Interaction of Vanadium Complexes with Proteins: Revisiting the Reported Structures in the Protein Data Bank (PDB) since 2015
Abstract
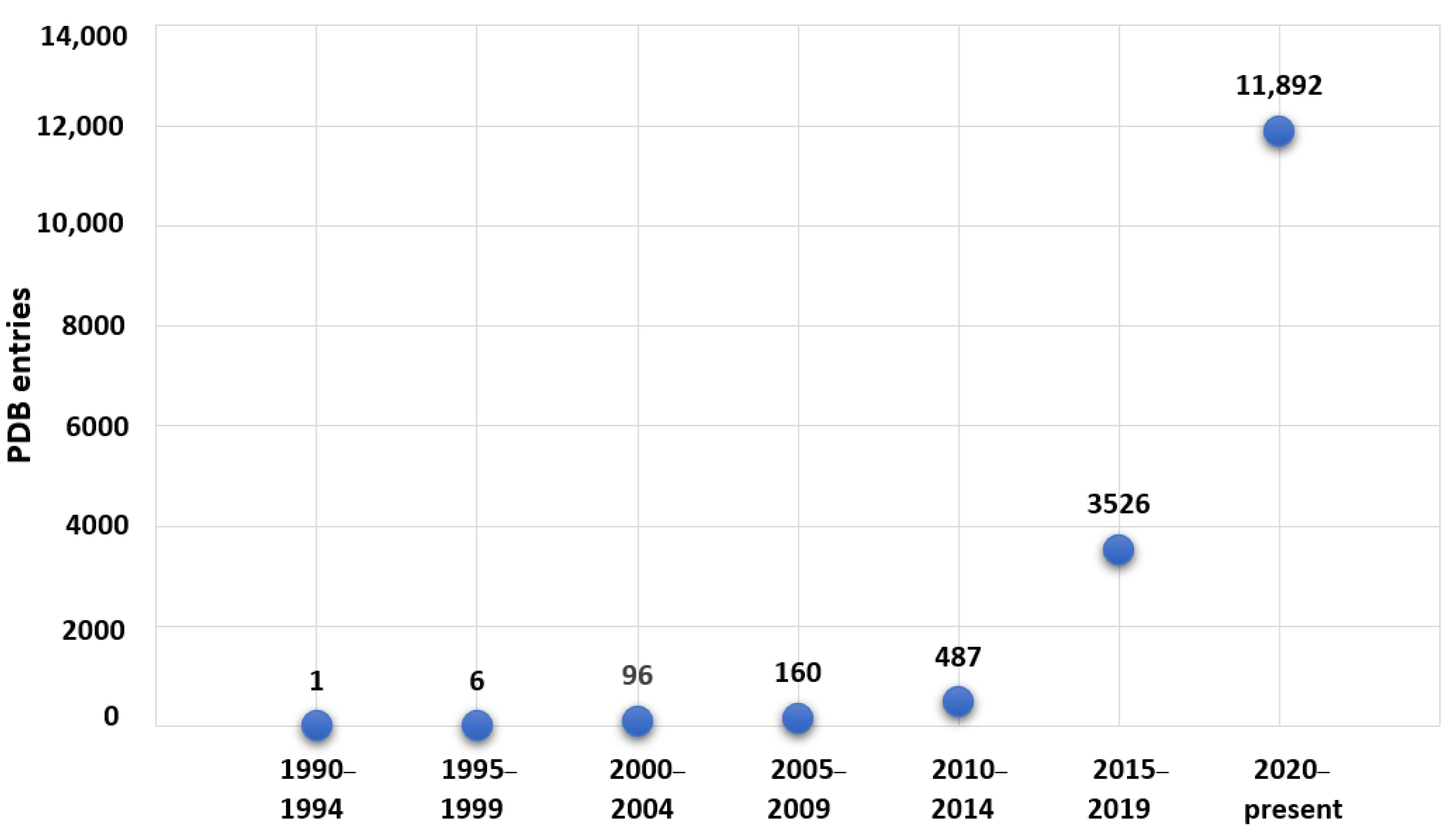
:1. Introduction
2. Vanadium-Containing Proteins and X-ray Crystallography
2.1. Inorganic-Based Vanadium Complexes
2.1.1. Monovanadates
2.1.2. Polyoxidovanadates
2.2. Organic-Based Vanadium Complexes
2.2.1. Nucleotides
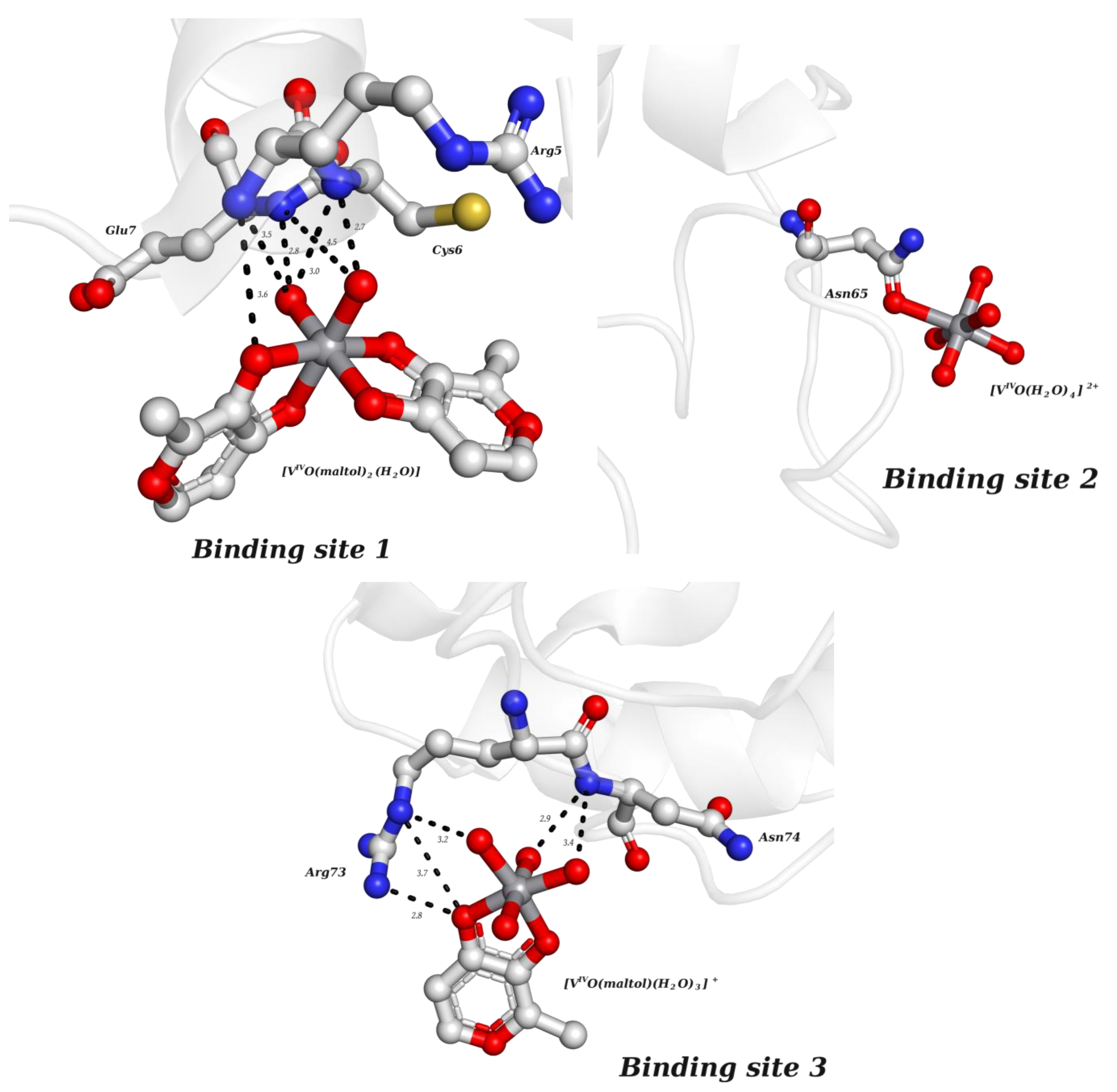
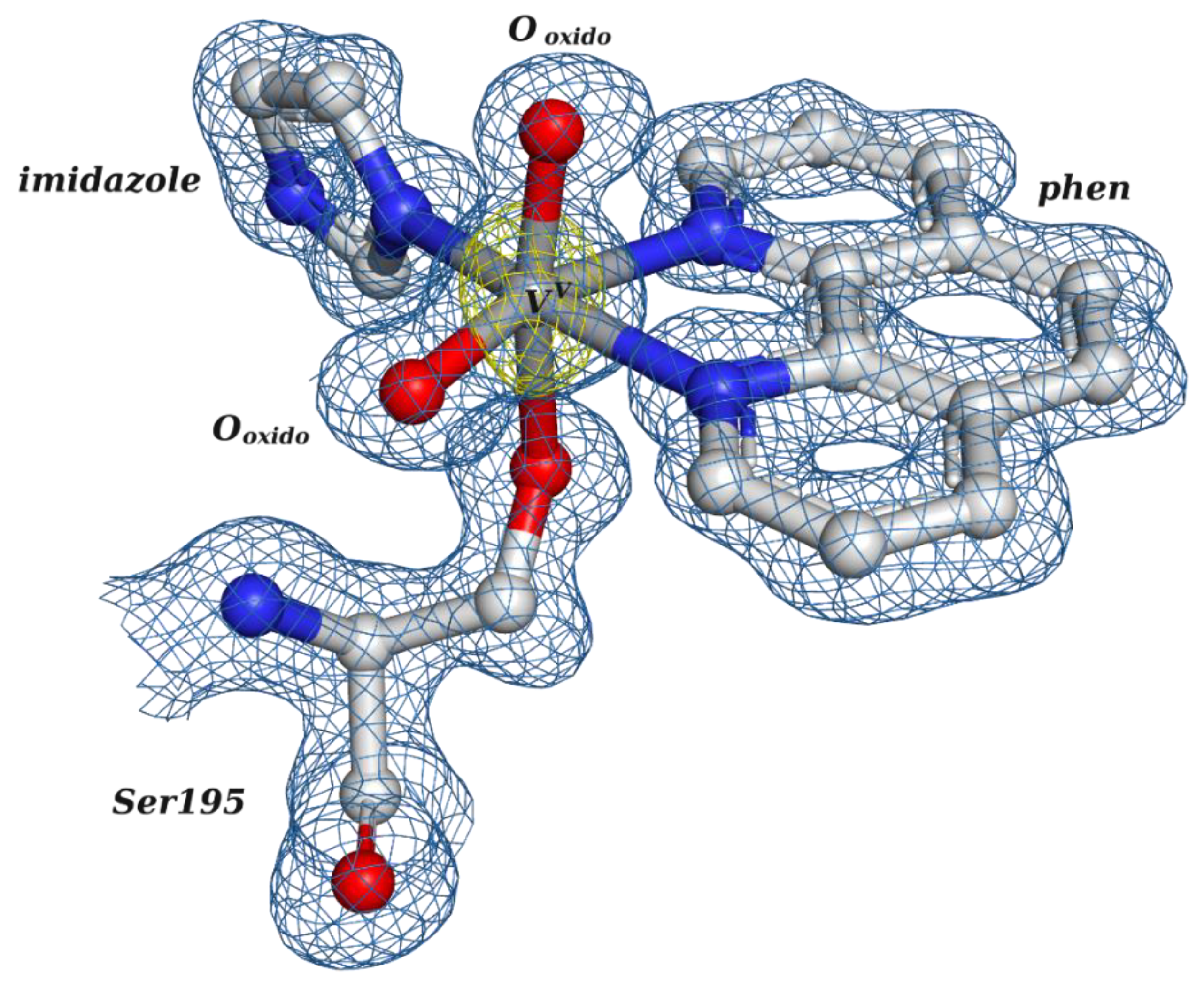
2.2.2. Putative Therapeutic V-Complexes and Model Proteins
Hen Egg White Lysozyme (HEWL)
Bovine Pancreatic Ribonuclease A (RNase A)
Bovine Trypsin
3. Vanadium-Containing Proteins and Cryo-EM
4. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Costa Pessoa, J.; Etcheverry, S.; Gambino, D. Vanadium compounds in medicine. Coord. Chem. Rev. 2015, 301–302, 24–48. [Google Scholar] [CrossRef] [PubMed]
- Rehder, D. Vanadium in biological systems and medicinal applications. Inorg. Chim. Acta 2023, 549, 121387. [Google Scholar] [CrossRef]
- Jakusch, T.; Kiss, T. In vitro study of the antidiabetic behavior of vanadium compounds. Coord. Chem. Rev. 2017, 351, 118–126. [Google Scholar] [CrossRef]
- Korbecki, J.; Baranowska-Bosiacka, I.; Gutowska, I.; Chlubek, D. Vanadium Compounds as Pro-Inflammatory Agents: Effects on Cyclooxygenases. Int. J. Mol. Sci. 2015, 16, 12648–12668. [Google Scholar] [CrossRef] [PubMed]
- Kioseoglou, E.; Petanidis, S.; Gabriel, C.; Salifoglou, A. The chemistry and biology of vanadium compounds in cancer therapeutics. Coord. Chem. Rev. 2015, 301–302, 87–105. [Google Scholar] [CrossRef]
- Scior, T.; Guevara-Garcia, J.A.; Do, Q.-T.; Bernard, P.; Laufer, S. Why Antidiabetic Vanadium Complexes are Not in the Pipeline of “Big Pharma” Drug Research? A Critical Review. Curr. Med. Chem. 2016, 23, 2874–2891. [Google Scholar] [CrossRef]
- Crans, D.C.; Smee, J.J.; Gaidamauskas, E.; Yang, L. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem. Rev. 2004, 104, 849–902. [Google Scholar] [CrossRef]
- Chasteen, N.D. Vanadium-protein interactions. In Metal Ions in Biological Systems; Sigel, H., Sigel, A., Eds.; Marcel Dekker Inc: New York, NY, USA, 1995; Volume 31, pp. 231–247. [Google Scholar]
- Korbecki, J.; Baranowska-Bosiacka, I.; Gutowska, I.; Chlubek, D. Biochemical and medical importance of vanadium compounds. Acta Chim. Pol. 2012, 59, 195–200. [Google Scholar] [CrossRef]
- Costa Pessoa, L.; Garribba, E.; Santos, M.F.A.; Santos-Silva, T. Vanadium and proteins: Uptake, transport, structure, activity and function. Coord. Chem. Rev. 2015, 301–302, 49–86. [Google Scholar] [CrossRef]
- Sciortino, G.; Garribba, E. The binding modes of VIVO2+ ions in blood proteins and enzymes. Chem. Commun. 2020, 56, 12218–12221. [Google Scholar] [CrossRef]
- Ugone, V.; Sanna, D.; Sciortino, G.; Maréchal, J.-D.; Garribba, E. Interaction of Vanadium(IV) Species with Ubiquitin: A Combined Instrumental and Computational Approach. Inorg. Chem. 2019, 58, 8064–8078. [Google Scholar] [CrossRef]
- Ugone, V.; Sanna, D.; Sciortino, G.; Crans, D.C.; Garribba, E. ESI-MS Study of the Interaction of Potential Oxidovanadium(IV) Drugs and Amavadin with Model Proteins. Inorg. Chem. 2020, 59, 9739–9755. [Google Scholar] [CrossRef]
- Ugone, V.; Sanna, D.; Ruggiu, S.; Sciortino, G.; Garribba, E. Covalent and non-covalent binding in vanadium–protein adducts. Inorg. Chem. Front. 2021, 8, 1189–1196. [Google Scholar] [CrossRef]
- Costa Pessoa, J.; Santos, M.F.A.; Correia, I.; Sanna, D.; Sciortino, G.; Garribba, E. Binding of vanadium ions and complexes to proteins and enzymes in aqueous solution. Coord. Chem. Rev. 2021, 449, 214192. [Google Scholar] [CrossRef]
- Santos, M.F.A.; Sciortino, G.; Correia, I.; Fernandes, A.C.P.; Santos-Silva, T.; Pisanu, F.; Garribba, E.; Costa Pessoa, J. Binding of VIVO2+, VIVOL+, VIVOL2 and VVO2L moieties to proteins: X-ray/theoretical characterization and biological implications. Chem. Eur. J. 2022, 28, e202200105. [Google Scholar] [CrossRef] [PubMed]
- Mehtab, S.; Gonçalves, G.; Roy, S.; Tomaz, A.I.; Santos-Silva, T.; Santos, M.F.A.; Romão, M.J.; Jakusch, T.; Kiss, T.; Costa Pessoa, J. Interaction of vanadium(IV) with human serum apo-transferrin. J. Inorg. Biochem. 2013, 121, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Sanna, D.; Micera, G.; Garribba, E. Interaction of VO2+ Ion and Some Insulin-Enhancing Compounds with Immunoglobulin G. Inorg. Chem. 2011, 50, 3717–3728. [Google Scholar] [CrossRef]
- Correia, I.; Jakusch, T.; Cobbinna, E.; Mehtab, S.; Tomaz, I.; Nagy, N.V.; Rockenbauer, A.; Costa Pessoa, J.; Kiss, T. Evaluation of the binding of oxovanadium(IV) to human serum albumin. Dalton Trans. 2012, 41, 6477–6487. [Google Scholar] [CrossRef]
- Sciortino, G.; Sanna, D.; Ugone, V.; Maréchal, J.-D.; Alemany-Chavarria, M.; Garribba, E. Effect of secondary interactions, steric hindrance and electric charge on the interaction of VIVO species with proteins. New J. Chem. 2019, 43, 17647–17660. [Google Scholar] [CrossRef]
- Sanna, D.; Serra, M.; Micera, G.; Garribba, E. Interaction of Antidiabetic Vanadium Compounds with Hemoglobin and Red Blood Cells and Their Distribution between Plasma and Erythrocytes. Inorg. Chem. 2014, 53, 1449–1464. [Google Scholar] [CrossRef]
- Jakusch, T.; Hollender, D.; Enyedy, É.A.; González, C.S.; Montes-Bayón, M.; Sanz-Medel, A.; Costa Pessoa, J.; Tomaz, I.; Kiss, T. Biospeciation of different antidiabetic VIVO compounds in serum. Dalton Trans. 2009, 13, 2428–2437. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Jakusch, T.; Hollender, D.; Dörnyei, A.; Enyedy, E.A.; Costa Pessoa, J.; Sakurai, H.; Sanz-Medel, A. Biospeciation of antidiabetic VO(IV) complexes. Coord. Chem. Rev. 2008, 252, 1153–1162. [Google Scholar] [CrossRef]
