossRef]
- Ventura, T.; Cassiano, L.P.S.; Souza, E.S.C.M.; Taira, E.A.; Leite, A.L.; Rios, D.; Buzalaf, M.A.R. The proteomic profile of the acquired enamel pellicle according to its location in the dental arches. Arch. Oral Biol. 2017, 79, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Burrow, M.F.; Leung, W.K. Proteomic profile of in situ acquired pellicle on tooth and restorative material surfaces. J. Dent. 2022, 129, 104389. [Google Scholar] [CrossRef] [PubMed]
- Hannig, C.; Wasser, M.; Becker, K.; Hannig, M.; Huber, K.; Attin, T. Influence of different restorative materials on lysozyme and amylase activity of the salivary pellicle in situ. J. Biomed. Mater. Res. A 2006, 78, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.; Helmerhorst, E.J.; Oppenheim, F.G. Saliva and Serum Protein Exchange at the Tooth Enamel Surface. J. Dent. Res. 2017, 96, 437–443. [Google Scholar] [CrossRef]
- Pelá, V.T.; Cassiano, L.P.S.; Ventura, T.; Souza, E.S.C.M.; Gironda, C.C.; Rios, D.; Buzalaf, M.A.R. Proteomic analysis of the acquired enamel pellicle formed on human and bovine tooth: A study using the Bauru in situ pellicle model (BISPM). J. Appl. Oral Sci. 2018, 27, e20180113. [Google Scholar] [CrossRef]
- Sterzenbach, T.; Helbig, R.; Hannig, C.; Hannig, M. Bioadhesion in the oral cavity and approaches for biofilm management by surface modifications. Clin. Oral Investig. 2020, 24, 4237–4260. [Google Scholar] [CrossRef]
- Wei, C.X.; Burrow, M.F.; Botelho, M.G.; Lam, H.; Leung, W.K. In Vitro Salivary Protein Adsorption Profile on Titanium and Ceramic Surfaces and the Corresponding Putative Immunological Implications. Int. J. Mol. Sci. 2020, 21, 3083. [Google Scholar] [CrossRef]
- Fischer, N.G.; Aparicio, C. The salivary pellicle on dental biomaterials. Colloids Surf. B Biointerfaces 2021, 200, 111570. [Google Scholar] [CrossRef]
- Göcke, R.; Gerath, F.; von Schwanewede, H. Quantitative determination of salivary components in the pellicle on PMMA denture base material. Clin. Oral Investig. 2002, 6, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Hannig, C.; Huber, K.; Lambrichts, I.; Gräser, J.; D’Haen, J.; Hannig, M. Detection of salivary alpha-amylase and lysozyme exposed on the pellicle formed in situ on different materials. J. Biomed. Mater. Res. A 2007, 83, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Hirota, M.; Hayakawa, T. Adsorption behaviors of salivary pellicle proteins onto denture base metals using 27-MHz quartz crystal microbalance. Biomed. Mater. Eng. 2022, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ozden, A.N.; Haghighat, N.; Al-Hashimi, I. Preliminary evaluation of salivary pellicle on nickel-chromium alloy in vivo. Quintessence Int. 2002, 33, 731–735. [Google Scholar] [PubMed]
