Response of Osteoblasts on Amine-Based Nanocoatings Correlates with the Amino Group Density
Abstract
:1. Introduction
2. Results
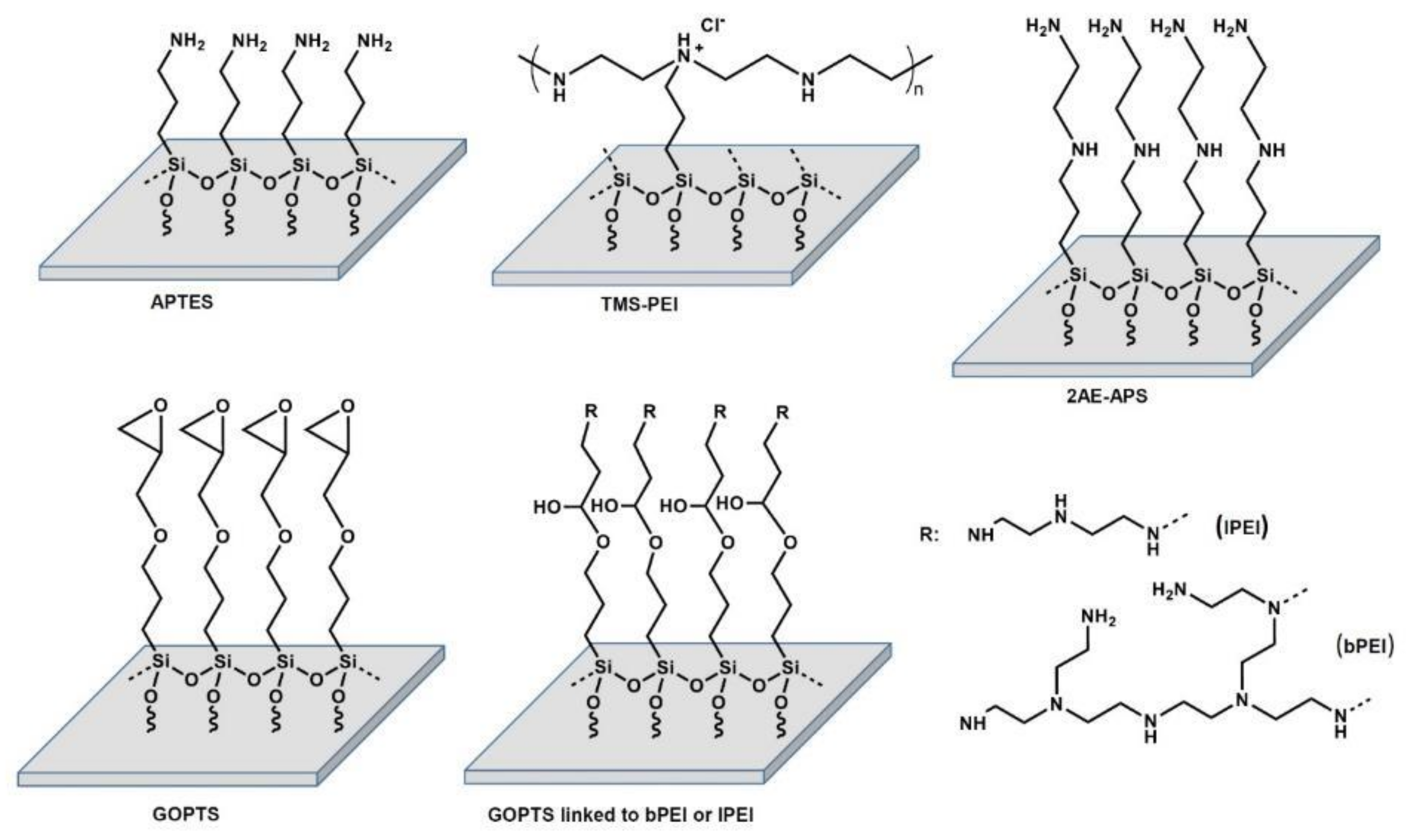
2.1. Chemical Characterization of Amine-Based Polymer Coatings
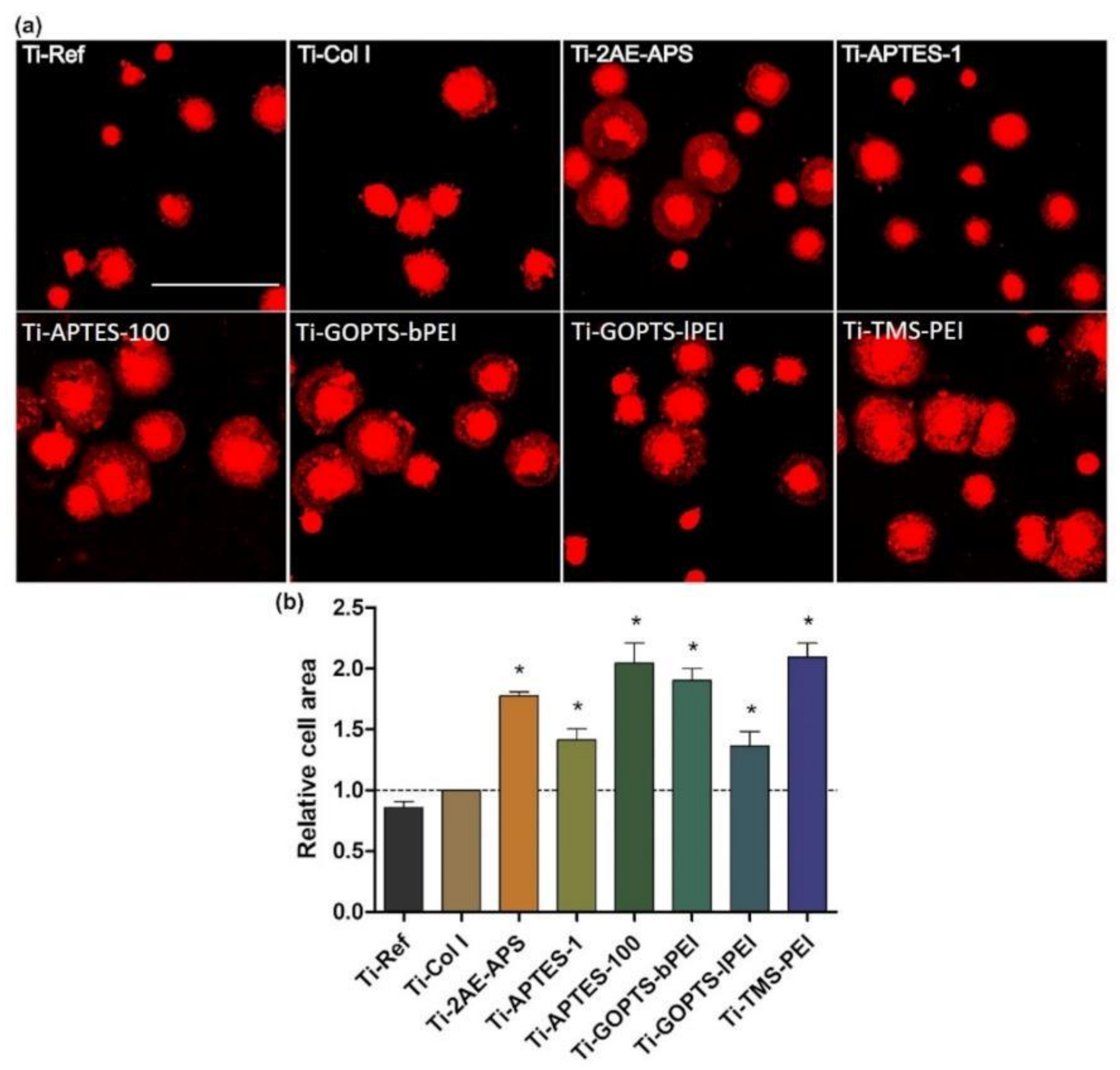
2.2. N-Dependent Cell Spreading
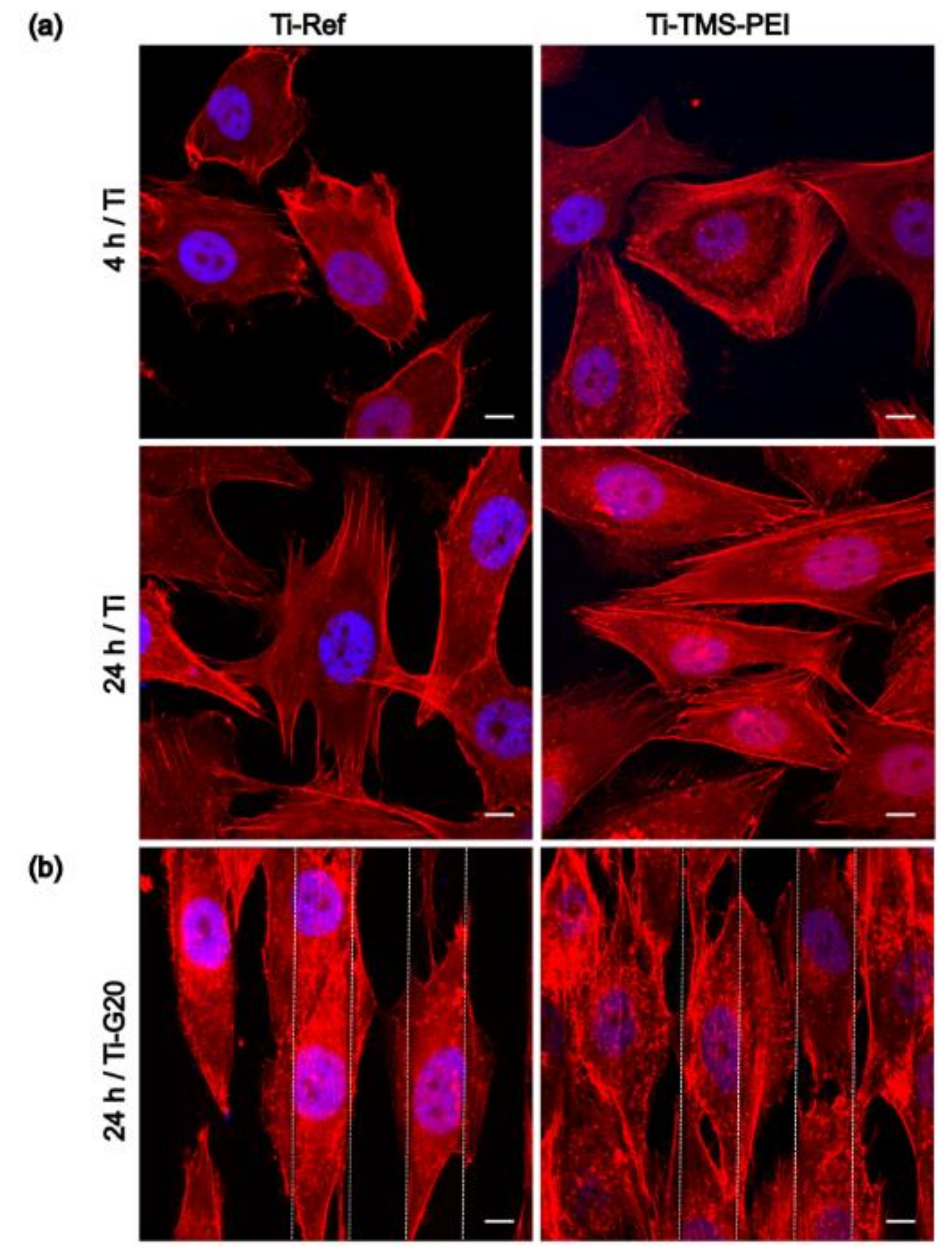
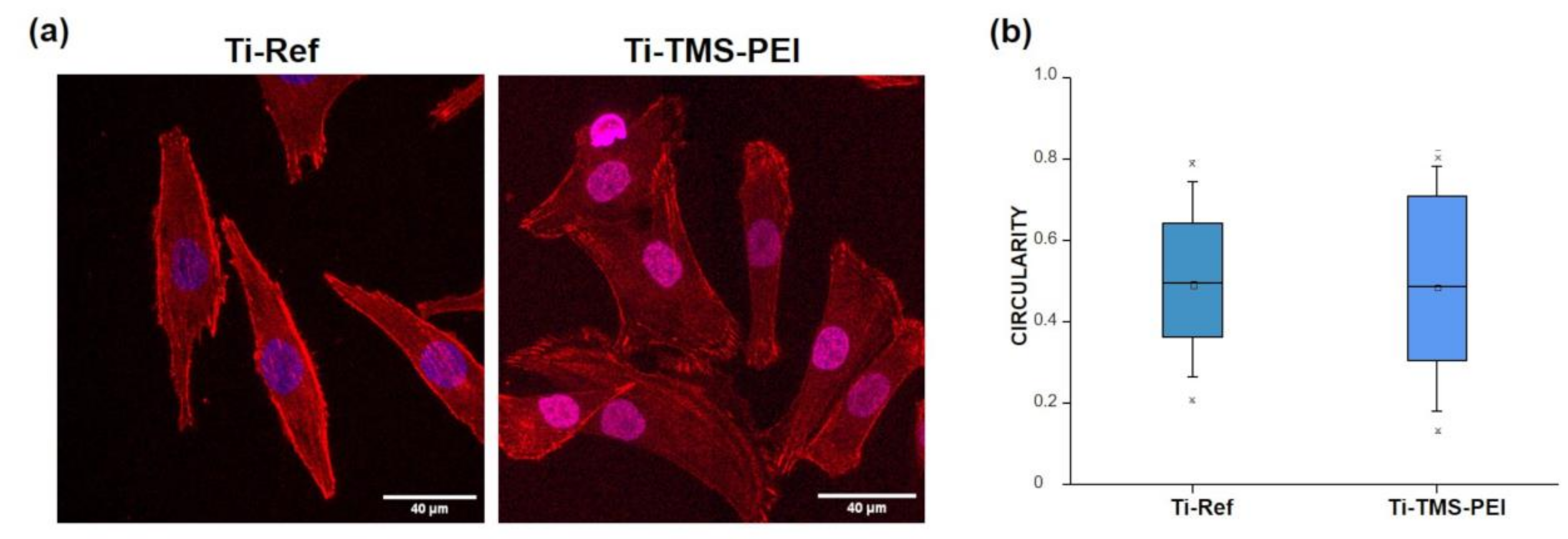
2.3. Cell Morphology on Ti-TMS-PEI
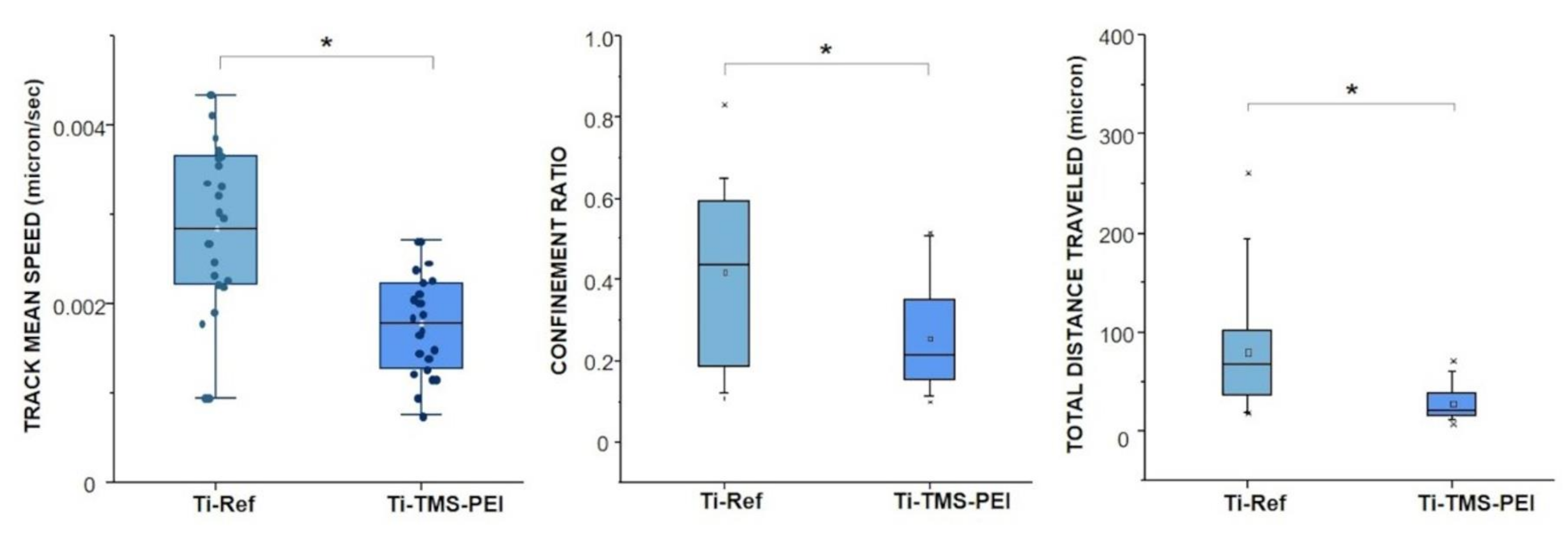
2.4. Migration Capacity of MG-63 Cells on Ti-TMS-PEI
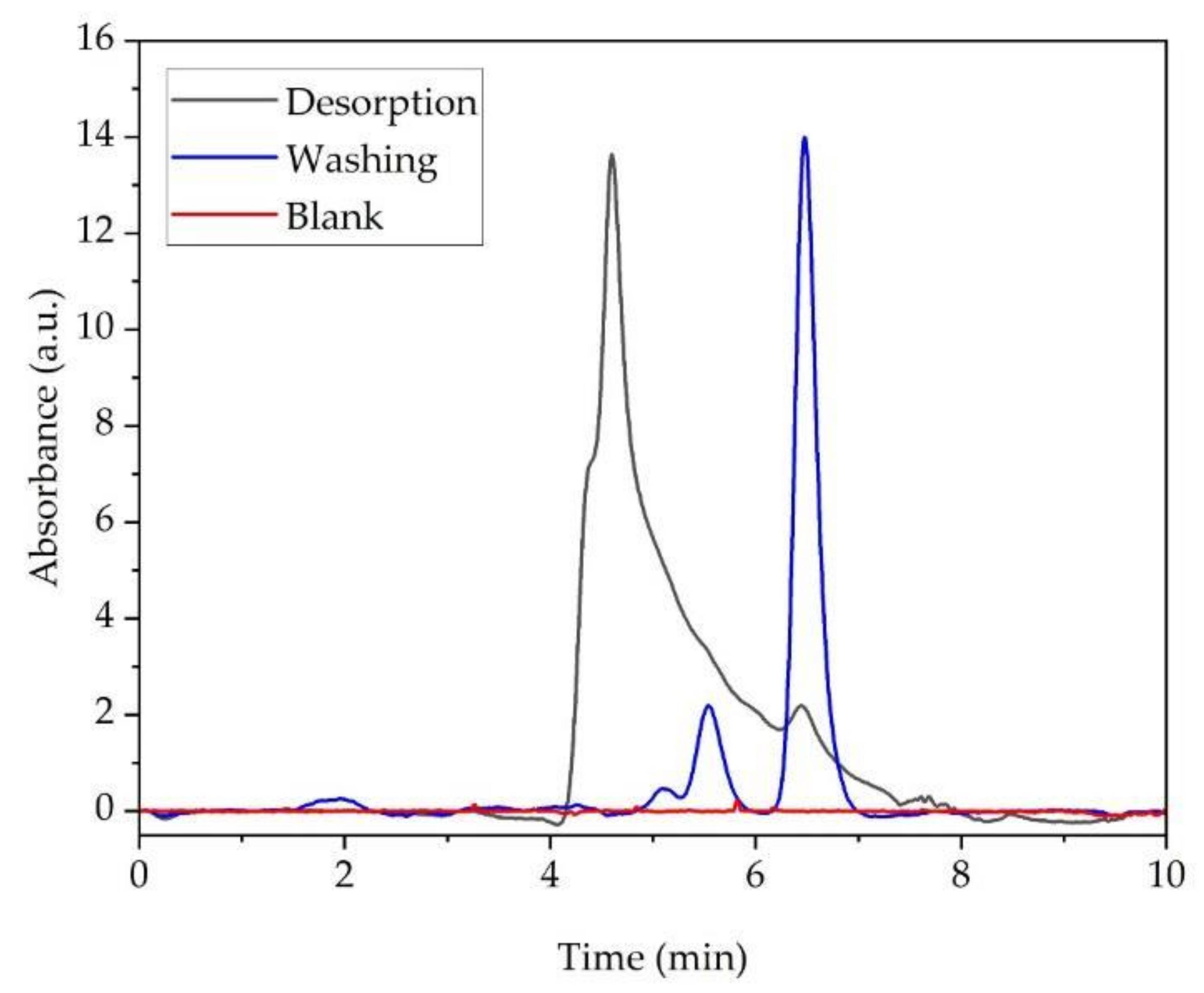
2.5. Bovine Serum Albumin (BSA) Adsorption and Desorption on Ti-TMS-PEI
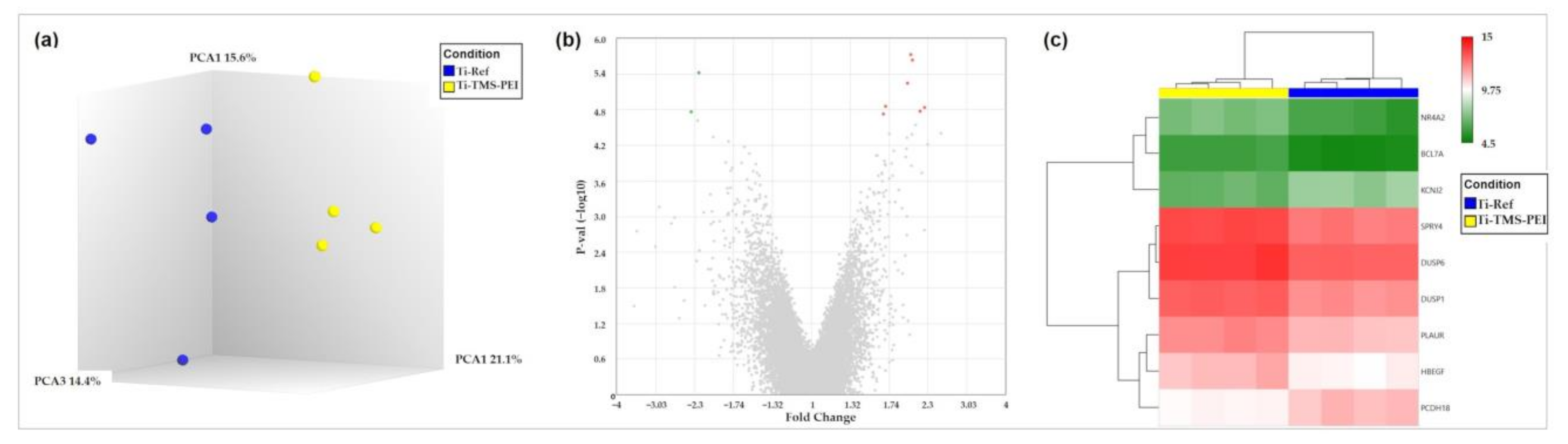
2.6. Gene Expression of MG-63 Cells on Amine-Based Polymer Coating Ti-TMS-PEI
2.6.1. Premature Activation of Erk1/2 Pathway after 4 h of MG-63 on Ti-TMS-PEI
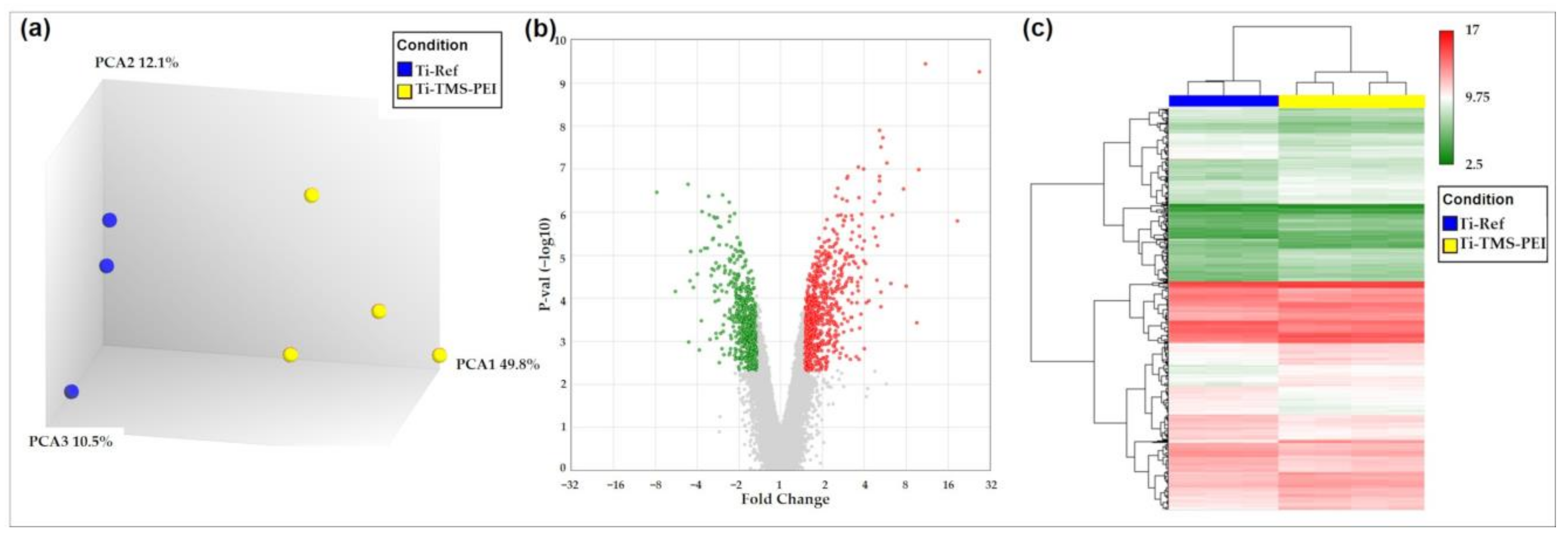
2.6.2. Premature Activation of Proliferation Pathways after 24 h Cultivation of MG-63s on Ti-TMS-PEI
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Titanium Arrays
5.2. Coating with Collagen Type I (Col I)
5.3. Amine-Based Nanocoatings
5.3.1. Cleaning and Activation Procedure
5.3.2. Trimethoxysilylpropyl Modified Poly(ethylenimine) (TMS-PEI)
5.3.3. Aminopropyltriethoxysilane (APTES)
5.3.4. (3-Glycidyloxypropyl)trimethoxysilane (GOPTS)
5.3.5. Linear and Branched (Poly(ethyleneimine)) (lPEI, bPEI)
5.3.6. N-(2-Aminoethyl)-3-Aminopropyltrimethoxysilane (2AE-APS)
5.4. Surface Characterization
5.4.1. Wettability
5.4.2. Zeta Potential
5.4.3. X-ray Photoelectron Spectroscopy (XPS)
5.5. Cell Culture
5.6. Cell Area Determination
5.7. Actin Cytoskeleton Staining
5.8. Scanning Electron Microscopy (SEM)
5.9. Gene Expression Profiling
5.10. Live Cell Imaging
5.11. Protein Adsorption
5.12. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gibon, E.; Lu, L.Y.; Nathan, K.; Goodman, S.B. Inflammation, ageing, and bone regeneration. J. Orthop. Translat. 2017, 10, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Mörke, C.; Rebl, H.; Finke, B.; Dubs, M.; Nestler, P.; Airoudj, A.; Roucoules, V.; Schnabelrauch, M.; Körtge, A.; Anselme, K.; et al. Abrogated Cell Contact Guidance on Amino-Functionalized Microgrooves. ACS Appl. Mater. Interfaces 2017, 9, 10461–10471. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Ke, D.; Sahasrabudhe, H.; Bandyopadhyay, A. Additive manufacturing of biomaterials. Prog. Mater. Sci. 2018, 93, 45–111. [Google Scholar] [CrossRef] [PubMed]
- Kunz, F.; Rebl, H.; Quade, A.; Matschegewski, C.; Finke, B.; Nebe, J.B. Osteoblasts with impaired spreading capacity benefit from the positive charges of plasma polymerised allylamine. Eur. Cell Mater. 2015, 29, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Albrektsson, T.; Chrcanovic, B.; Östman, P.O.; Sennerby, L. Initial and long-term crestal bone responses to modern dental implants. Periodontol. 2000 2017, 73, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Reyes, C.D.; Petrie, T.A.; Burns, K.L.; Schwartz, Z.; García, A.J. Biomolecular surface coating to enhance orthopaedic tissue healing and integration. Biomaterials 2007, 28, 3228–3235. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; García, A.J. Biomaterial strategies for engineering implants for enhanced osseointegration and bone repair. Adv. Drug Deliv. Rev. 2015, 94, 53–62. [Google Scholar] [CrossRef]