- Hu, Y.; Ribbe, M.W. Nitrogenases-A Tale of Carbon Atom(s). Angew. Chem. Int. Ed. 2016, 55, 8216–8226. [Google Scholar] [CrossRef]
- Sippel, D.; Schlesier, J.; Rohde, M.; Trncik, C.; Decamps, L.; Djurdjevic, I.; Spatzal, T.; Andrade, S.L.A.; Einsle, O. Production and isolation of vanadium nitrogenase from Azotobacter vinelandii by molybdenum depletion. J. Biol. Inorg. Chem. 2017, 22, 161–168. [Google Scholar] [CrossRef]
- Wever, R.; Barnett, P. Vanadium Chloroperoxidases: The Missing Link in the Formation of Chlorinated Compounds and Chloroform in the Terrestrial Environment? Chem. Asian J. 2017, 12, 1997–2007. [Google Scholar] [CrossRef]
- McLauchlan, C.C.; Murakami, H.A.; Wallace, C.A.; Crans, D.C. Coordination environment changes of the vanadium in vanadium-dependent haloperoxidase enzymes. J. Inorg. Biochem. 2018, 186, 267–279. [Google Scholar] [CrossRef]
- McLauchlan, C.C.; Peters, B.J.; Willsky, G.R.; Crans, D.C. Vanadium–phosphatase complexes: Phosphatase inhibitors favor the trigonal bipyramidal transition state geometries. Coord. Chem. Rev. 2015, 301–302, 163–199. [Google Scholar] [CrossRef]
- Leblanc, C.; Vilter, H.; Fournier, J.-B.; Delage, L.; Potin, P.; Rebuffet, E.; Michel, G.; Solari, P.L.; Feiters, M.C.; Czjzek, M. Vanadium haloperoxidases: From the discovery 30 years ago to X-ray crystallographic and V K-edge absorption spectroscopic studies. Coord. Chem. Rev. 2015, 301–302, 134–146. [Google Scholar] [CrossRef]
- Ueki, T.; Michibata, H. Molecular mechanism of the transport and reduction pathway of vanadium in ascidians. Coord. Chem. Rev. 2011, 255, 2249–2257. [Google Scholar] [CrossRef]
- Ueki, T.; Yamaguchi, N.; Romaidi; Isago, Y.; Tanahashi, H. Vanadium accumulation in ascidians: A system overview. Coord. Chem. Rev. 2015, 301–302, 300–308. [Google Scholar] [CrossRef]
- Akabayov, S.R.; Akabayov, B. Vanadate in structural biology. Inorg. Chim. Acta 2014, 420, 16–23. [Google Scholar] [CrossRef]
- Costa Pessoa, J.; Gonçalves, G.; Roy, S.; Correia, I.; Mehtab, S.; Santos, M.F.A.; Santos-Silva, T. New insights on vanadium binding to human serum transferrin. Inorg. Chim. Acta 2014, 420, 60–68. [Google Scholar] [CrossRef]
- Correia, I.; Chorna, I.; Cavaco, I.; Roy, S.; Kuznetsov, M.L.; Ribeiro, N.; Justino, G.; Marques, F.; Santos-Silva, T.; Santos, M.F.A.; et al. Interaction of [VIVO(acac)2] with Human Serum Transferrin and Albumin. Chem. Asian J. 2017, 12, 2062–2084. [Google Scholar] [CrossRef]
- Azevedo, C.G.; Correia, I.; Santos, M.M.C.; Santos, M.F.A.; Santos-Silva, J.; Doutch, J.; Fernandes, L.; Santos, H.M.; Capelo, J.L.; Costa Pessoa, J. Binding of vanadium to human serum transferrin—Voltammetric and spectrometric studies. J. Inorg. Biochem. 2018, 180, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography: Methods in Molecular Biology; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1607, pp. 627–641. [Google Scholar] [CrossRef]
- Velankar, S.; Burley, S.K.; Kurisu, G.; Hoch, J.C.; Markley, J.L. The Protein Data Bank Archive. In Structural Proteomics: Methods in Molecular Biology; Owens, R.J., Ed.; Humana Press: New York, NY, USA, 2021; Volume 2305, pp. 3–21. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 2022, 31, e4482. [Google Scholar] [CrossRef]
- Kearns, S. Structure of the Pandemic. Structure 2020, 28, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Bank, P.D. Crystallography: Protein Data Bank. Nat. New Biol. 1971, 233, 223. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank: A historical perspective. Acta Crystallogr. 2008, A64, 88–95. [Google Scholar] [CrossRef]
- Berman, H.M.; Gierasch, L.M. How the Protein Data Bank changed biology: An introduction to the JBC Reviews thematic series, part 1. J. Biol. Chem. 2021, 296, 100608. [Google Scholar] [CrossRef]
- Gierasch, L.M.; Berman, H.M. How the Protein Data Bank changed biology: An introduction to the JBC Reviews thematic series, part 2. J. Biol. Chem. 2021, 296, 100748. [Google Scholar] [CrossRef]
- Levina, A.; Crans, D.C.; Lay, P.A. Speciation of metal drugs, supplements and toxins in media and bodily fluids controls in vitro activities. Coord. Chem. Rev. 2017, 352, 473–498. [Google Scholar] [CrossRef]
- Levina, A.; Lay, P.A. Stabilities and Biological Activities of Vanadium Drugs: What is the Nature of the Active Species? Chem. Asian J. 2017, 12, 1692–1699. [Google Scholar] [CrossRef] [PubMed]
- Nunes, P.; Correia, I.; Cavaco, I.; Marques, F.; Pinheiro, T.; Avecilla, F.; Costa Pessoa, J. Therapeutic potential of vanadium complexes with 1,10-phenanthroline ligands, quo vadis? Fate of complexes in cell media and cancer cells. Inorg. Biochem. 2021, 217, 111350. [Google Scholar] [CrossRef] [PubMed]
- Sanna, D.; Ugone, V.; Micera, G.; Buglyó, P.; Bíró, L.; Garribba, E. Speciation in human blood of Metvan, a vanadium based potential anti-tumor drug. Dalton Trans. 2017, 46, 8950–8967. [Google Scholar] [CrossRef]
- Sanna, D.; Ugone, V.; Serra, M.; Garribba, E. Speciation of potential anti-diabetic vanadium complexes in real serum samples. J. Inorg. Biochem. 2017, 173, 52–65. [Google Scholar] [CrossRef]
- Nunes, P.; Correia, I.; Marques, F.; Matos, A.P.; Santos, M.M.C.; Azevedo, C.G.; Capelo, J.-L.; Santos, H.M.; Gama, S.; Pinheiro, T.; et al. Copper complexes with 1,10-phenanthroline derivatives: Underlying factors affecting their cytotoxicity. Inorg. Chem. 2020, 59, 9116–9134. [Google Scholar] [CrossRef]
- Thomas, J.M. The birth of X-ray crystallography. Nature 2012, 491, 186–187. [Google Scholar] [CrossRef]
- Helliwell, J.R. New developments in crystallography: Exploring its technology, methods and scope in the molecular biosciences. Biosci. Rep. 2017, 37, BSR20170204. [Google Scholar] [CrossRef]