- Svendsen, I.E.; Lindh, L. The composition of enamel salivary films is different from the ones formed on dental materials. Biofouling 2009, 25, 255–261. [Google Scholar] [CrossRef]
- Trautmann, S.; Barghash, A.; Fecher-Trost, C.; Schalkowsky, P.; Hannig, C.; Kirsch, J.; Rupf, S.; Keller, A.; Helms, V.; Hannig, M. Proteomic Analysis of the Initial Oral Pellicle in Caries-Active and Caries-Free Individuals. Proteom. Clin. Appl. 2019, 13, e1800143. [Google Scholar] [CrossRef]
- Li, J.; Helmerhorst, E.J.; Leone, C.W.; Troxler, R.F.; Yaskell, T.; Haffajee, A.D.; Socransky, S.S.; Oppenheim, F.G. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 2004, 97, 1311–1318. [Google Scholar] [CrossRef]
- Marin, L.M.; Xiao, Y.; Cury, J.A.; Siqueira, W.L. Modulation of Streptococcus mutans Adherence to Hydroxyapatite by Engineered Salivary Peptides. Microorganisms 2022, 10, 223. [Google Scholar] [CrossRef]
- Lie, T. Scanning and transmission electron microscope study of pellicle morphogenesis. Scand. J. Dent. Res. 1977, 85, 217–231. [Google Scholar] [CrossRef]
- Nyvad, B.; Fejerskov, O. Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 1987, 95, 287–296. [Google Scholar] [CrossRef]
- Peckys, D.B.; N, D.E.J.; Hannig, M. Oil droplet formation on pellicle covered tooth surfaces studied with environmental scanning electron microscopy. J. Microsc. 2019, 274, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Deimling, D.; Breschi, L.; Hoth-Hannig, W.; Ruggeri, A.; Hannig, C.; Nekrashevych, Y.; Prati, C.; Hannig, M. Electron microscopic detection of salivary alpha-amylase in the pellicle formed in situ. Eur. J. Oral Sci. 2004, 112, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, T.T.; Anderson Matias da Rocha, B.; Alves Carneiro, V.; Vassiliepe Sousa Arruda, F.; Fernandes do Nascimento, A.S.; Cardoso Sá, N.; do Nascimento, K.S.; Sousa Cavada, B.; Holanda Teixeira, E. Effect of lectins from Diocleinae subtribe against oral Streptococci. Molecules 2011, 16, 3530–3543. [Google Scholar] [CrossRef]
- Kozmos, M.; Virant, P.; Rojko, F.; Abram, A.; Rudolf, R.; Raspor, P.; Zore, A.; Bohinc, K. Bacterial Adhesion of Streptococcus mutans to Dental Material Surfaces. Molecules 2021, 26, 1152. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, S.; Sandberg, A.L.; Cisar, J.O. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J. Dent. Res. 2004, 83, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Lapinska, B.; Rogowski, J.; Nowak, J.; Nissan, J.; Sokolowski, J.; Lukomska-Szymanska, M. Effect of Surface Cleaning Regimen on Glass Ceramic Bond Strength. Molecules 2019, 24, 389. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, Y.W.; Soare, R.V.; Leite Assis, M.A.; Zenóbio, E.G.; Girundi, F.M. Titanium Surface Roughing Treatments contribute to Higher Interaction with Salivary Proteins MG2 and Lactoferrin. J. Contemp. Dent. Pract. 2015, 16, 141–146. [Google Scholar] [CrossRef]