- Chen, S.; Guo, Y.; Lu, R.; Wu, S.; Fang, J.; Huang, B.; Li, Z.; Chen, Z.; Chen, Z. Tuning surface properties of bone biomaterials to manipulate osteoblastic cell adhesion and the signaling pathways for the enhancement of early osseointegration. Colloids Surf. B Biointerfaces 2018, 164, 58–69. [Google Scholar] [CrossRef]
- Finke, B.; Luethen, F.; Schroeder, K.; Mueller, P.D.; Bergemann, C.; Frant, M.; Ohl, A.; Nebe, J.B. The effect of positively charged plasma polymerization on initial osteoblastic focal adhesion on titanium surfaces. Biomaterials 2007, 28, 4521–4534. [Google Scholar] [CrossRef]
- Anselme, K.; Ponche, A.; Bigerelle, M. Relative influence of surface topography and surface chemistry on cell response to bone implant materials. Part 2: Biological aspects. Proc. Inst. Mech. Eng. H 2010, 224, 1487–1507. [Google Scholar] [CrossRef]
- Nebe, J.B.; Rebl, H.; Schlosser, M.; Staehlke, S.; Gruening, M.; Weltmann, K.D.; Walschus, U.; Finke, B. Plasma Polymerized Allylamine-The Unique Cell-Attractive Nanolayer for Dental Implant Materials. Polymers 2019, 11, 1004. [Google Scholar] [CrossRef] [PubMed]
- Rebl, H.; Finke, B.; Lange, R.; Weltmann, K.D.; Nebe, J.B. Impact of plasma chemistry versus titanium surface topography on osteoblast orientation. Acta Biomater. 2012, 8, 3840–3851. [Google Scholar] [CrossRef]
- Liu, X.; Feng, Q.; Bachhuka, A.; Vasilev, K. Surface modification by allylamine plasma polymerization promotes osteogenic differentiation of human adipose-derived stem cells. ACS Appl. Mater. Interfaces 2014, 6, 9733–9741. [Google Scholar] [CrossRef] [PubMed]
- Gruening, M.; Neuber, S.; Nestler, P.; Lehnfeld, J.; Dubs, M.; Fricke, K.; Schnabelrauch, M.; Helm, C.A.; Müller, R.; Staehlke, S.; et al. Enhancement of Intracellular Calcium Ion Mobilization by Moderately but Not Highly Positive Material Surface Charges. Front. Bioeng. Biotechnol. 2020, 8, 1016. [Google Scholar] [CrossRef]
- Cohen, M.; Joester, D.; Geiger, B.; Addadi, L. Spatial and temporal sequence of events in cell adhesion: From molecular recognition to focal adhesion assembly. Chembiochem 2004, 5, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Kao, W.L.; Chang, H.Y.; Lin, K.Y.; Lee, Y.W.; Shyue, J.J. Effect of Surface Potential on the Adhesion Behavior of NIH3T3 Cells Revealed by Quartz Crystal Microbalance with Dissipation Monitoring (QCM-D). J. Phys. Chem. C 2017, 121, 533–541. [Google Scholar] [CrossRef]
- Gabler, C.; Zietz, C.; Göhler, R.; Fritsche, A.; Lindner, T.; Haenle, M.; Finke, B.; Meichsner, J.; Lenz, S.; Frerich, B.; et al. Evaluation of osseointegration of titanium alloyed implants modified by plasma polymerization. Int. J. Mol. Sci. 2014, 15, 2454–2464. [Google Scholar] [CrossRef] [PubMed]
- Nebe, B.; Lüthen, F.; Finke, B.; Bergemann, C.; Schröder, K.; Rychly, J.; Liefeith, K.; Ohl, A. Improved initial osteoblast’s functions on amino-functionalized titanium surfaces. Biomol. Eng. 2007, 24, 447–454. [Google Scholar] [CrossRef]
- Arima, Y.; Iwata, H. Preferential adsorption of cell adhesive proteins from complex media on self-assembled monolayers and its effect on subsequent cell adhesion. Acta Biomater. 2015, 26, 72–81. [Google Scholar] [CrossRef]
- Lin, M.; Wang, H.; Ruan, C.; Xing, J.; Wang, J.; Li, Y.; Wang, Y.; Luo, Y. Adsorption force of fibronectin on various surface chemistries and its vital role in osteoblast adhesion. Biomacromolecules 2015, 16, 973–984. [Google Scholar] [CrossRef]