- Rathore, I.; Mishra, V.; Bhaumik, P. Advancements in macromolecular crystallography: From past to present. Emerg. Top. Life Sci. 2021, 5, 127–149. [Google Scholar] [CrossRef]
- Kermani, A.A.; Aggarwal, S.; Ghanbarpour, A. Advances in X-ray crystallography methods to study structural dynamics of macromolecules. In Advanced Spectroscopic Methods to Study Biomolecular Structure and Dynamics; Saudagar, P., Tripathi, T., Eds.; Academic Press: London, UK, 2023; Volume 2305, pp. 309–355. [Google Scholar] [CrossRef]
- Cohen, A.E. A new era of synchrotron-enabled macromolecular crystallography. Nat. Methods 2021, 18, 433–434. [Google Scholar] [CrossRef]
- Rittle, J.; Field, M.J.; Green, M.T.; Tezcan, F.A. An efficient, step-economical strategy for the design of functional metalloproteins. Nat. Chem. 2019, 11, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-E.; Li, M.-Y.; Chou, C.-C.; Ho, M.-R.; Chen, G.-C.; Meng, T.-C.; Wang, A. H-J. Substrate Specificity and Plasticity of FERM-Containing Protein Tyrosine Phosphatases. Structure 2015, 23, 653–664. [Google Scholar] [CrossRef]
- Wulf, S.F.; Ropars, V.; Fujita-Becker, S.; Oster, M.; Hofhaus, G.; Trabuco, L.G.; Pylypenko, O.; Sweeney, H.L.; Houdusse, A.M.; Schröder, R.R. Force-producing ADP state of myosin bound to actin. Proc. Natl. Acad. Sci. USA 2016, 113, E1844–E1852. [Google Scholar] [CrossRef]
- Moise, G.; Gallup, N.M.; Alexandrova, A.N.; Hengge, A.C.; Johnson, S.J. Conservative Tryptophan Mutants of the Protein Tyrosine Phosphatase YopH Exhibit Impaired WPD-Loop Function and Crystallize with Divanadate Esters in Their Active Sites. Biochemistry 2015, 54, 6490–6500. [Google Scholar] [CrossRef]
- Lee, C.-U.; Hahne, G.; Hanske, J.; Bange, T.; Bier, D.; Rademacher, C.; Hennig, S.; Grossmann, T.N. Redox Modulation of PTEN Phosphatase Activity by Hydrogen Peroxide and Bisperoxidovanadium Complexes. Angew. Chem. Int. Ed. 2015, 54, 13796–13800. [Google Scholar] [CrossRef]
- Ropars, V.; Yang, Z.; Isabet, T.; Blanc, F.; Zhou, K.; Lin, T.; Liu, X.; Hissier, P.; Samazan, F.; Amigues, B.; et al. The myosin X motor is optimized for movement on actin bundles. Nat. Commun. 2016, 7, 12456. [Google Scholar] [CrossRef] [PubMed]
- Stanford, S.M.; Aleshin, A.E.; Zhang, V.; Ardecky, R.J.; Hedrick, M.P.; Zou, J.; Ganji, S.R.; Bliss, M.R.; Yamamoto, F.; Bobkov, A.A.; et al. Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase. Nat. Chem. Biol. 2017, 13, 624–632. [Google Scholar] [CrossRef]
- Planelles-Herrero, V.J.; Hartman, J.J.; Robert-Paganin, J.; Malik, F.I.; Houdusse, A. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 2017, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Bountra, K.; Hagelueken, G.; Choudhury, H.G.; Corradi, V.; El Omari, K.; Wagner, A.; Mathavan, I.; Zirah, S.; Wahlgren, W.Y.; Tieleman, D.P.; et al. Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 2017, 36, 3062–3079. [Google Scholar] [CrossRef]
- Moise, G.; Morales, Y.; Beaumont, V.; Caradonna, T.; Loria, J.P.; Johnson, S.J.; Hengge, A.C. YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases. Biochemistry 2018, 57, 5315–5326. [Google Scholar] [CrossRef]
- Robert-Paganin, J.; Robblee, J.P.; Auguin, D.; Blake, T.C.A.; Bookwalter, C.S.; Krementsova, E.B.; Moussaoui, D.; Previs, M.J.; Jousset, G.; Baum, J.; et al. Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun. 2019, 10, 3286. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-H.; Chang, C.-C.; Chuang, H.-C.; Tan, T.-H.; Lyu, P.-C. Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases. Int. J. Mol. Sci. 2020, 21, 7515.e14. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.; Salguero, A.L.; Liu, A.Z.; Chen, Z.; Dempsey, D.R.; Ficarro, S.B.; Alexander, W.M.; Marto, J.A.; Li, Y.; Amzel, L.M.; et al. Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell 2018, 174, 897–907.e14. [Google Scholar] [CrossRef]
- Cui, D.S.; Lipchock, J.M.; Brookner, D.; Loria, J.P. Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B. J. Am. Chem. Soc. 2019, 141, 12634–12647. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.X.; Chen, W.F.; Liu, N.N.; Teng, F.Y.; Guo, H.L.; Hou, X.M.; Dou, S.X.; Rety, S.; Xi, X.G. Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition. Nucleic Acids Res. 2021, 49, 4129–4143. [Google Scholar] [CrossRef]
- Shen, R.; Crean, R.M.; Olsen, K.J.; Corbella, M.; Calixto, A.R.; Richan, T.; Brandão, T.A.S.; Berry, R.D.; Tolman, A.; Loria, J.P.; et al. Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases. Chem. Sci. 2022, 13, 13524–13540. [Google Scholar] [CrossRef]
- Moussaoui, D.; Robblee, J.P.; Auguin, D.; Krementsova, E.B.; Haase, S.; Blake, T.C.A.; Baum, J.; Robert-Paganin, J.; Trybus, K.M.; Houdusse, A. Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets. eLife 2020, 9, e60581. [Google Scholar] [CrossRef]
- Gyimesi, M.; Horvath, A.I.; Turos, D.; Suthar, S.K.; Penzes, M.; Kurdi, C.; Canon, L.; Kikuti, C.; Ruppel, K.M.; Trivedi, D.V.; et al. Single Residue Variation in Skeletal Muscle Myosin Enables Direct and Selective Drug Targeting for Spasticity and Muscle Stiffness. Cell 2020, 183, 335–346.e13. [Google Scholar] [CrossRef]
- Shen, R.; Crean, R.M.; Johnson, S.J.; Kamerlin, S.C.L.; Hengge, A.C. Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases. JACS Au 2021, 1, 646–659. [Google Scholar] [CrossRef]
- Pinkston, J.; Jo, J.; Olsen, K.J.; Comer, D.; Glaittli, C.A.; Loria, J.P.; Johnson, S.J.; Hengge, A.C. Significant Loop Motions in the SsoPTP Protein Tyrosine Phosphatase Allow for Dual General Acid Functionality. Biochemistry 2021, 60, 2888–2901. [Google Scholar] [CrossRef]