- Zuanazzi, D.; Xiao, Y.; Siqueira, W.L. Evaluating protein binding specificity of titanium surfaces through mass spectrometry-based proteomics. Clin. Oral Investig. 2021, 25, 2281–2296. [Google Scholar] [CrossRef]
- AlAli, M.; Silikas, N.; Satterthwaite, J. The Effects of Toothbrush Wear on the Surface Roughness and Gloss of Resin Composites with Various Types of Matrices. Dent. J. 2021, 9, 8. [Google Scholar] [CrossRef]
- Amari, Y.; Takamizawa, T.; Kawamoto, R.; Namura, Y.; Murayama, R.; Yokoyama, M.; Tsujimoto, A.; Miyazaki, M. Influence of one-step professional mechanical tooth cleaning pastes on surface roughness and morphological features of tooth substrates and restoratives. J. Oral Sci. 2021, 63, 133–138. [Google Scholar] [CrossRef]
- Gjorgievska, E.; Oh, D.S.; Haam, D.; Gabric, D.; Coleman, N.J. Evaluation of Efficiency of Polymerization, Surface Roughness, Porosity and Adaptation of Flowable and Sculptable Bulk Fill Composite Resins. Molecules 2021, 26, 5202. [Google Scholar] [CrossRef]
- Matzinger, M.; Hahnel, S.; Preis, V.; Rosentritt, M. Polishing effects and wear performance of chairside CAD/CAM materials. Clin. Oral Investig. 2019, 23, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Schipper, R.; Loof, A.; de Groot, J.; Harthoorn, L.; Dransfield, E.; van Heerde, W. SELDI-TOF-MS of saliva: Methodology and pre-treatment effects. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 847, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Aita, A.; Battisti, I.; Contran, N.; Furlan, S.; Padoan, A.; Franchin, C.; Barbaro, F.; Cattelan, A.M.; Zambon, C.F.; Plebani, M.; et al. Salivary proteomic analysis in asymptomatic and symptomatic SARS-CoV-2 infection: Innate immunity, taste perception and FABP5 proteins make the difference. Clin. Chim. Acta 2022, 537, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.P.; Buzalaf, M.A.R.; Silva, N.C.; Ventura, T.M.O.; Toniolo, J.; Rodrigues, J.A. Saliva proteomic profile of early childhood caries and caries-free children. Acta Odontol. Scand 2022, 81, 216–226. [Google Scholar] [CrossRef]
- Simões, C.; Caeiro, I.; Carreira, L.; Silva, F.C.E.; Lamy, E. How Different Snacks Produce a Distinct Effect in Salivary Protein Composition. Molecules 2021, 26, 2403. [Google Scholar] [CrossRef]
- Trautmann, S.; Künzel, N.; Fecher-Trost, C.; Barghash, A.; Dudek, J.; Flockerzi, V.; Helms, V.; Hannig, M. Is the proteomic composition of the salivary pellicle dependent on the substrate material? Proteom. Clin. Appl. 2022, 16, e2100109. [Google Scholar] [CrossRef]