- Phan, H.T.; Bartelt-Hunt, S.; Rodenhausen, K.B.; Schubert, M.; Bartz, J.C. Investigation of Bovine Serum Albumin (BSA) Attachment onto Self-Assembled Monolayers (SAMs) Using Combinatorial Quartz Crystal Microbalance with Dissipation (QCM-D) and Spectroscopic Ellipsometry (SE). PLoS ONE 2015, 10, e0141282. [Google Scholar] [CrossRef] [PubMed]
- Schvartz, M.; Saudrais, F.; Devineau, S.; Aude, J.C.; Chédin, S.; Henry, C.; Millán-Oropeza, A.; Perrault, T.; Pieri, L.; Pin, S.; et al. A proteome scale study reveals how plastic surfaces and agitation promote protein aggregation. Sci. Rep. 2023, 13, 1227. [Google Scholar] [CrossRef] [PubMed]
- Borzova, V.A.; Markossian, K.A.; Chebotareva, N.A.; Kleymenov, S.Y.; Poliansky, N.B.; Muranov, K.O.; Stein-Margolina, V.A.; Shubin, V.V.; Markov, D.I.; Kurganov, B.I. Kinetics of Thermal Denaturation and Aggregation of Bovine Serum Albumin. PLoS ONE 2016, 11, e0153495. [Google Scholar] [CrossRef] [PubMed]
- Arima, Y.; Iwata, H. Effects of surface functional groups on protein adsorption and subsequent cell adhesion using self-assembled monolayers. J. Mater. Chem. 2007, 17, 4079–4087. [Google Scholar] [CrossRef]
- Toledano, M.; Carrasco-Carmona, Á.; Medina-Castillo, A.L.; Toledano-Osorio, M.; Osorio, R. Protein adsorption and bioactivity of functionalized electrospun membranes for bone regeneration. J. Dent. 2020, 102, 103473. [Google Scholar] [CrossRef] [PubMed]
- Vassaux, M.; Pieuchot, L.; Anselme, K.; Bigerelle, M.; Milan, J.L. A biophysical model for curvature-guided cell migration. Biophys. J. 2019, 117, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Devreotes, P.; Janetopoulos, C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J. Biol. Chem. 2003, 278, 20445–20448. [Google Scholar] [CrossRef]
- Faucheux, N.; Tzoneva, R.; Nagel, M.D.; Groth, T. The dependence of fibrillar adhesions in human fibroblasts on substratum chemistry. Biomaterials 2006, 27, 234–245. [Google Scholar] [CrossRef]
- Linke, P.; Suzuki, R.; Yamamoto, A.; Nakahata, M.; Kengaku, M.; Fujiwara, T.; Ohzono, T.; Tanaka, M. Dynamic contact guidance of myoblasts by feature size and reversible switching of substrate topography: Orchestration of cell shape, orientation, and nematic ordering of actin cytoskeletons. Langmuir 2019, 35, 7538–7551. [Google Scholar] [CrossRef]
- Pieuchot, L.; Marteau, J.; Guignandon, A.; Dos Santos, T.; Brigaud, I.; Chauvy, P.F.; Cloatre, T.; Ponche, A.; Petithory, T.; Rougerie, P.; et al. Curvotaxis directs cell migration through cell-scale curvature landscapes. Nat. Commun. 2018, 9, 3995. [Google Scholar] [CrossRef]
- Böhmler, J.; Ploux, L.; Ball, V.; Anselme, K.; Ponche, A. Necessity of a Thorough Characterization of Functionalized Silicon Wafers before Biointerface Studies. J. Phys. Chem. C 2011, 115, 11102–11111. [Google Scholar] [CrossRef]
- Chen, Z.; Lv, Z.; Sun, Y.; Chi, Z.; Qing, G. Recent advancements in polyethyleneimine-based materials and their biomedical, biotechnology, and biomaterial applications. J. Mater. Chem. B 2020, 8, 2951–2973. [Google Scholar] [CrossRef] [PubMed]
- McConnell, K.I.; Shamsudeen, S.; Meraz, I.M.; Mahadevan, T.S.; Ziemys, A.; Rees, P.; Summers, H.D.; Serda, R.E. Reduced Cationic Nanoparticle Cytotoxicity Based on Serum Masking of Surface Potential. J. Biomed. Nanotechnol. 2016, 12, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, T.; Neshati, Z. Role of microRNAs in osteogenesis of stem cells. J. Cell Biochem. 2019, 120, 14136–14155. [Google Scholar] [CrossRef] [PubMed]
