Chiral Recognition of D/L-Ribose by Visual and SERS Assessments
Abstract
:1. Introduction
2. Results and Discussion
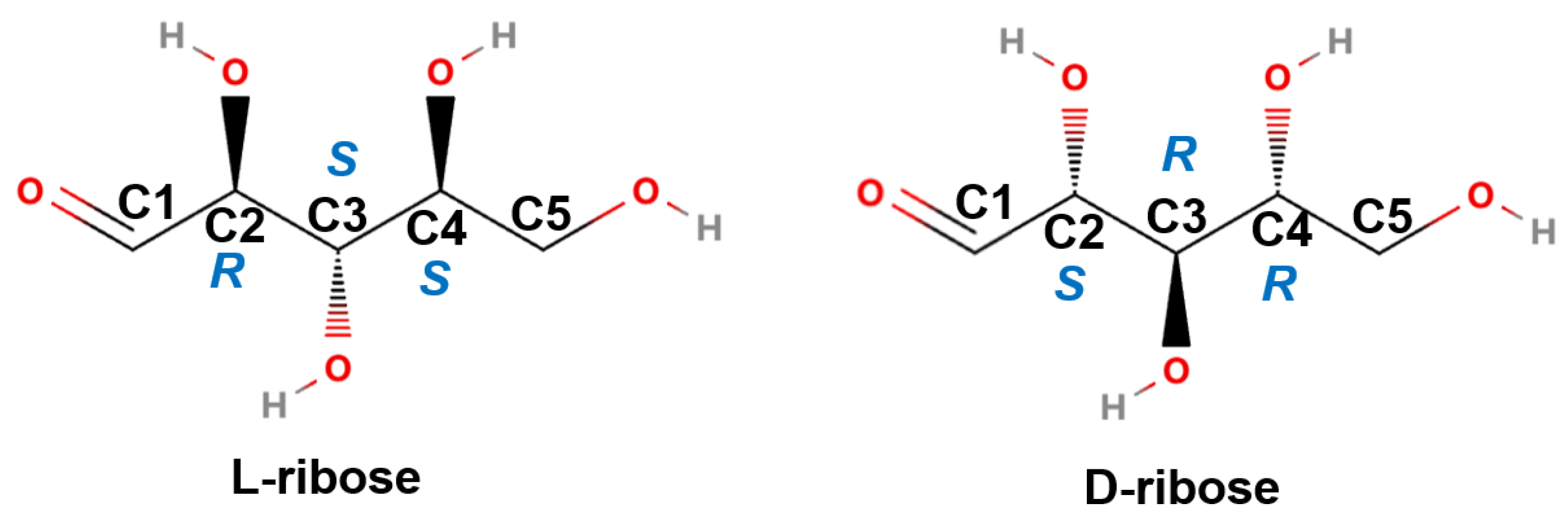
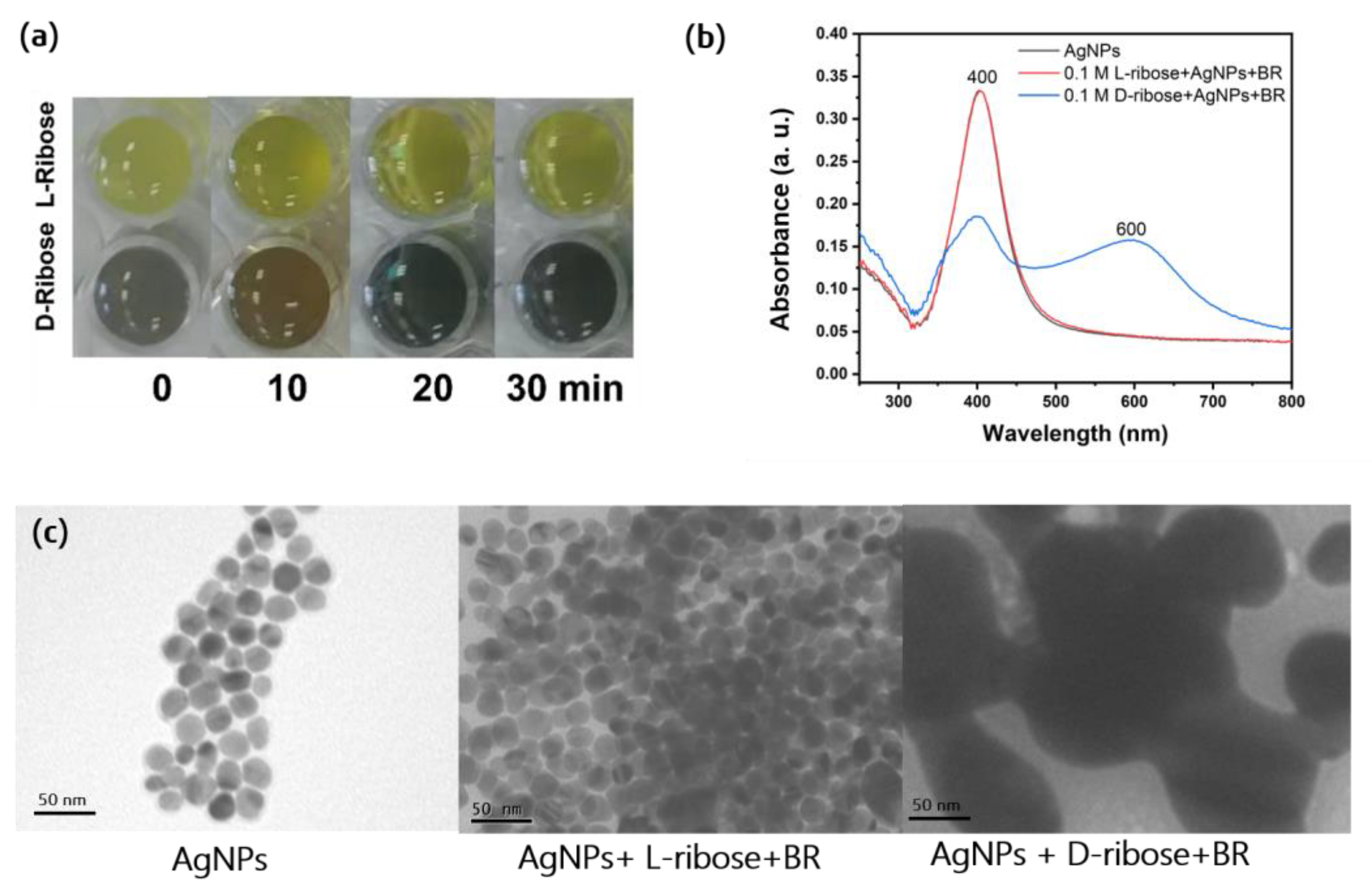
2.1. Chiral Recognition by Visual Assessment
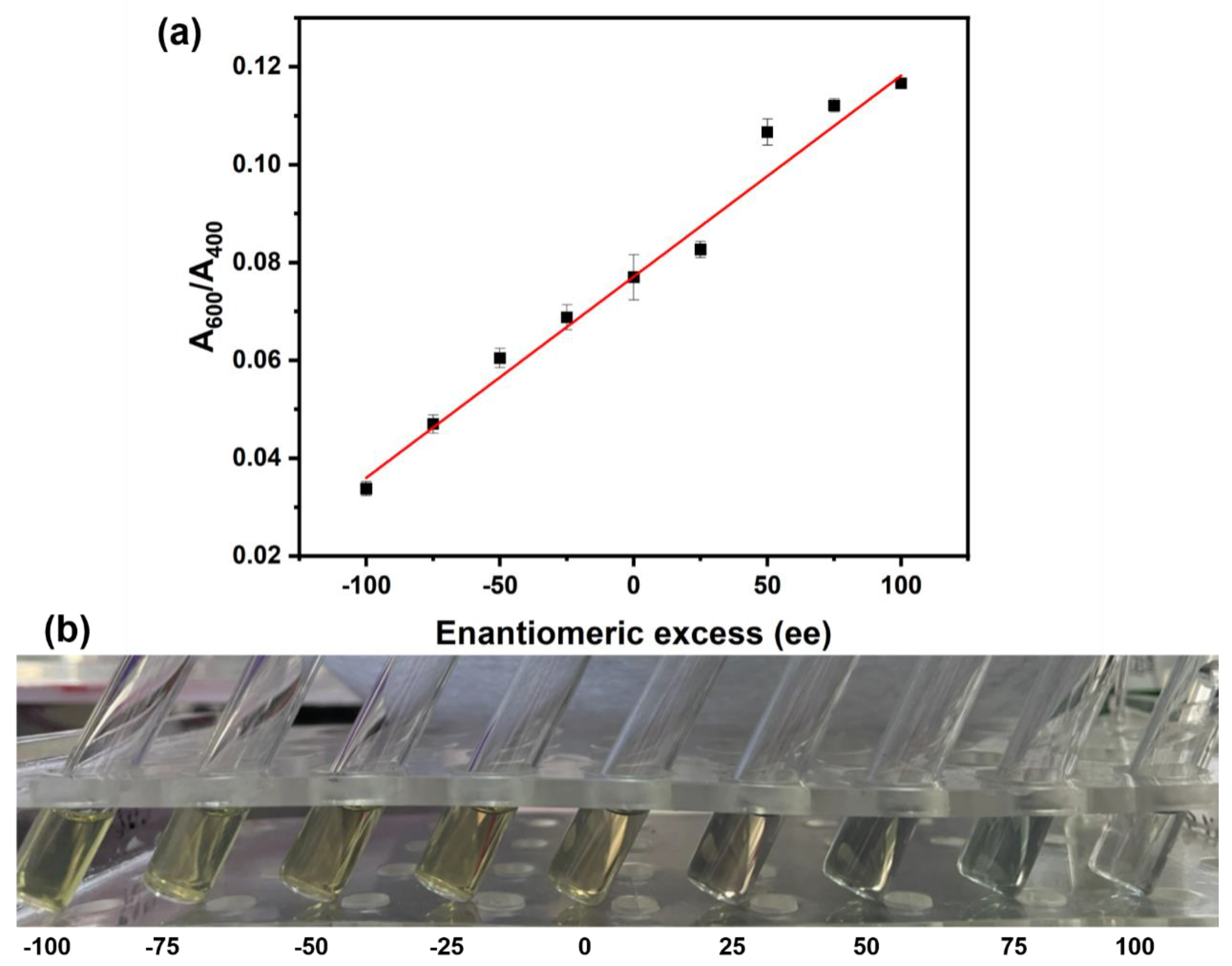
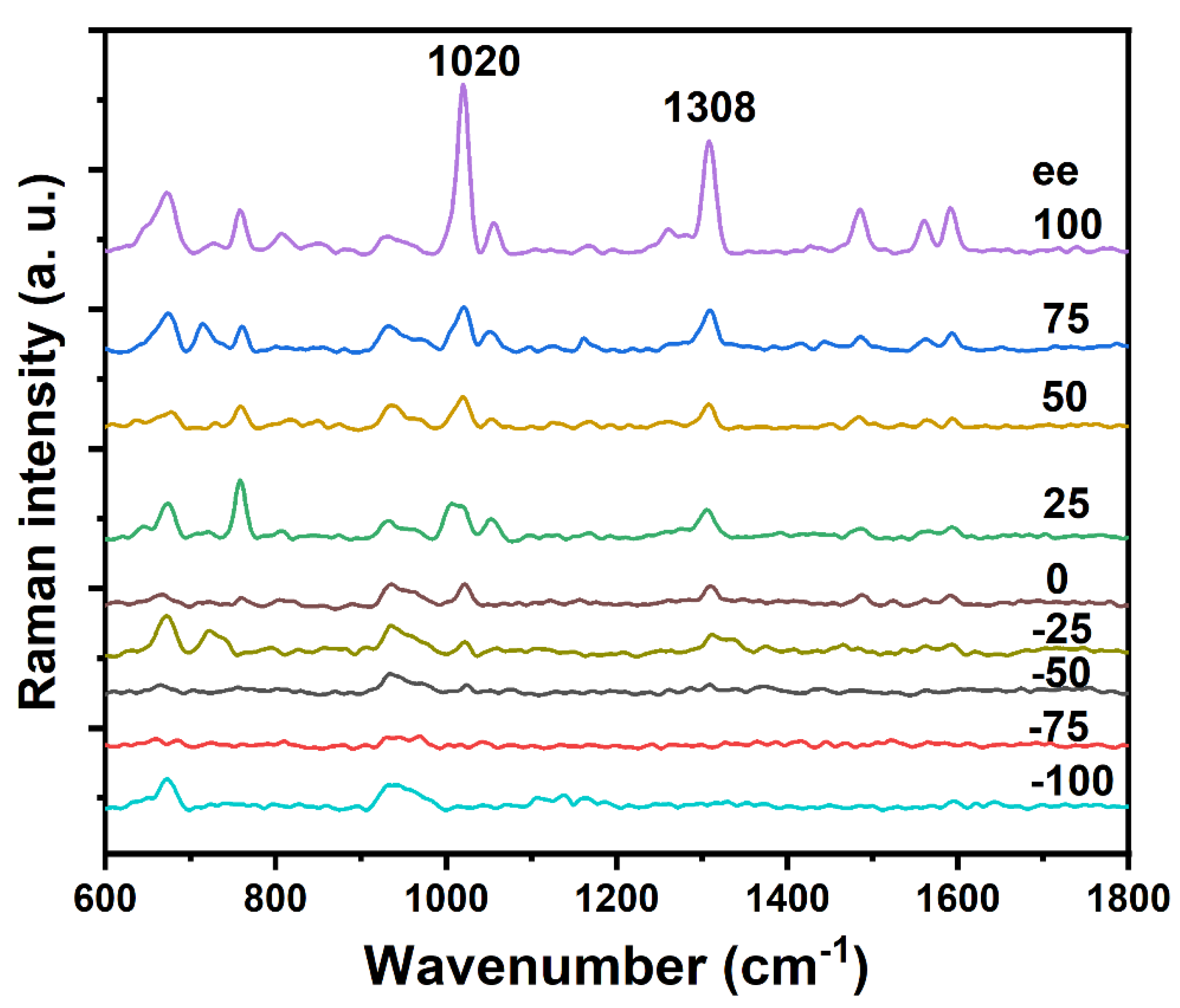
2.2. SERS Chiral Recognition
2.3. DFT Calculation and Analysis
3. Conclusions
4. Materials and Methods
4.1. Reagents and Materials
4.2. Preparation of Ag@CD NPs (β-cyclodextrin Coated Ag Nanoparticles)
4.3. Experimental Processing of Chiral Recognition
4.4. UV–vis Absorption Spectra Measurements
4.5. SERS Measurements
4.6. DFT Calculation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Meinert, C.; Myrgorodska, I.; de Marcellus, P.; Buhse, T.; Nahon, L.; Hoffmann, S.V.; d’Hendecourt, L.L.S.; Meierhenrich, U.J. Ribose and related sugars from ultraviolet irradiation of interstellar ice analogs. Science 2016, 352, 208–212. [Google Scholar] [CrossRef]
- Ricardo, A.; Carrigan, M.A.; Olcott, A.N.; Benner, S.A. Borate Minerals Stabilize Ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef]