- Ferraro, G.; Paolillo, M.; Sciortino, G.; Garribba, E.; Merlino, A. Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme. Inorg. Chem. 2022, 61, 16458–16467. [Google Scholar] [CrossRef]
- Hausmann, J.; Keune, W.J.; Hipgrave Ederveen, A.L.; van Zeijl, L.; Joosten, R.P.; Perrakis, A. Structural snapshots of the catalytic cycle of the phosphodiesterase Autotaxin. J. Struct. Biol. 2016, 195, 199–206. [Google Scholar] [CrossRef]
- Clausen, J.D.; Bublitz, M.; Arnou, B.; Olesen, C.; Andersen, J.P.; Moller, J.V.; Nissen, P. Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase. Structure 2016, 24, 617–623. [Google Scholar] [CrossRef]
- Bartlett, E.J.; Brissett, N.C.; Plocinski, P.; Carlberg, T.; Doherty, A.J. Molecular basis for DNA strand displacement by NHEJ repair polymerases. Nucleic Acids Res. 2016, 44, 2173–2186. [Google Scholar] [CrossRef] [PubMed]
- El Omari, K.; Mohamad, N.; Bountra, K.; Duman, R.; Romano, M.; Schlegel, K.; Kwong, H.-S.; Mykhaylyk, V.; Olesen, C.; Moller, J.V.; et al. Experimental phasing with vanadium and application to nucleotide-binding membrane proteins. IUCrJ 2020, 7, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.Y.; Lee, J.; Kim, H.; Ryu, H.; Shin, H.C.; Oh, B.H.; Ku, B.; Kim, S.J. Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1. PLoS ONE 2018, 13, e0197635. [Google Scholar] [CrossRef]
- Feder, D.; Gahan, L.R.; McGeary, R.P.; Guddat, L.W.; Schenk, G. The binding mode of an ADP analog to a metallohydrolase mimics the likely transition state. Chembiochem 2019, 20, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Chekan, J.R.; Ongpipattanakul, C.; Wright, T.R.; Zhang, B.; Bollinger, J.M., Jr.; Rajakovich, L.J.; Krebs, C.; Cicchillo, R.M.; Nair, S.K. Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants. Proc. Natl. Acad. Sci. USA 2019, 116, 13299–13304. [Google Scholar] [CrossRef]
- Mitchell, A.J.; Dunham, N.P.; Martinie, R.J.; Bergman, J.A.; Pollock, C.J.; Hu, K.; Allen, B.D.; Chang, W.C.; Silakov, A.; Bollinger, J.M.; et al. Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase. J. Am. Chem. Soc. 2017, 139, 13830–13836. [Google Scholar] [CrossRef] [PubMed]
- Dunham, N.P.; Chang, W.C.; Mitchell, A.J.; Martinie, R.J.; Zhang, B.; Bergman, J.A.; Rajakovich, L.J.; Wang, B.; Silakov, A.; Krebs, C.; et al. Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance. J. Am. Chem. Soc. 2018, 140, 7116–7126. [Google Scholar] [CrossRef]
- Davis, K.M.; Altmyer, M.; Martinie, R.J.; Schaperdoth, I.; Krebs, C.; Bollinger, J.M., Jr.; Boal, A.K. Structure of a Ferryl Mimic in the Archetypal Iron(II)- and 2-(Oxo)-glutarate-Dependent Dioxygenase, TauD. Biochemistry 2019, 58, 4218–4223. [Google Scholar] [CrossRef]
- Jonnalagadda, R.; Del Rio Flores, A.; Cai, W.; Mehmood, R.; Narayanamoorthy, M.; Ren, C.; Zaragoza, J.P.T.; Kulik, H.J.; Zhang, W.; Drennan, C.L. Biochemical and crystallographic investigations into isonitrile formation by a nonheme iron-dependent oxidase/decarboxylase. J. Biol. Chem. 2021, 296, 100231. [Google Scholar] [CrossRef]
- Sippel, D.; Rohde, M.; Netzer, J.; Trncik, C.; Gies, J.; Grunau, K.; Djurdjevic, I.; Decamps, L.; Andrade, S.L.A.; Einsle, O. A bound reaction intermediate sheds light on the mechanism of nitrogenase. Science 2018, 359, 1484–1489. [Google Scholar] [CrossRef]
- Rohde, M.; Grunau, K.; Einsle, O. CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution. Angew. Chem. Int. Ed. 2020, 59, 23626–23630. [Google Scholar] [CrossRef]
- Rohde, M.; Laun, K.; Zebger, I.; Stripp, S.T.; Einsle, O. Two ligand-binding sites in CO-reducing V nitrogenase reveal a general mechanistic principle. Sci. Adv. 2021, 7, eabg4474. [Google Scholar] [CrossRef] [PubMed]
- Sippel, D.; Einsle, O. The structure of vanadium nitrogenase reveals an unusual bridging ligand. Nat. Chem. Biol. 2017, 13, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Maccari, R.; Ottanà, R. Low molecular weight phosphotyrosine protein phosphatases as emerging targets for the design of novel therapeutic agents. J. Med. Chem. 2012, 55, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Muzio, L. The crucial of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Feng, B.; Dong, Y.; Shang, B.; Zhang, B.; Crans, D.C.; Yang, X. Convergent Protein Phosphatase Inhibitor Design for PTP1B and TCPTP: Exchangeable Vanadium Coordination Complexes on Graphene Quantum Dots. Adv. Funct. Mater. 2021, 32, 2108645. [Google Scholar] [CrossRef]
- Chiarugi, P.; Cirri, P.; Marra, F.; Raugei, G.; Camici, G.; Manao, G.; Ramponi, G. LMW-PTP is a Negative Regulator of Insulin-mediated Mitotic and Metabolic Signaling. Biochem. Biophys. Res. Commun. 1997, 238, 676–682. [Google Scholar] [CrossRef]
- Crans, D.C.; Zhang, B.; Gaidamauskas, E.; Keramidas, A.D.; Willsky, G.R.; Roberts, C.R. Is vanadate reduced by thiols under biological conditions? Changing the redox potential of V(V)/V(IV) by complexation in aqueous solution. Inorg. Chem. 2010, 49, 4245–4256. [Google Scholar] [CrossRef]
- Evans, P.; McCoy, A. An introduction to molecular replacement. Acta Crystallogr. 2008, D64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dodson, E. Introduction to molecular replacement: A time perspective. Acta Crystallogr. 2021, 77, 867–879. [Google Scholar] [CrossRef]
- Hendrickson, W.A. Anomalous diffraction in crystallographic phase evaluation. Q. Rev. Biophys. 2014, 47, 49–93. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.P.; Wang, B.-C. SAD phasing: History, current impact and future opportunities. Arch. Biochem. Biophys. 2016, 602, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.L. Introduction to phasing. Acta Crystallogr. 2010, 66, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Modis, Y. Exploiting subtle structural differences in heavy-atom derivatives for experimental phasing. Acta Crystallogr. 2014, 70, 1873–1883. [Google Scholar] [CrossRef] [PubMed]