- Suthon, S.; Perkins, R.S.; Bryja, V.; Miranda-Carboni, G.A.; Krum, S.A. WNT5B in Physiology and Disease. Front. Cell Dev. Biol. 2021, 9, 667581. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Tao, Y.; Lu, J.; Jones, J.R.; Fowler, L.; Weick, J.P.; Zhang, S.C. Time-Course Gene Expression Profiling Reveals a Novel Role of Non-Canonical WNT Signaling During Neural Induction. Sci. Rep. 2016, 6, 32600. [Google Scholar] [CrossRef] [PubMed]
- Dupree, M.A.; Pollack, S.R.; Levine, E.M.; Laurencin, C.T. Fibroblast growth factor 2 induced proliferation in osteoblasts and bone marrow stromal cells: A whole cell model. Biophys. J. 2006, 91, 3097–3112. [Google Scholar] [CrossRef] [PubMed]
- Agas, D.; Sabbieti, M.G.; Marchetti, L.; Xiao, L.; Hurley, M.M. FGF-2 enhances Runx-2/Smads nuclear localization in BMP-2 canonical signaling in osteoblasts. J. Cell Physiol. 2013, 228, 2149–2158. [Google Scholar] [CrossRef]
- Han, D.; Wang, M.; Yu, Z.; Yin, L.; Liu, C.; Wang, J.; Liu, Y.; Jiang, S.; Ren, Z.; Yin, J. FGF5 promotes osteosarcoma cells proliferation via activating MAPK signaling pathway. Cancer Manag. Res. 2019, 11, 6457–6466. [Google Scholar] [CrossRef]
- Sharff, K.A.; Song, W.X.; Luo, X.; Tang, N.; Luo, J.; Chen, J.; Bi, Y.; He, B.C.; Huang, J.; Li, X.; et al. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J. Biol. Chem. 2009, 284, 649–659. [Google Scholar] [CrossRef]
- Vlashi, R.; Zhang, X.; Wu, M.; Chen, G. Wnt signaling: Essential roles in osteoblast differentiation, bone metabolism and therapeutic implications for bone and skeletal disorders. Genes Dis. 2023, 10, 1291–1317. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, K.; Bakiri, L.; Wolff, L.I.; Linder, M.; Mikels-Vigdal, A.; Patiño-García, A.; Lecanda, F.; Hartmann, C.; Sibilia, M.; Wagner, E.F. Wnt signaling and Loxl2 promote aggressive osteosarcoma. Cell Res. 2020, 30, 885–901. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Udagawa, N.; Uehara, S.; Maeda, K.; Yamashita, T.; Nakamichi, Y.; Kato, H.; Saito, N.; Minami, Y.; Takahashi, N.; et al. Noncanonical Wnt5a enhances Wnt/β-catenin signaling during osteoblastogenesis. Sci. Rep. 2014, 4, 4493. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Tian, H.; Zhang, K.; Chen, D.; Chen, D.; Wang, X.; Zhao, J. Wnt5a/FZD4 Mediates the Mechanical Stretch-Induced Osteogenic Differentiation of Bone Mesenchymal Stem Cells. Cell Physiol. Biochem. 2018, 48, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Li, X.; Bai, M.R.; Chen, X.; Wang, C.L.; Xie, J.; Ye, L. FGF-7 Dictates Osteocyte Cell Processes Through Beta-Catenin Transduction. Sci. Rep. 2018, 8, 14792. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, N.; Shibayama, M.; Kurotaki, Y.; Imanishi, M.; Fujimori, T.; Itoh, N.; Takada, S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002, 16, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Shemirani, R.; Lin, G.; Abduweli Uyghurturk, D.; Le, M.; Nakano, Y. An miRNA derived from amelogenin exon4 regulates expression of transcription factor Runx2 by directly targeting upstream activators Nfia and Prkch. J. Biol. Chem. 2022, 298, 101807. [Google Scholar] [CrossRef]
- Dowling, D.P.; Miller, I.S.; Ardhaoui, M.; Gallagher, W.M. Effect of surface wettability and topography on the adhesion of osteosarcoma cells on plasma-modified polystyrene. J. Biomater. Appl. 2011, 26, 327–347. [Google Scholar] [CrossRef]