- Reid, C.; Orgel, L.E.; Ponnamperuma, C. Nucleoside Synthesis under Potentially Prebiotic Conditions. Nature 1967, 216, 936. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Robertson, M.P.; Joyce, G.F. The Origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4, 3608. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.-R.; Wang, X. A plausible prebiotic selection of ribose for RNA—Formation, dynamic isolation, and nucleotide synthesis based on metal-doped-clays. Chem 2021, 7, 3292–3308. [Google Scholar] [CrossRef]
- Devínsky, F. Chirality and the Origin of Life. Symmetry 2021, 13, 2277. [Google Scholar] [CrossRef]
- Cowan, J.A.; Furnstahl, R.J. Origin of Chirality in the Molecules of Life. ACS Earth Space Chem. 2022, 6, 2575–2581. [Google Scholar] [CrossRef]
- Blackmond, D.G. Asymmetric autocatalysis and its implications for the origin of homochirality. Proc. Natl. Acad. Sci. USA 2004, 101, 5732–5736. [Google Scholar] [CrossRef] [PubMed]
- Gusev, G.A.; Guseva, Z.G. Type 1a Supernova Explosion and the Origin of Sugar Chiral Asymmetry in Biological Systems. Bull. Lebedev Phys. Inst. 2018, 45, 145–148. [Google Scholar] [CrossRef]