- Dauter, M.; Dauter, Z. Many Ways to Derivatize Macromolecules and Their Crystals for Phasing. In Protein Crystallography. Methods in Molecular Biology; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1607, pp. 349–356. [Google Scholar] [CrossRef]
- Vilas Boas, L.F.; Costa Pessoa, J. Vanadium. In Comprehensive Coordination Chemistry; Wilkinson, G., Gillard, R.D., McCleverty, J.A., Eds.; Pergamon: Oxford, UK, 1987; Volume 3, pp. 453–583. [Google Scholar]
- Santos, M.F.A.; Correia, I.; Oliveira, A.R.; Garribba, E.; Costa Pessoa, J.; Santos-Silva, T. Vanadium Complexes as Prospective Therapeutics: Structural Characterization of a VIV Lysozyme Adduct. Eur. J. Inorg. Chem. 2014, 2014, 3293–3297. [Google Scholar] [CrossRef]
- Lee, C.C.; Hu, Y.; Ribbe, M.W. Tracing the Hydrogen Source of Hydrocarbons Formed by Vanadium Nitrogenase. Angew. Chem. Int. Ed. 2011, 50, 5545–5547. [Google Scholar] [CrossRef]
- Eady, R.R. Current status of structure function relationships of vanadium nitrogenase. Coord. Chem. Rev. 2003, 237, 23–30. [Google Scholar] [CrossRef]
- Aureliano, M. The Future Is Bright for Polyoxometalates. BioChem 2022, 2, 8–26. [Google Scholar] [CrossRef]
- Pereira, M.J.; Carvalho, E.; Eriksson, J.W.; Crans, D.C.; Aureliano, M. Effects of decavanadate and insulin enhancing vanadium compounds on glucose uptake in isolated rat adipocytes. J. Inorg. Biochem. 2009, 103, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Treviño, S.; Sánchez-Lara, E.; Sarmiento-Ortega, V.E.; Sánchez-Lombardo, I.; Flores-Hernández, J.Á.; Pérez-Benítez, A.; Brambila-Colombres, E.; González-Vergara, E. Hypoglycemic, lipid-lowering and metabolic regulation activities of metforminium decavanadate (H2Metf)3[V10O28]⋅8H2O using hypercaloric-induced carbohydrate and lipid deregulation in Wistar rats as biological model. J. Inorg. Biochem. 2015, 147, 85–92. [Google Scholar] [CrossRef]
- Aureliano, M.; Gumerova, N.I.; Sciortino, G.; Garribba, E.; Rompel, A.; Crans, D.C. Polyoxovanadates with emerging biomedical activities. Coord. Chem. Rev. 2021, 447, 214143. [Google Scholar] [CrossRef]
- Aureliano, M.; Gumerova, N.I.; Sciortino, G.; Garribba, E.; McLauchlan, C.C.; Rompel, A.; Crans, D.C. Polyoxidovanadates’ interactions with proteins: An overview. Coord. Chem. Rev. 2022, 454, 214344. [Google Scholar] [CrossRef]
- Locher, K.P.; Lee, A.T.; Rees, D.C. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science 2002, 296, 1091–1098. [Google Scholar] [CrossRef]
- Evans, H.R.; Holloway, D.E.; Sutton, J.M.; Ayriss, J.; Shone, C.C.; Acharya, K.R. C3 Exoenzyme from Clostridium botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure. Acta Crystallogr. 2004, D60, 1502–1505. [Google Scholar] [CrossRef]
- Pinto, P.H.; Kroupova, A.; Schleiffer, A.; Mechtler, K.; Jinek, M.; Weitzer, S.; Martinez, J. ANGEL2 is a member of the CCR4 family of deadenylases with 2′,3′-cyclic phosphatase activity. Science 2020, 369, 524–530. [Google Scholar] [CrossRef]
- Kim, Y.; Wower, J.; Maltseva, N.; Chang, C.; Jedrzejczak, R.; Wilamowski, M.; Kang, S.; Nicolaescu, V.; Randall, G.; Michalska, K.; et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Chinthalapudi, K.; Heissler, S.M.; Preller, M.; Sellers, J.R.; Manstein, D.J. Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C. eLife 2017, 6, e32742. [Google Scholar] [CrossRef] [PubMed]
- Feder, D.; McGeary, R.P.; Mitić, N.; Lonhienne, T.; Furtado, A.; Schulz, B.L.; Henry, R.J.; Schmidt, S.; Guddat, L.W.; Schenk, G. Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops. Plant Sci. 2020, 294, 110445. [Google Scholar] [CrossRef] [PubMed]
- Franz, P.; Ewert, W.; Preller, M.; Tsiavaliaris, G. Unraveling a Force-Generating Allosteric Pathway of Actomyosin Communication Associated with ADP and Pi Release. Int. J. Mol. Sci. 2021, 22, 104. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.; Golden, B.L. Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 2016, 55, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Teplova, M.; Falschlunger, C.; Krasheninina, O.; Egger, M.; Ren, A.; Patel, D.J.; Micura, R. Crucial Roles of Two Hydrated Mg2+ Ions in Reaction Catalysis of the Pistol Ribozyme. Angew. Chem. Int. Ed. 2020, 59, 2837–2843. [Google Scholar] [CrossRef]
- Elvingson, K.; Keramidas, A.D.; Crans, D.C.; Pettersson, L. Speciation in Vanadium Bioinorganic Systems. 5. Interactions between Vanadate, Uridine and Imidazole—An Aqueous Potentiometric, 51V, 17O and 13C NMR Study. Inorg. Chem. 1998, 37, 6153–6160. [Google Scholar] [CrossRef]
- Angus-Dunne, S.J.; Batchelor, R.J.; Tracey, A.S.; Einstein, F.W.B. The Crystal and Solution Structures of the Major Products of the Reaction of Vanadate with Adenosine. J. Am. Chem. Soc. 1995, 117, 5292–5296. [Google Scholar] [CrossRef]
- McNeill, J.H.; Yuen, V.G.; Dai, S.; Orvig, C. Increased potency of vanadium using organic ligands. Mol. Cell. Biochem. 1995, 153, 175–180. [Google Scholar] [CrossRef]
- Thompson, K.H.; Liboiron, B.D.; Sun, Y.; Bellman, K.D.D.; Setyawati, I.A.; Patrick, B.O.; Karunaratne, V.; Rawji, G.; Wheeler, J.; Sutton, K.; et al. Preparation and characterization of vanadyl complexes with bidentate maltol type ligands; in vivo comparisons of anti-diabetic therapeutic potential. J. Biol. Inorg. Chem. 2003, 8, 66–74. [Google Scholar] [CrossRef]
- Sakurai, H.; Yoshikawa, Y.; Yasui, H. Current state for the development of metallopharmaceutics and anti-diabetic metal complexes. Chem. Soc. Rev. 2008, 37, 2383–2392. [Google Scholar] [CrossRef]