- Xie, X.; Mahmood, S.R.; Gjorgjieva, T.; Percipalle, P. Emerging roles of cytoskeletal proteins in regulating gene expression and genome organization during differentiation. Nucleus 2020, 11, 53–65. [Google Scholar] [CrossRef]
- Uray, I.P.; Uray, K. Mechanotransduction at the Plasma Membrane-Cytoskeleton Interface. Int. J. Mol. Sci. 2021, 22, 11566. [Google Scholar] [CrossRef]
- Su, Y.; Besner, G.E. Heparin-binding EGF-like growth factor (HB-EGF) promotes cell migration and adhesion via focal adhesion kinase. J. Surg. Res. 2014, 189, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Finke, B.; Rebl, H.; Hempel, F.; Schäfer, J.; Liefeith, K.; Weltmann, K.D.; Nebe, J.B. Aging of plasma-polymerized allylamine nanofilms and the maintenance of their cell adhesion capacity. Langmuir 2014, 30, 13914–13924. [Google Scholar] [CrossRef] [PubMed]
- Rauner, M.; Sipos, W.; Pietschmann, P. Age-dependent Wnt gene expression in bone and during the course of osteoblast differentiation. Age 2008, 30, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Dar, M.S.; Gesty-Palmer, D.; El-Shewy, H.M.; Robinson, K.M.; Haycraft, C.J.; Barth, J.L. Transcriptomic characterization of signaling pathways associated with osteoblastic differentiation of MC-3T3E1 cells. PLoS ONE 2019, 14, e0204197. [Google Scholar] [CrossRef] [PubMed]
- Martocq, L.; Douglas, T.E.L. Amine-Rich Coatings to Potentially Promote Cell Adhesion, Proliferation and Differentiation, and Reduce Microbial Colonization: Strategies for Generation and Characterization. Coatings 2021, 11, 983. [Google Scholar] [CrossRef]
- Huang, T.; Anselme, K.; Sarraih, S.; Ponche, A. High-performance liquid chromatography as a technique to determine the protein adsorption onto hydrophilic/hydrophobic surfaces. Inter. J. Pharm. 2016, 497, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Gruening, M.; Dawson, J.E.; Voelkner, C.; Neuber, S.; Fricke, K.; van Rienen, U.; Speller, S.; Helm, C.A.; Nebe, J.B. Automatic actin filament quantification and cell shape modeling of osteoblasts on charged Ti surfaces. Appl. Sci. 2021, 11, 5689. [Google Scholar] [CrossRef]
- Rebl, H.; Finke, B.; Rychly, J.; Schröder, K.; Nebe, J.B. Positively charged material surfaces generated by plasma polymerized allylamine enhance vinculin mobility in vital human osteoblasts. Adv. Eng. Mater. 2010, 12, 356–364. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef]
- Gorejová, R.; Oriňaková, R.; Macko, J.; Oriňak, A.; Kupková, M.; Hrubovčáková, M.; Džupon, M.; Sopčák, T.; Ševc, J.; Maskaľová, I.; et al. Electrochemical behavior, biocompatibility and mechanical performance of biodegradable iron with PEI coating. J. Biomed. Mater. Res. A 2022, 110, 659–671. [Google Scholar] [CrossRef]
- Matschegewski, C.; Staehlke, S.; Birkholz, H.; Lange, R.; Beck, U.; Engel, K.; Nebe, J.B. Automatic Actin Filament Quantification of Osteoblasts and their Morphometric Analysis on Microtextured Silicon-Titanium Arrays. Materials 2012, 5, 1176–1195. [Google Scholar] [CrossRef]