- Wagner, A.J.; Zubarev, D.Y.; Aspuru-Guzik, A.; Blackmond, D.G. Chiral Sugars Drive Enantioenrichment in Prebiotic Amino Acid Synthesis. ACS Cent. Sci. 2017, 3, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Zor, E.; Bingol, H.; Ersoz, M. Chiral sensors. TrAC Trends Anal. Chem. 2019, 121, 115662. [Google Scholar] [CrossRef]
- Mahmood, S.; Iqbal, M.W.; Zhang, W.; Mu, W. A review on l-ribose isomerases for the biocatalytic production of l-ribose and l-ribulose. Food Res. Int. 2021, 145, 110409. [Google Scholar] [CrossRef]
- Khose, V.N.; John, M.E.; Pandey, A.D.; Borovkov, V.; Karnik, A.V. Chiral Heterocycle-Based Receptors for Enantioselective Recognition. Symmetry 2018, 10, 34. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, B.-C. Colorimetric Chiral Recognition of Enantiomers Using the Nucleotide-Capped Silver Nanoparticles. Anal. Chem. 2011, 83, 1504–1509. [Google Scholar] [CrossRef]
- Zor, E.; Bekar, N. Lab-in-a-syringe using gold nanoparticles for rapid colorimetric chiral discrimination of enantiomers. Biosens. Bioelectron. 