- Pelá, V.T.; Lunardelli, J.G.Q.; Ventura, T.M.O.; Camiloti, G.D.; Baumann, T.; Carvalho, T.S.; Lussi, A.; Buzalaf, M.A.R. Proteomic profiles of the acquired enamel pellicle formed in vitro, in situ, or in vivo. Eur. J. Oral Sci. 2020, 128, 487–494. [Google Scholar] [CrossRef]
- PelÁ, V.T.; Ventura, T.M.O.; Buzalaf, M.A.R. Optimizing the formation of the acquired enamel pellicle in vitro for proteomic analysis. J. Appl. Oral Sci. 2020, 28, e20200189. [Google Scholar] [CrossRef]
- Trautmann, S.; Künzel, N.; Fecher-Trost, C.; Barghash, A.; Schalkowsky, P.; Dudek, J.; Delius, J.; Helms, V.; Hannig, M. Deep Proteomic Insights into the Individual Short-Term Pellicle Formation on Enamel-An In Situ Pilot Study. Proteom. Clin. Appl. 2020, 14, e2070054. [Google Scholar] [CrossRef]
- Pelá, V.T.; Prakki, A.; Wang, L.; Ventura, T.M.S.; de Souza, E.S.C.M.; Cassiano, L.P.S.; Brianezzi, L.F.F.; Leite, A.L.; Buzalaf, M.A.R. The influence of fillers and protease inhibitors in experimental resins in the protein profile of the acquired pellicle formed in situ on enamel-resin specimens. Arch. Oral Biol. 2019, 108, 104527. [Google Scholar] [CrossRef] [PubMed]
- Carlén, A.; Nikdel, K.; Wennerberg, A.; Holmberg, K.; Olsson, J. Surface characteristics and in vitro biofilm formation on glass ionomer and composite resin. Biomaterials 2001, 22, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Spencer, P.; Ye, Q.; Misra, A.; Goncalves, S.E.; Laurence, J.S. Proteins, pathogens, and failure at the composite-tooth interface. J. Dent. Res. 2014, 93, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Delius, J.; Trautmann, S.; Médard, G.; Kuster, B.; Hannig, M.; Hofmann, T. Label-free quantitative proteome analysis of the surface-bound salivary pellicle. Colloids Surf. B Biointerfaces 2017, 152, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Hannig, M. Transmission electron microscopy of early plaque formation on dental materials in vivo. Eur. J. Oral Sci. 1999, 107, 55–64. [Google Scholar] [CrossRef]

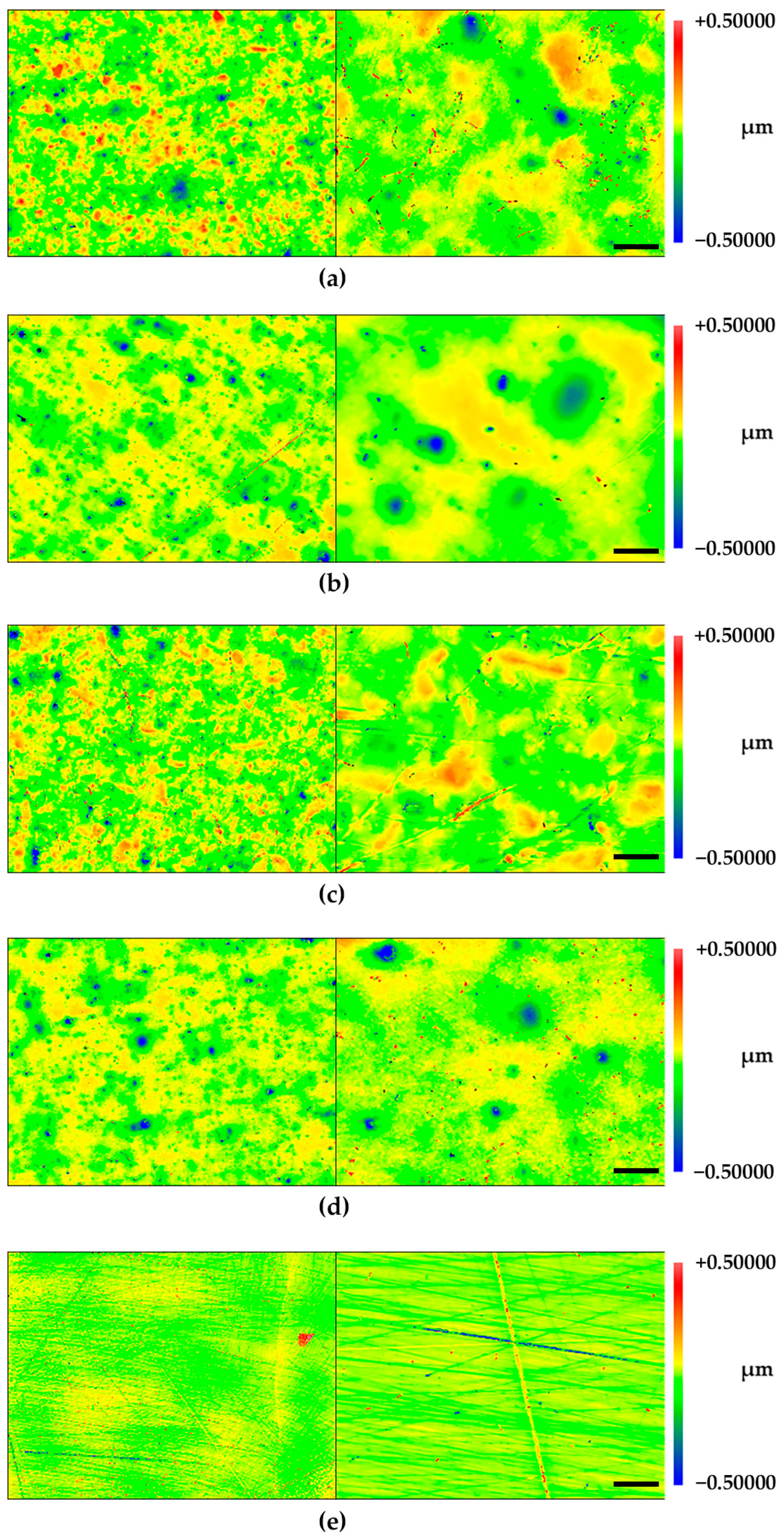
| Rq (nm) | Ra (nm) | ||
|---|---|---|---|
| 10× Magnification | 50× Magnification | ||
| Dyract® eXtra | 73 ± 8 | 78 ± 14 | 68 ± 8.4 |
| Estelite Σ Quick | 95 ± 9 | 66 ± 13 | 80 ± 15.8 |
| GrandioSO | 92 ± 19 | 56 ± 17 | 72 ± 13.0 |
| Venus® Diamond | 93 ± 6 | 78 ± 4 | 74 ± 11.4 |
| enamel control | 71 ± 23 | 46 ± 16 | 50 ± 23.5 |
| CM10 Array | Q10 Array | CM10 and Q10 Array | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Intensity | Total | Intensity | Total | Intensity | Total | ||||
| I > 1 | I < 1 | I > 1 | I < 1 | I > 1 | I < 1 | ||||
| Saliva [Da] | |||||||||
| 1.500–25.000 | 63 | 4 | 67 | 54 | 3 | 57 | 53 | 3 | 56 |
| 25.000–300.000 | 7 | 28 | 35 | 2 | 31 | 33 | 3 | 31 | 34 |