- Staehlke, S.; Rebl, H.; Finke, B.; Mueller, P.; Gruening, M.; Nebe, J.B. Enhanced calcium ion mobilization in osteoblasts on amino group containing plasma polymer nanolayer. Cell Biosci. 2018, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Staehlke, S.; Rebl, H.; Nebe, B. Phenotypic stability of the human MG-63 osteoblastic cell line at different passages. Cell Biol. Int. 2019, 43, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Koczan, D.; Fitzner, B.; Zettl, U.K.; Hecker, M. Microarray data of transcriptome shifts in blood cell subsets during S1P receptor modulator therapy. Sci. Data 2018, 24, 180145. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hartmann, A.; Nürnberg, G.; Repsilber, D.; Janczyk, P.; Walz, C.; Ponsuksili, S.; Souffrant, W.B.; Schwerin, M. Effects of threshold choice on the results of gene expression profiling, using microarray analysis, in a model feeding experiment with rat. Arch. Tierzucht. 2009, 52, 65–78. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 8, W83–W89. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 8, D605–D612. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 20, e47. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 2, W191–W198. [Google Scholar] [CrossRef]



| Surface Modification | WCA [θ in °] | Zeta Potential [ζ in mV] |
|---|---|---|
| Ti-Ref (titanium reference) | 94.6° ± 3.5° | −81.2 ± 6.3 mV |
| Ti-Col I (collagen I) | 53.1° ± 4.3° | −2.8 ± 1.5 mV [14] |
| Ti-2AE-APS (N-(2-aminoethyl)-3-aminopropyltrimethoxysilane) | 95.2° ± 0.4° | −11.5 ± 10.2 mV |
| Ti-APTES-1 (1 mM aminopropyltriethoxysilane) | 68.9° ± 6.3° | −20.6 ± 10.4 mV |
| Ti-APTES-100 (100 mM aminopropyltriethoxysilane) | 76.8° ± 1.7° | +51.4 ± 22.1 mV |
| Ti-GOPTS-bPEI (branched (poly(ethyleneimine)) on 3-glycidyloxypropyl)trimethoxysilane) | 54.7° ± 2.0° | −34.2 ± 9.4 mV |
| Ti-GOPTS-lPEI (linear (poly(ethyleneimine)) on 3-glycidyloxypropyl)trimethoxysilane) | 50.7° ± 3.8° | −48.9 ± 2.8 mV |
| Ti-TMS-PEI (trimethoxysilylpropyl modified poly(ethyleneimine)) | 53.6° ± 5.4° | +11.7 ± 14.1 mV |
| Coating | Ti [at-%] | N [at-%] | C [at-%] | N/C [%] | Spreading Fold Change of Col I | ||
|---|---|---|---|---|---|---|---|
| Ti-APTES-1 | 14.13 | 3.40 | 40.56 | 8.38 |  | 1.42 |  |
| Ti-APTES-100 | 0.00 | 10.36 | 55.60 | 18.63 | 2.05 | ||
| Ti-2AE-APS-1 | 0.52 | 13.11 | 59.96 | 21.86 | 1.78 | ||
| Ti-GOPTS-l-PEI | 22.21 | 4.14 | 18.16 | 22.80 | 1.37 | ||
| Ti-GOPTS-b-PEI | 21.58 | 5.58 | 20.58 | 27.11 | 1.90 | ||
| Ti-TMS-PEI | 21.39 | 7.38 | 23.04 | 32.03 | 2.29 |
| Substrates | BSA Concentration [µg/mL] | Adsorbed Amount | ||
|---|---|---|---|---|
| Adsorption | Washing | Desorption | [µg/cm2] | |
| Ti-Ref | 189.1 ± 4.3 | 2.3 ± 0.1 | 0.6 ± 0.0 | 1.7 ± 0.0 |
| Ti-TMS-PEI | 181.8 ± 0.7 | 2.7 ± 0.0 | 2.4 ± 0.2 | 7.2 ± 0.7 |
| Gene | Fold Change * | p-Value | FDR p-Value | Description | Entrez ID |
|---|---|---|---|---|---|
| SPRY4 | 2.06 | 2.31 × 10−6 | 0.0247 | sprouty RTK signaling antagonist 4 | 81848 |
| DUSP1 | 2.03 | 1.87 × 10−6 | 0.0247 | dual specificity phosphatase 1 | 1843 |
| PCDH18 | −2.23 | 3.77 × 10−6 | 0.0269 | protocadherin 18 | 54510 |
| PLAUR | 1.99 | 5.60 × 10−6 | 0.0300 | plasminogen activator, urokinase receptor | 5329 |
| NR4A2 | 2.17 | 1.66 × 10−5 | 0.0441 | nuclear receptor subfamily 4, group A, member 2 | 4929 |
| HBEGF | 2.24 | 1.45 × 10−5 | 0.0441 | heparin-binding EGF-like growth factor | 1839 |
| BCL7A | 1.70 | 1.38 × 10−5 | 0.0441 | B-cell CLL/lymphoma 7A | 605 |
| DUSP6 | 1.67 | 1.85 × 10−5 | 0.0441 | dual specificity phosphatase 6 | 1848 |
| KCNJ2 | −2.36 | 1.71 × 10−5 | 0.0441 | potassium channel, inwardly rectifying subfamily J, member 2 | 3759 |