- Sakurai, H.; Fugono, J.; Yasui, H. Pharmacokinetic Study and Trial for Preparation of Enteric-Coated Capsule Containing Insulinomimetic Vanadyl Compounds: Implications for Clinical Use. Mini-Rev. Med. Chem. 2004, 4, 41–48. [Google Scholar] [CrossRef]
- Rehder, D. Perspectives for vanadium in health issues. Future Med. Chem. 2016, 8, 325–338. [Google Scholar] [CrossRef]
- Selvaraj, S.; Krishnan, U.M. Vanadium-Flavonoid Complexes: A Promising Class of Molecules for Therapeutic Applications. J. Med. Chem. 2021, 64, 12435–12452. [Google Scholar] [CrossRef]
- Sharfalddin, A.A.; Al-Younis, I.M.; Mohammed, H.A.; Dhahri, M.; Mouffouk, F.; Ali, H.A.; Anwar, M.J.; Qureshi, K.A.; Hussien, M.A.; Alghrably, M.; et al. Therapeutic Properties of Vanadium Complexes. Inorganics 2022, 10, 244. [Google Scholar] [CrossRef]
- Jakusch, T.; Costa Pessoa, J.; Kiss, T. The Speciation of Vanadium in Human Serum. Coord. Chem. Rev. 2011, 255, 2218–2226. [Google Scholar] [CrossRef]
- Costa Pessoa, J.; Tomaz, I. Transport of Therapeutic Vanadium and Ruthenium Complexes by Blood Plasma Components. Curr. Med. Chem. 2010, 17, 3701–3738. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Demitri, N.; Vitale, L.; Sciortino, G.; Sanna, D.; Ugone, V.; Garribba, E.; Merlino, A. Spectroscopic/Computational Characterization and the X-ray Structure of the Adduct of the VIVO−Picolinato Complex with Rnase A. Inorg. Chem. 2021, 60, 19098–19109. [Google Scholar] [CrossRef]
- Ferraro, G.; Paolillo, M.; Sciortino, G.; Pisanu, F.; Garribba, E.; Merlino, A. Implications of Protein Interaction in the Speciation of Potential VIVO–Pyridinone Drugs. Inorg. Chem. 2023, 62, 8407–8417. [Google Scholar] [CrossRef]
- Ferraro, G.; Vitale, L.; Sciortino, G.; Pisanu, F.; Garribba, E.; Merlino, A. Interaction of VIVO-8-hydroxyquinoline species with RNase A: The effect of metal ligands in the protein adduct stabilization. Inorg. Chem. Front. 2023, 10, 5186–5198. [Google Scholar] [CrossRef]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef]
- Razavet, M.; Artero, V.; Cavazza, C.; Oudart, Y.; Lebrun, C.; Fontecilla-Camps, J.C.; Fontecave, M. Tricarbonylmanganese(I)–lysozyme complex: A structurally characterized organometallic protein. Chem. Commun. 2007, 27, 2805–2807. [Google Scholar] [CrossRef]
- Casini, A.; Mastrobuoni, G.; Temperini, C.; Gabbiani, C.; Francese, S.; Moneti, G.; Supuran, C.T.; Scozzafava, A.; Messori, L. ESI mass spectrometry and X-ray diffraction studies of adducts between anticancer platinum drugs and hen egg white lysozyme. Chem. Commun. 2007, 2, 156–158. [Google Scholar] [CrossRef]
- Binkley, S.L.; Ziegler, C.J.; Herrick, R.S.; Rowlett, R.S. Specific derivatization of lysozyme in aqueous solution with Re(CO)3(H2O)3+. Chem. Commun. 2010, 46, 1203–1205. [Google Scholar] [CrossRef]
- Santos, M.F.A.; Seixas, J.D.; Coelho, A.C.; Mukhopadhyay, A.; Reis, P.M.; Romão, M.J.; Romão, C.C.; Santos-Silva, T. New insights into the chemistry of fac-[Ru(CO)3]2+ fragments in biologically relevant conditions: The CO releasing activity of [Ru(CO)3Cl2(1,3-thiazole)], and the X-ray crystal structure of its adduct with lysozyme. J. Inorg. Biochem. 2012, 117, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Seixas, J.D.; Santos, M.F.A.; Mukhopadhyay, A.; Coelho, A.C.; Reis, P.M.; Veiros, L.F.; Marques, A.R.; Penacho, N.; Gonçalves, A.M.L.; Romão, M.J.; et al. A contribution to the rational design of Ru(CO)3Cl2L complexes for in vivo delivery of CO. Dalton Trans. 2015, 44, 5058–5075. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Giorgio, A.; Mansour, A.M.; Merlino, A. Protein-mediated disproportionation of Au(i): Insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme. Dalton Trans. 2019, 48, 14027–14035. [Google Scholar] [CrossRef]
- Gambino, D. Potentiality of vanadium compounds as anti-parasitic agents. Coord. Chem. Rev. 2011, 255, 2193–2203. [Google Scholar] [CrossRef]
- Benitez, J.; Becco, L.; Correia, I.; Leal, S.M.; Guiset, H.; Costa Pessoa, J.; Lorenzo, J.; Tanco, S.; Escobar, P.; Moreno, V.; et al. Vanadium polypyridyl compounds as potential antiparasitic and antitumoral agents: New achievements. J. Inorg. Biochem. 2011, 105, 303–312. [Google Scholar] [CrossRef]
- Thompson, K.H.; Orvig, C. Vanadium in diabetes: 100 years from Phase 0 to Phase I. J. Inorg. Biochem. 2006, 100, 1925–1935. [Google Scholar] [CrossRef]
- Costa Pessoa, J.; Marques, R.L.; Vilas Boas, L.F.; Gillard, R.D. Oxovanadium(IV) and Aminoacids—III. The system L-Aspartic Acid + VO2+; A Potentiometric and Spectroscopic Study. Polyhedron 1990, 9, 81–99. [Google Scholar] [CrossRef]
- Thompson, K.H.; Lichter, J.; LeBel, C.; Scaife, M.C.; McNeill, J.H.; Orvig, C. Vanadium treatment of type 2 diabetes: A view to the future. J. Inorg. Biochem. 2009, 103, 554–558. [Google Scholar] [CrossRef]
- Liboiron, B.D.; Thompson, K.H.; Hanson, G.R.; Lam, E.; Aebischer, N.; Orvig, C. New insights into the Interactions of Serum Proteins with Bis(maltolato)oxovanadium(IV): Transport and Biotransformation of Insulin-Enhancing Vanadium Pharmaceuticals. J. Am. Chem. Soc. 2005, 127, 5104–5115. [Google Scholar] [CrossRef] [PubMed]
- Rangel, M.; Tamura, A.; Fukushima, C.; Sakurai, H. In vitro study of the insulin-like action of vanadyl-pyrone and -pyridinone complexes with a VO(O4) coordination mode. J. Biol. Inorg. Chem. 2001, 6, 128–132. [Google Scholar] [CrossRef]
- Raines, R.T. Ribonuclease A. Chem. Rev. 1998, 98, 1045–1065. [Google Scholar] [CrossRef]
- Marshall, G.R.; Feng, J.A.; Kuster, D.J. Back to the Future: Ribonuclease A. Pept. Sci. 2008, 90, 259–277. [Google Scholar] [CrossRef]
- Cuchillo, C.M.; Nogués, M.V.; Raines, R.T. Bovine Pancreatic Ribonuclease: 50 Years of the First Enzymatic Reaction Mechanism. Biochemistry 2011, 50, 7835–7841. [Google Scholar] [CrossRef]