| 102 | 90 | 90 | |||||||
| Peliclle [Da] | |||||||||
| 1.500–25.000 | 18 | 6 | 24 | 13 | 5 | 18 | 16 | 5 | 21 |
| 25.000–300.000 | 0 | 22 | 22 | 0 | 24 | 24 | 0 | 18 | 18 |
| 46 | 42 | 39 | |||||||
| Proteinn | Dental | Exposure | Mean | Standard | Bonferroni | Array |
|---|---|---|---|---|---|---|
| (m/z) | Composite | Time [min] | Difference | Error | Sequence | Chip |
| (Sequential) | ||||||
| 1634 | Venus® Diamond | 30 | 5.194 | 1.555 | 0.008 | Q10 |
| GrandioSO | 6.682 | 2.312 | 0.031 | Q10 | ||
| Dyract® eXtra | 5.209 | 1.022 | 0 | Q10 | ||
| 1661 | Dyract® eXtra | 30 | 8.059 | 2.485 | 0.012 | Q10 |
| 1678 | GrandioSO | 30 | 4.92 | 1.321 | 0.002 | Q10 |
| Dyract® eXtra | 2.877 | 0.6 | 0 | Q10 | ||
| 1634 | Dyract® eXtra | 90 | 7.33 | 2.720 | 0.042 | Q10 |
| 1678 | Dyract® eXtra | 90 | 11.631 | 3.639 | 0.014 | Q10 |
| 1843 | Dyract® eXtra | 90 | 10.386 | 3.336 | 0.018 | CM10 |
| 2069 | Dyract® eXtra | 90 | 17.156 | 5.829 | 0.029 | CM10 |
| 6965 | Dyract® eXtra | 90 | 0.676 | 0.237 | 0.043 | CM10 |
| 1634 | Venus® Diamond | 120 | 7.798 | 1.407 | 0 | Q10 |
| Estelite Σ Quick | 10.422 | 2.931 | 0.003 | Q10 | ||
| Dyract® eXtra | 7.51 | 1.439 | 0 | Q10 | ||
| 1661 | Venus® Diamond | 120 | 3.063 | 1.139 | 0.05 | Q10 |
| Estelite Σ Quick | 4.236 | 0.607 | 0 | Q10 | ||
| Dyract® eXtra | 1.135 | 0.39 | 0.029 | Q10 | ||
| 1678 | Venus® Diamond | 120 | 10.349 | 1.718 | 0 | Q10 |
| Estelite Σ Quick | 7.367 | 1.812 | 0 | CM10 | ||
| Dyract® eXtra | 9.152 | 2.044 | 0 | Q10 | ||
| 1843 | Venus® Diamond | 120 | 12.829 | 2.903 | 0 | Q10 |
| Estelite Σ Quick | 7.965 | 2.385 | 0.008 | CM10 | ||
| Dyract® eXtra | 15.184 | 4.619 | 0.009 | Q10 | ||
| 2045 | Venus® Diamond | 120 | 16.336 | 3.589 | 0 | Q10 |
| Dyract® eXtra | 18.686 | 5.976 | 0.016 | Q10 | ||
| Dyract® eXtra | 16.746 | 4.32 | 0.001 | CM10 | ||
| 2069 | Venus® Diamond | 120 | 20.723 | 4.004 | 0 | Q10 |
| Estelite Σ Quick | 14.692 | 4.186 | 0.004 | CM10 | ||
| Dyract® eXtra | 19.665 | 6.362 | 0.018 | CM10 | ||
| Dyract® eXtra | 23.883 | 7.313 | 0.009 | Q10 |
| Composite A | Composite B | Protein (m/z) | Exposure Time | Mean Difference | Standard Error | Bonferroni Sequence (Sequential) | Array |
|---|---|---|---|---|---|---|---|
| Venus® Diamond | Dyract® eXtra | 1843 | 30 | 8.526 | 1.479 | 0 | CM10 |
| Venus® Diamond | Dyract® eXtra | 2069 | 30 | 11.999 | 2.998 | 0.001 | CM10 |
| GrandioSO | Dyract® eXtra | 1938 | 30 | 1.491 | 0.372 | 0.001 | Q10 |
| Estelite Σ Quick | GrandioSO | 1661 | 30 | 7.596 | 2.315 | 0.010 | CM10 |
| Venus® Diamond | GrandioSO | 1634 | 90 | 9.306 | 1.718 | 0 | Q10 |
| Venus® Diamond | GrandioSO | 1843 | 90 | 13.822 | 4.78 | 0.031 | CM10 |
| Venus® Diamond | GrandioSO | 2260 | 90 | 14.779 | 5.209 | 0.045 | CM10 |