| Gene * | Gene Ontology Annotation |
|---|---|
| SPRY4 | negative regulation of ERK1 and ERK2 cascade; regulation of signal transduction; negative regulation of MAP kinase activity |
| DUSP1 | inactivation of MAPK activity, negative regulation of ERK1 and ERK2 cascade; mitotic cell cycle arrest; regulation of mitotic cell cycle spindle assembly checkpoint; negative regulation of DNA biosynthetic process |
| PCDH18 | Cadherin, homophilic cell adhesion via plasma membrane adhesion molecules; cell adhesion |
| PLAUR | plasminogen activator, urokinase receptor, CD59 antigen |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
| HBEGF | HBEGF: epidermal growth factor receptor signaling pathway; Fc-epsilon receptor signaling pathway; vascular endothelial growth factor receptor signaling pathway; neurotrophin TRK receptor signaling pathway; phosphatidylinositol-mediated signaling; MAPK cascade; activation of MAPKK activity; Ras protein signal transduction; insulin receptor signaling pathway; fibroblast growth factor receptor signaling pathway; positive regulation of cell migration; spreading of epidermal cells; cell chemotaxis; positive regulation of cell growth, positive regulation of cell proliferation |
| BCL7A | negative regulation of transcription |
| DUSP6 | phosphatase activity; negative regulation in the MAPK pathway |
| KCNJ2 | Kir2.1 inward-rectifier potassium channel; potassium ion transmembrane transport; magnesium ion transport; relaxation of cardiac muscle; relaxation of skeletal muscle; cellular response to mechanical stimulus |
| GO.ID | Description | p-Value | Genes |
|---|---|---|---|
| GO:0008330 | protein tyrosine/threonine phosphatase activity | 0.001 | DUSP1, DUSP6 |
| GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity | 0.002 | DUSP1, DUSP6 |
| GO:0033549 | MAP kinase phosphatase activity | 0.004 | DUSP1, DUSP6 |
| GO:0008138 | protein tyrosine/serine/threonine phosphatase activity | 0.028 | DUSP1, DUSP6 |
| GO:0051172 | negative regulation of nitrogen compound metabolic process | 0.008 | SPRY4, DUSP1, PLAUR, NR4A2, HBEGF, BCL7A, DUSP6 |
| GO:0031324 | negative regulation of cellular metabolic process | 0.011 | SPRY4, DUSP1, PLAUR, NR4A2, HBEGF, BCL7A, DUSP6 |
| GO:0051248 | negative regulation of protein metabolic process | 0.036 | SPRY4, DUSP1, PLAUR, HBEGF, DUSP6 |
| GO:1902532 | negative regulation of intracellular signal transduction | 0.045 | SPRY4, DUSP1, PLAUR, DUSP6 |
| GO:0001932 | regulation of protein phosphorylation | 0.047 | SPRY4, DUSP1, PLAUR, HBEGF, DUSP6 |
| GO:0043409 | negative regulation of MAPK cascade | 0.049 | SPRY4, DUSP1, DUSP6 |
| REAC:R-HSA-112409 | RAF-independent MAPK1/3 activation | 0.012 | DUSP1, DUSP6 |
| REAC:R-HSA-5675221 | Negative regulation of MAPK pathway | 0.045 | DUSP1, DUSP6 |
| Gene | Fold Change * | Description | Role in Osteoblast Differentiation | References | Entrez ID |
|---|---|---|---|---|---|
| WNT5B | −1.68 | wingless-type MMTV integration site family, member 5B | WNT5B often functions as an antagonist of canonical WNT signaling; indication that WNT5B suppresses osteoblast differentiation | [35] | 16265 |
| WNT2B | −1.78 | wingless-type MMTV integration site family, member 2B | increases during differentiation; promotes osteogenesis and regulates expression of Osterix and RUNX2, which drive differentiation | [36] | 12781 |