- Merlino, A. Recent advances in protein metalation: Structural studies. Chem. Commun. 2021, 57, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Merlino, A. Protein metalation by metal-based drugs: X-ray crystallography and mass spectrometry studies. Chem. Commun. 2017, 53, 11622–11633. [Google Scholar] [CrossRef]
- Kowacz, M.; Mukhopadhyay, A.; Carvalho, A.L.; Esperança, J.M.S.S.; Romão, M.J.; Rebelo, L.P.N. Hofmeister effects of ionic liquids in protein crystallization: Direct and water-mediated interactions. CrystEngComm 2012, 14, 4912–4921. [Google Scholar] [CrossRef]
- Costa Pessoa, J.; Antunes, J.L.; Vilas Boas, L.F.; Gillard, R.D. Oxovanadium(IV) and aminoacids—V. The System L-Glutamic Acid + VO2+; A Potentiometric and Spectroscopic Study. Polyhedron 1992, 11, 1449–1461. [Google Scholar] [CrossRef]
- Page, M.J.; Di Cera, E. Serine peptidases: Classification, structure and function. Cell. Mol. Life Sci. 2008, 65, 1220–1236. [Google Scholar] [CrossRef]
- Di Cera, E. Serine Proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Turk, V. Protease signaling: The cutting edge. EMBO J. 2012, 31, 1630–1643. [Google Scholar] [CrossRef]
- Bond, J.S. Proteases: History, discovery, and roles in health and disease. J. Biol. Chem. 2019, 294, 1643–1651. [Google Scholar] [CrossRef]
- Moulin, A.; Bell, J.H.; Pratt, R.F.; Ringe, D. Inhibition of Chymotrypsin by a Complex of Ortho-Vanadate and Benzohydroxamic Acid: Structure of the Inert Complex and Its Mechanistic Interpretation. Biochemistry 2007, 46, 5982–5990. [Google Scholar] [CrossRef] [PubMed]
- Costa Pessoa, J.; Vilas Boas, L.F.; Gillard, R.D. Oxovanadium(IV) and Aminoacids—II. The systems L-Serine and L-Threonine+VO2+. A Potentiometric and Spectroscopic Study. Polyhedron 1989, 8, 1173–1199. [Google Scholar] [CrossRef]
- Chiu, W.; Schmid, M.F.; Pintilie, G.D.; Lawson, C.L. Evolution of standardization and dissemination of cryo-EM structures and data jointly by the community, PDB, and EMDB. J. Biol. Chem. 2021, 296, 100560. [Google Scholar] [CrossRef] [PubMed]
- Guaita, M.; Watters, S.C.; Loerch, S. Recent advances and current trends in cryo-electron Microscopy. Curr. Opin. Struct. Biol. 2022, 77, 102484. [Google Scholar] [CrossRef]
- Chua, E.Y.D.; Mendez, J.H.; Rapp, M.; Ilca, S.L.; Tan, Y.Z.; Maruthi, K.; Kuang, H.; Zimanyi, C.M.; Chen, A.; Eng, E.T.; et al. Better, faster, cheaper: Recent advances in cryo–electron microscopy. Annu. Rev. Biochem. 2022, 91, 1–32. [Google Scholar] [CrossRef]
- Lees, J.A.; Dias, J.M.; Han, S. Applications of Cryo-EM in small molecule and biologics drug design. Biochem. Soc. Trans. 2021, 49, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.-F.; Yuan, C.; Du, Y.-M.; Sun, K.-L.; Zhang, X.-K.; Vogel, H.; Jia, X.-D.; Gao, Y.-Z.; Zhang, Q.-F.; Wang, D.-P.; et al. Applications and prospects of cryo-EM in drug discovery. Mil. Med. Res. 2023, 10, 10. [Google Scholar] [CrossRef]
- Fan, C.; Kaiser, J.T.; Rees, D.C. A structural framework for unidirectional transport by a bacterial ABC exporter. Proc. Natl. Acad. Sci. USA 2020, 117, 19228–19236. [Google Scholar] [CrossRef] [PubMed]
- Kehlenbeck, D.M.; Traore, D.A.K.; Josts, I.; Sander, S.; Moulin, M.; Haertlein, M.; Prevost, S.; Forsyth, V.T.; Tidow, H. Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination. FEBS J. 2022, 289, 2959–2970. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Q.; Chi, X.; Zhou, Q.; Li, Y. Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter. Cell Discov. 2020, 6, 86. [Google Scholar] [CrossRef]
- Cui, Z.; Li, X.; Shin, J.; Gamper, H.; Hou, Y.M.; Sacchettini, J.C.; Zhang, J. Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 432. [Google Scholar] [CrossRef]
- Fan, C.; Rees, D.C. Glutathione binding to the plant AtAtm3 transporter and implications for the conformational coupling of ABC transporters. eLife 2022, 11, e76140. [Google Scholar] [CrossRef]
- Borsellini, A.; Kunetsky, V.; Friedhoff, P.; Lamers, M.H. Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair. Nat. Struct. Mol. Biol. 2022, 29, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Sedzicki, J.; Ni, D.; Lehmann, F.; Wu, N.; Zenobi, R.; Jung, S.; Stahlberg, H.; Dehio, C. Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella. Nat. Struct. Mol. Biol. 2022, 29, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Silberberg, J.M.; Stock, C.; Hielkema, L.; Corey, R.A.; Rheinberger, J.; Wunnicke, D.; Dubach, V.R.A.; Stansfeld, P.J.; Hänelt, I.; Paulino, C. Inhibited KdpFABC transitions into an E1 off-cycle state. eLife 2022, 11, e80988. [Google Scholar] [CrossRef]
- Winkler, P.A.; Huang, Y.; Sun, W.; Du, J.; Lü, W. Electron cryo-microscopy structure of a human TRPM4 channel. Nature 2017, 552, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Orlando, B.J.; Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 2019, 567, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Januliene, D.; Mehdipour, A.R.; Thomas, C.; Stefan, E.; Brüchert, S.; Kuhn, B.T.; Geertsma, E.R.; Hummer, G.; Tampé, R.; et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 2019, 580, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhou, R.; Wan, L.; Feng, S.; Song, K.; Xu, C.; Li, Y.; Liao, M. Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat. Commun. 2021, 12, 4687. [Google Scholar] [CrossRef]
- Harris, A.; Wagner, M.; Du, D.; Raschka, S.; Nentwig, L.-M.; Gohlke, H.; Smits, S.H.J.; Luisi, B.F.; Schmitt, L. Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat. Commun. 2021, 12, 5254. [Google Scholar] [CrossRef]