| Venus® Diamond | Estelite Σ Quick | 1634 | 90 | 8.138 | 2.574 | 0.011 | Q10 |
| GrandioSO | Estelite Σ Quick | 5784 | 90 | 2.68 | 0.944 | 0.045 | CM10 |
| GrandioSO | Dyract® eXtra | 3364 | 90 | 10.726 | 3.745 | 0.038 | Q10 |
| Estelite Σ Quick | GrandioSO | 1661 | 90 | 6.215 | 1.869 | 0.009 | CM10 |
| Estelite Σ Quick | Dyract® eXtra | 3364 | 90 | 18.356 | 5.608 | 0.011 | Q10 |
| Estelite Σ Quick | Dyract® eXtra | 3436 | 90 | 34.798 | 9.988 | 0.005 | Q10 |
| Estelite Σ Quick | Dyract® eXtra | 3708 | 90 | 2.144 | 0.666 | 0.013 | Q10 |
| Dyract® eXtra | Venus® Diamond | 1661 | 90 | 11.751 | 3.954 | 0.024 | Q10 |
| Dyract® eXtra | Venus® Diamond | 5424 | 90 | 0.572 | 0.179 | 0.014 | CM10 |
| Dyract® eXtra | GrandioSO | 1634 | 90 | 10.618 | 2.827 | 0.001 | Q10 |
| Dyract® eXtra | GrandioSO | 1661 | 90 | 10.337 | 2.934 | 0.004 | Q10 |
| Dyract® eXtra | GrandioSO | 1843 | 90 | 17.881 | 5.723 | 0.018 | CM10 |
| Dyract® eXtra | GrandioSO | 2069 | 90 | 27.639 | 8.461 | 0.011 | CM10 |
| Dyract® eXtra | Estelite Σ Quick | 1634 | 90 | 9.45 | 2.264 | 0 | Q10 |
| Dyract® eXtra | Estelite Σ Quick | 1661 | 90 | 11.441 | 3.571 | 0.012 | Q10 |
| Dyract® eXtra | Estelite Σ Quick | 1678 | 90 | 8.661 | 3.067 | 0.043 | Q10 |
| Estelite Σ Quick | GrandioSO | 1634 | 120 | 8.445 | 1.689 | 0 | Q10 |
| Estelite Σ Quick | GrandioSO | 1843 | 120 | 11.532 | 3.79 | 0.019 | Q10 |
| Estelite Σ Quick | GrandioSO | 1843 | 120 | 6.762 | 1.683 | 0.001 | CM10 |
| Estelite Σ Quick | GrandioSO | 2069 | 120 | 17.889 | 5.271 | 0.006 | Q10 |
| Estelite Σ Quick | GrandioSO | 2260 | 120 | 15.317 | 5.142 | 0.023 | Q10 |
| Estelite Σ Quick | GrandioSO | 2481 | 120 | 20.661 | 6.425 | 0.01 | Q10 |
| Estelite Σ Quick | Dyract® eXtra | 1661 | 120 | 3.101 | 0.617 | 0 | Q10 |
| MW [Da] | Assinged Proteins | UniProt Registration |
|---|---|---|
| 1678 | 2′-5′ oligoadenylate synthetase 1 protein | C6EMZ7_HUMAN |
| Anaplastic lymphoma kinase | B2MXD8_HUMAN | |
| Himp | A0A140KRR5_HUMAN | |
| Protein FAM114A2 | E5RIK7_HUMAN | |
| 1703 | Uncharacterized protein | Q69YS1_HUMA |
| 1938 | Ax glycosyltransferase | G9HR99_HUMAN |
| NADH dehydrogenase subunit 1 | Q5Q8C5_HUMAN | |
| 2481 | Mitogen-activated protein kinase 10 | A0A1W2PNF5_HUMAN |
| 3364 | cAMP-regulated phosphoprotein 21 | C9J2U3_HUMAN |
| M3 muscarinic receptor | Q8NG01_HUMAN | |
| Mitogen-activated protein kinase 3 | J3QS54_HUMAN | |
| Retina-specific ABC transporter | Q86V62_HUMAN | |
| 3480 | ABL1 protein | Q86Y36_HUMAN |
| Cellular tumor antigen p53 | I3L0W9_HUMAN | |
| ETB1 protein | Q16261_HUMAN | |
| Glycophorin B | A0A3G2LR13_HUMAN | |
| A0A346RF30_HUMAN | ||
| Protein yippee-like 3 | H3BNP5_HUMAN | |
| 3708 | Tal-1 product | Q9UE36_HUMAN |
| Trimeric intracellular cation channel typ B | X6RGH1_HUMAN | |