| FGF2 | −2.18 | fibroblast growth factor 2 | enhances the osteogenic potential of BMSCs and their proliferative capacity. FGF2 induces bone and BMSC proliferation; proliferation factor for bone cells in cell culture; fibroblast growth factor 2 (FGF2) positively modulates osteoblast differentiation and bone formation | [37,38] | 3676 |
| FGF5 | −1.60 | fibroblast growth factor 5 | promotes OS cell proliferation via MAPK signaling pathway; closely associated with poor differentiation | [39] | 3683 |
| HEY1 | 1.68 | hes-related family bHLH transcription factor with YRPW motif 1 | upregulated expression at the immediate early stage of BMP9-induced osteogenic differentiation; acts synergistically with Runx2 in BMP9-induced osteogenic differentiation | [40] | 4880 |
| WNT9A | 3.23 | wingless-type MMTV integration site family, member 9A | expression decreases during osteoblastic differentiation; Wnt9a knockout mice display bone malformations (low levels of bone ossification, indications of skeletal dysplasia); activates Loxl2 (collagen-cross-linking enzyme) which activates the biogenesis of connective tissue | [36,41,42] | 12778 |
| WNT7B | 2.41 | wingless-type MMTV integration site family, member 7B | enhancing factor for osteoblast differentiation, induces the non-canonical activation of mTORC1 and PKC δ signaling | [41] | 12787 |
| WNT5A | 1.64 | wingless-type MMTV integration site family, member 5A | involved in osteoblast differentiation; upregulates Lrp5/6; mediates the mechanical stretch-induced osteogenic differentiation of BMSCs | [43,44] | 12784 |
| FGF7 | 1.79 | fibroblast growth factor 7 | facilitates osteogenic differentiation of embryonic stem cells; activates ERK/Runx2 | [45] | 3685 |
| FGF18 | 1.92 | fibroblast growth factor 18 | expressed in osteogenic mesenchymal cells and differentiating osteoblasts | [46] | 3674 |
| PRKCH | 2.59 | protein kinase C, eta | upstream activator of RUNX2 (essential transcription factor for osteoblast differentiation) | [47] | 9403 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seemann, S.; Dubs, M.; Koczan, D.; Salapare, H.S., III; Ponche, A.; Pieuchot, L.; Petithory, T.; Wartenberg, A.; Staehlke, S.; Schnabelrauch, M.; et al. Response of Osteoblasts on Amine-Based Nanocoatings Correlates with the Amino Group Density. Molecules 2023, 28, 6505. https://doi.org/10.3390/molecules28186505
Seemann S, Dubs M, Koczan D, Salapare HS III, Ponche A, Pieuchot L, Petithory T, Wartenberg A, Staehlke S, Schnabelrauch M, et al. Response of Osteoblasts on Amine-Based Nanocoatings Correlates with the Amino Group Density. Molecules. 2023; 28(18):6505. https://doi.org/10.3390/molecules28186505
Chicago/Turabian StyleSeemann, Susanne, Manuela Dubs, Dirk Koczan, Hernando S. Salapare, III, Arnaud Ponche, Laurent Pieuchot, Tatiana Petithory, Annika Wartenberg, Susanne Staehlke, Matthias Schnabelrauch, and et al. 2023. "Response of Osteoblasts on Amine-Based Nanocoatings Correlates with the Amino Group Density" Molecules 28, no. 18: 6505. https://doi.org/10.3390/molecules28186505
APA StyleSeemann, S., Dubs, M., Koczan, D., Salapare, H. S., III, Ponche, A., Pieuchot, L., Petithory, T., Wartenberg, A., Staehlke, S., Schnabelrauch, M., Anselme, K., & Nebe, J. B. (2023). Response of Osteoblasts on Amine-Based Nanocoatings Correlates with the Amino Group Density. Molecules, 28(18), 6505. https://doi.org/10.3390/molecules28186505