- Lyu, J.; Liu, C.; Zhang, T.; Schrecke, S.; Elam, N.P.; Packianathan, C.; Hochberg, G.K.A.; Russell, D.; Zhao, M.; Laganowsky, A. Structural basis for lipid and copper regulation of the ABC transporter MsbA. Nat. Commun. 2022, 13, 7291. [Google Scholar] [CrossRef]
- Zhou, C.; Shi, H.; Zhang, M.; Zhou, L.; Xiao, L.; Feng, S.; Im, W.; Zhou, M.; Zhang, X.; Huang, Y. Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa. J. Mol. Biol. 2021, 433, 166986. [Google Scholar] [CrossRef]
- Aureliano, M.; André Ohlin, C. Decavanadate in vitro and in vivo effects: Facts and opinions. J. Inorg. Biochem. 2014, 137, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Samart, N.; Althumairy, D.; Zhang, D.; Roess, D.A.; Crans, D.C. Initiation of a novel mode of membrane signaling: Vanadium facilitated signal transduction. Coord. Chem. Rev. 2020, 416, 213286. [Google Scholar] [CrossRef]
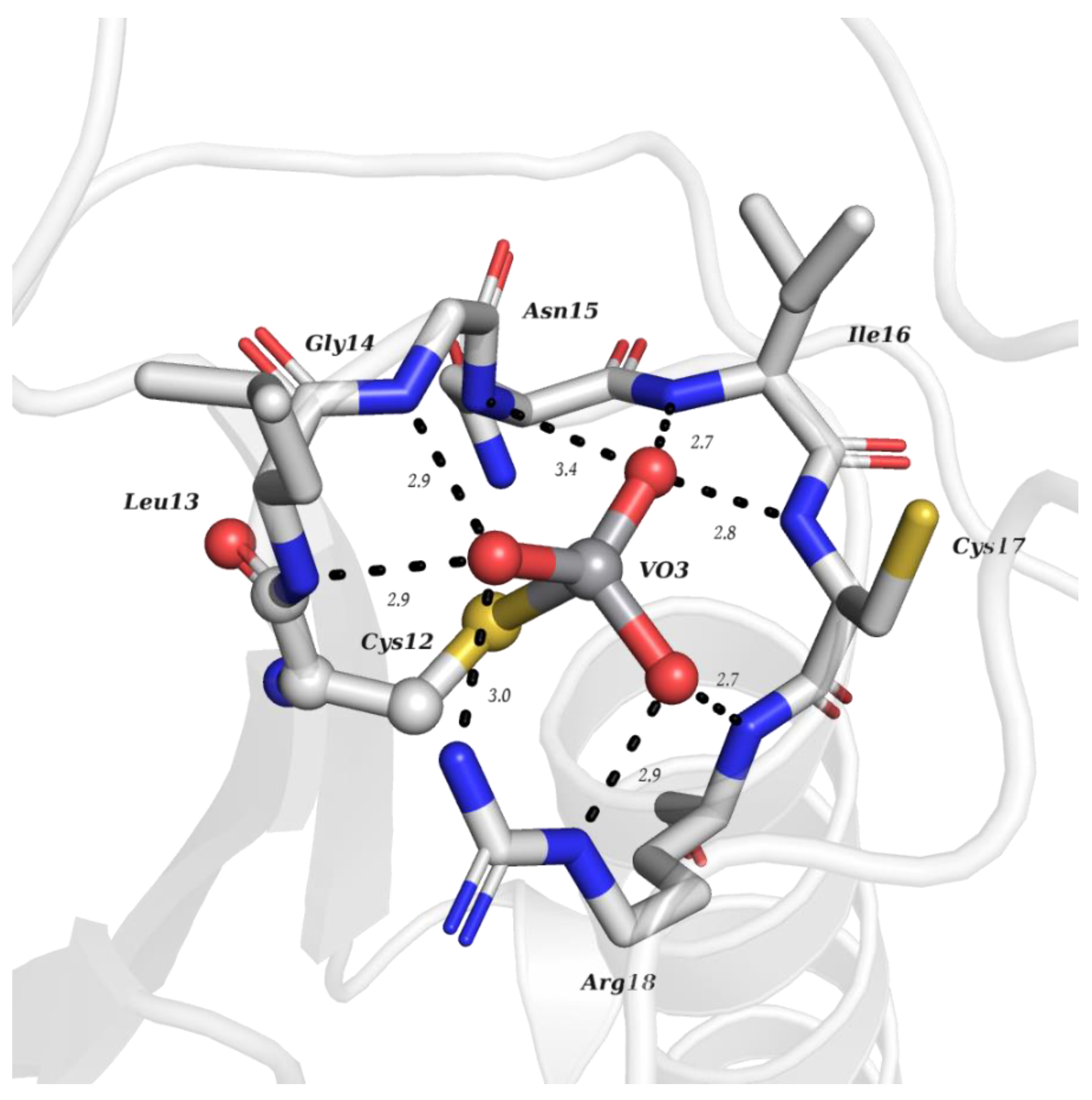
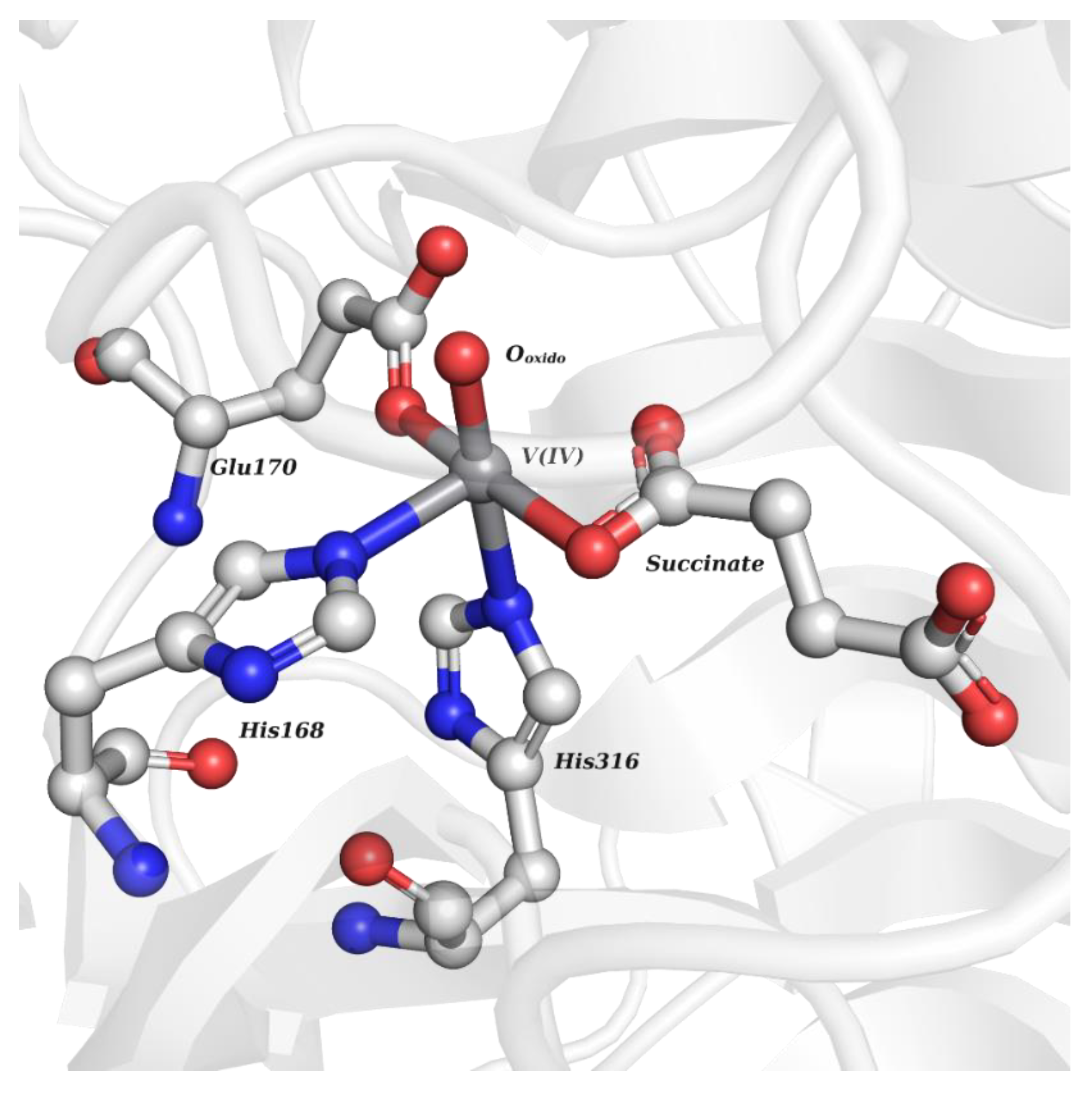
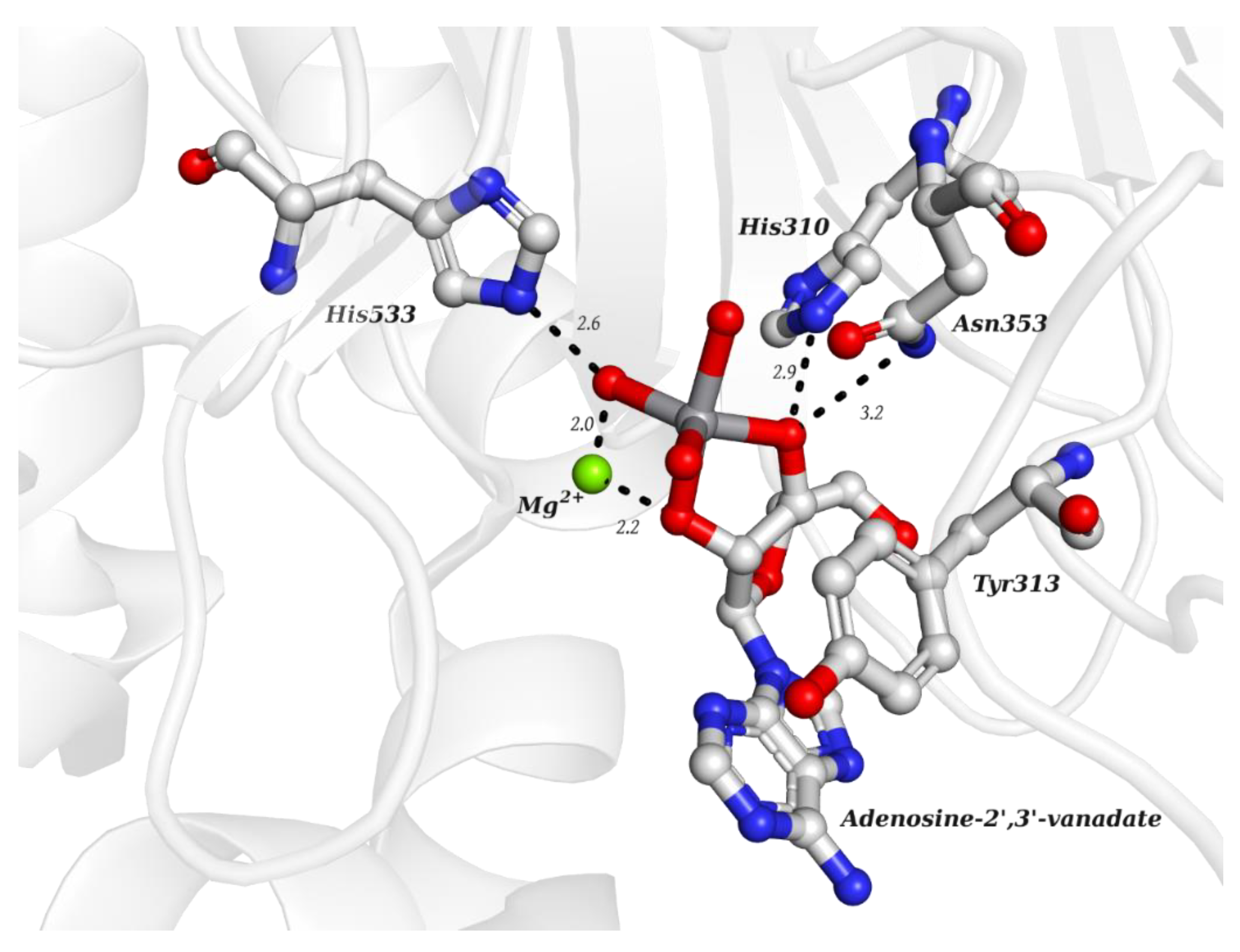
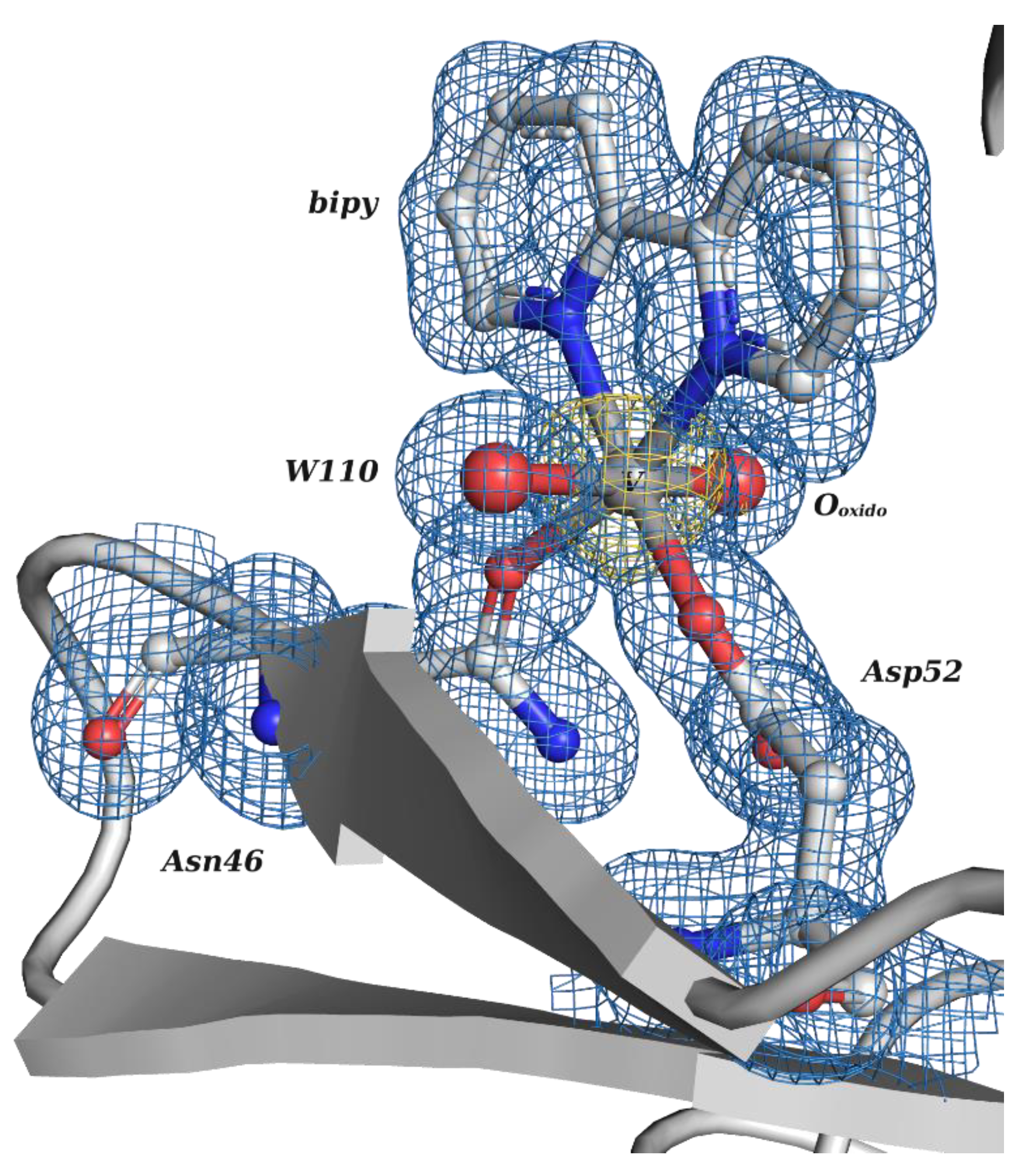
| V Species Identifier | V Species Name/Chemical Structure | PDB Codes |
|---|---|---|
| V | Vanadium ion | 6DYH [55], 6DYL [55], 7Q0T [16] |
| VO4 | Orthovanadate VVO43−  | 4RI4 [56], 4ZG4 [57], 4ZI4 [58], 4ZN5 [58], 5AA6 a, 5BZX [59], 5HMP [60], 5I0I [60], 5JNW [61], 5N69 [62], 5OFR [63], 6DR7 [64], 6DT6 [64], 6I7E [65], 6LVQ [66], 6NPZ [67], 6PG0 [68], 6PGT [68], 6PHA [68], 6PHS [68], 6PM8 [68], 6S3N [69], 6XEA [70], 6XEF [70], 6YCX [71], 6YSY [72], 7EZN a, 7L0H [73], 7L0M [73], 7MPC [74], 7QWI a, 8AJ5 [75] |
| 6BR | Threonine-vanadate VVC4H8NO7 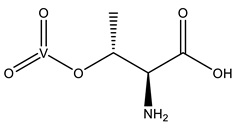 | 5IJS [76] |
| VN4 | Metavanadate VVO3−  | 4RI5 [56], 5A3Q [77], 5A3S [77], 5DMP [78], 6YSO [79] |
| VN3 | Metavanadate VVO3− 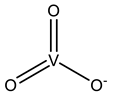 | 5Z5A [80], 6HWR [81] |
| VVO | Oxidovanadium(2+) VIVO2+  | 5BK9 [82], 6ALR [83], 6DB2 [84], 6EDH [85], 6VWQ a, 6VWR a, 6XPA [86], 6ZAK a, 8AJ5 [75] |
| VVB | Bis(oxidanyl)vanadium VII(OH)(OH) 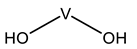 | 8AJ5 [75] |
| H1W | Pentakis(oxidanyl)vanadium VVH5O5 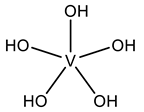 | 6HWR [81], 8AJ3 [75] |
| D6N | FeV-Center VCFe7NS7 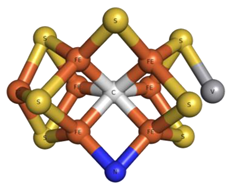 | 6FEA [87], 7ADR [88], 7ADY [88], 7AIZ [89] |
| 8P8 | FeV-Center VCFe7S8 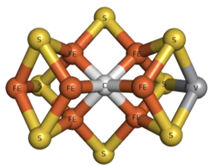 | 5N6Y [90] |
| V Species Identifier | V Species Name/Chemical Structure | PDB Codes |
|---|---|---|
| DVG | Divanadate Glycerol ester (DGV) V2C3H10O8 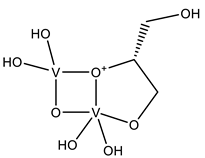 | 4ZI4 [58], 4ZN5 [58] |
| V4O | Cyclo-tetrametavanadate V4O12  | 7ZU6 a |
| VV6 | Hexavanadate V6H12O17  | 6HWR [81] |
| H1T | Heptavanadate V7O20  | 6HWR [81] |
| V Species Identifier | V Species Name/Chemical Structure | PDB Codes |
|---|---|---|
| KL2 | Adenosine-2′,3′-vanadate VC10H14N5O7 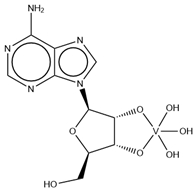 | 6RVZ [114] |
| H1Q | Adenosine divanadate V2C10H13N5O11  | 6HWR [81] |
| UVC | Uridine-2’,3’-vanadate VC9H12N2O9 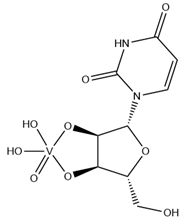 | 6YO1 [79], 7K1L [115] |
| AOV | ADP-vanadate VC10H17N5O14P2 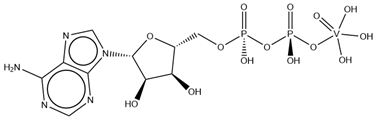 | 5I4E [116] |
| AD9 | ADP Metavanadate VC10H16N5O13P2 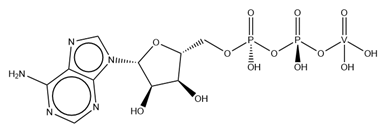 | 6PY9 [117], 6Z2S a, 7B19 [118], 7B1A [118] |
| CVC | Cytidine-5′-monophosphate-2′,3′-vanadate VC9H14N3O10P 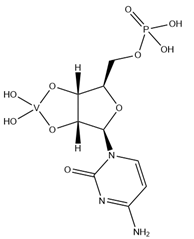 | 5EAO [119], 5EAQ [119] |
| Q61 | Guanosine-5′-monophosphate-2′,3′-vanadate VC10H12N5O10P 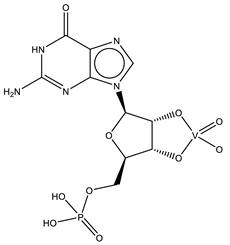 | 6UEY [120], 6UF1 [120] |
| Organic Molecule Name/Chemical Structure | PDB Codes |
|---|---|
Picolinato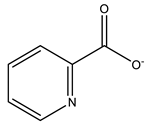 | 7P8R [132], 7Q0X [16] |
Maltol 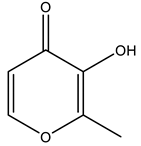 | 8AJ3 [75], 8AJ4 [75] |
2,2′-bipyridine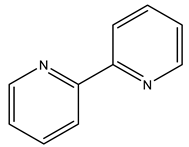 | 7Q0U [16] |
1,10-phenanthroline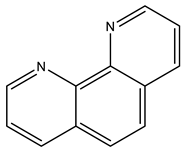 | 7Q0V [16], 7Q0W [16] |
1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone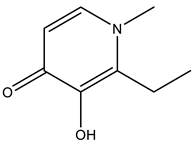 | 8OM8 [133], 8OMS [133], 8OMT [133] |
8-hydroxyquinolinato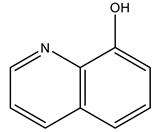 | 7QWH [134] |
| V Species Identifier | V Species Name/Chemical Structure | PDB Codes |
|---|---|---|
| VO4 | Orthovanadate VO43− 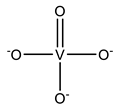 | 6VQT [167], 7BCW [168], 7D0A [169], 7MSM [170], 7MSZ [170], 7N5A [171], 7N5B [171], 7OU0 [172], 7ZNU [173], 7ZO9 [173], 7ZRD [174] |
| DVT | Decavanadate V10O28 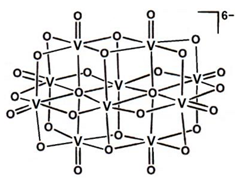 | 5WP6 [175] |
| AOV | ADP Orthovanadate VC10H17N5O14P2 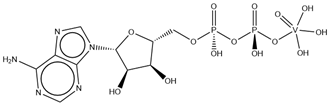 | 6MHZ [176], 6MI8 [176], 6RAJ [177], 6RAK [177], 7MDY [178], 7P06 [179], 8DMM [180] |
| AD9 | ADP Metavanadate VC10H16N5O13P2 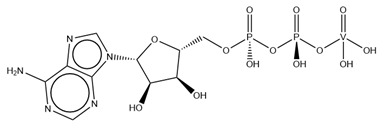 | 7CH8 [181] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.F.A.; Pessoa, J.C. Interaction of Vanadium Complexes with Proteins: Revisiting the Reported Structures in the Protein Data Bank (PDB) since 2015. Molecules 2023, 28, 6538. https://doi.org/10.3390/molecules28186538
Santos MFA, Pessoa JC. Interaction of Vanadium Complexes with Proteins: Revisiting the Reported Structures in the Protein Data Bank (PDB) since 2015. Molecules. 2023; 28(18):6538. https://doi.org/10.3390/molecules28186538
Chicago/Turabian StyleSantos, Marino F. A., and João Costa Pessoa. 2023. "Interaction of Vanadium Complexes with Proteins: Revisiting the Reported Structures in the Protein Data Bank (PDB) since 2015" Molecules 28, no. 18: 6538. https://doi.org/10.3390/molecules28186538
APA StyleSantos, M. F. A., & Pessoa, J. C. (2023). Interaction of Vanadium Complexes with Proteins: Revisiting the Reported Structures in the Protein Data Bank (PDB) since 2015. Molecules, 28(18), 6538. https://doi.org/10.3390/molecules28186538