| 4567 | Methyl-CpG-binding domain protein 3 | A0A087WVG6_HUMAN |
| STK4 protein | Q9BS84_HUMAN | |
| 5424 | Guanine nucleotide-binding protein G(i) subunit alpha-1 | A0A3B3IS42_HUMAN |
| Microcephalin | Q6RB59_HUMAN | |
| Zinc finger protein 385B | C9J0U3_HUMAN | |
| 2-hydroxy-3-oxopropionate reductase | A0A3D2YA96_PSESP | |
| 5784 | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2 | F8WAU0_HUMAN |
| Tryptophan--tRNA ligase, cytoplasmic | G3V2F2_HUMAN | |
| 6965 | DNA cytosine-5 methyltransferase 2 isoform D | A0A0U1SZ86_HUMAN |
| Putative deoxyribonuclease TATDN1 | E5RID7_HUMAN | |
| 10865 | Cytoskeleton-associated protein 2 | C9J7Y4_HUMAN |
| HCG1748409, isoform CRA_a | A0A024QZ00_HUMAN | |
| Mediator of RNA polymerase II transcription subunit 15 | C9JM58_HUMAN | |
| MHC class II antigen | A0A1X9I4S0_HUMANA0A6C0TJ11_HUMAN |
| Trade Name | Category | Content | Mean Particle Size (µm) | |
|---|---|---|---|---|
| Organic Matrix | Inorganic Matrix (Filler) | |||
| Venus® Diamond | nano-hybrid composite | TCD–DI–HEA, UDMA | barium–aluminium–fluoride–glass, nano–SiO2–particles | 0.005–20 |
| GrandioSO | nano-hybrid composite | Bis–GMA, Bis–EMA, TEGDMA | nano–SiO2–particles | 0.020–0.040 |
| glass ceramic | 1.0 | |||
| Estelite Σ Quick | sub-microfiller composite | Bis–GMA, TEGDMA | SiO2–ZrO2 | 0.2 |
| Dyract® eXtra | compomer | Bis–EDMA, UDMA, TEGDMA, TMPTMA, TCB | strontium–fluoride–glass | 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reise, M.; Kranz, S.; Heyder, M.; Beck, J.; Roth, C.; Guellmar, A.; von Eggeling, F.; Schubert, U.; Löffler, B.; Sigusch, B. Salivary Pellicle Formed on Dental Composites Evaluated by Mass Spectrometry—An In Situ Study. Molecules 2023, 28, 6804. https://doi.org/10.3390/molecules28196804
Reise M, Kranz S, Heyder M, Beck J, Roth C, Guellmar A, von Eggeling F, Schubert U, Löffler B, Sigusch B. Salivary Pellicle Formed on Dental Composites Evaluated by Mass Spectrometry—An In Situ Study. Molecules. 2023; 28(19):6804. https://doi.org/10.3390/molecules28196804
Chicago/Turabian StyleReise, Markus, Stefan Kranz, Markus Heyder, Julius Beck, Christian Roth, André Guellmar, Ferdinand von Eggeling, Ulrich Schubert, Bettina Löffler, and Bernd Sigusch. 2023. "Salivary Pellicle Formed on Dental Composites Evaluated by Mass Spectrometry—An In Situ Study" Molecules 28, no. 19: 6804. https://doi.org/10.3390/molecules28196804
APA StyleReise, M., Kranz, S., Heyder, M., Beck, J., Roth, C., Guellmar, A., von Eggeling, F., Schubert, U., Löffler, B., & Sigusch, B. (2023). Salivary Pellicle Formed on Dental Composites Evaluated by Mass Spectrometry—An In Situ Study. Molecules, 28(19), 6804. https://doi.org/10.3390/molecules28196804