2017, 91, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.F.; Gaspar, E.M.S.M. Simultaneous chromatographic separation of enantiomers, anomers and structural isomers of some biologically relevant monosaccharides. J. Chromatogr. A 2008, 1188, 34–42. [Google Scholar] [CrossRef]
- Xu, D.; Lin, Q.L.; Chang, H.T. Chiral Ag and Au Nanomaterials Based Optical Approaches for Analytical Applications. Part. Part. Syst. Charact. 2019, 36, 1800552. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Zhao, P.-Y.; Yu, L.-P. Highly-sensitive and selective colorimetric sensor for amino acids chiral recognition based on molecularly imprinted photonic polymers. Sens. Actuators B Chem. 2013, 181, 850–857. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, L.; Liu, C.; Zhang, J.; He, Y.; Yang, H.; Liu, X. Chiral Detection of Glucose: An Amino Acid-Assisted Surface-Enhanced Raman Scattering Strategy Showing Opposite Enantiomeric Effects on SERS Signals. Anal. Chem. 2022, 94, 14565–14572. [Google Scholar] [CrossRef]
- Sandilya, A.A.; Natarajan, U.; Priya, M.H. Molecular View into the Cyclodextrin Cavity: Structure and Hydration. ACS Omega 2020, 5, 25655–25667. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Huang, Q. DFT and SERS Study of (L)-Cysteine Adsorption on the Surface of Gold Nanoparticles. J. Phys. Chem. C 2018, 122, 15241–15251. [Google Scholar] [CrossRef]
- Olson, J.; Manjavacas, A.; Liu, L.; Chang, W.-S.; Foerster, B.; King, N.S.; Knight, M.W.; Nordlander, P.; Halas, N.J.; Link, S. Vivid, full-color aluminum plasmonic pixels. Proc. Natl. Acad. Sci. USA 2014, 111, 14348–14353. [Google Scholar] [CrossRef]
- Srinivasan, V.; Manne, A.K.; Patnaik, S.G.; Ramamurthy, S.S. Cellphone Monitoring of Multi-Qubit Emission Enhancements from Pd-Carbon Plasmonic Nanocavities in Tunable Coupling Regimes with Attomolar Sensitivity. Acs. Appl. Mater. Inter. 2016, 8, 23281–23288. [Google Scholar] [CrossRef] [PubMed]
- Carmona, P.; Molina, M. Raman and infrared spectra of D-ribose and D-ribose 5-phosphate. J. Raman Spectrosc. 1990, 21, 395–400. [Google Scholar] [CrossRef]
- Pour, S.O.; Rocks, L.; Faulds, K.; Graham, D.; Parchansky, V.; Bour, P.; Blanch, E.W. Through-space transfer of chiral information mediated by a plasmonic nanomaterial. Nat. Chem. 2015, 7, 591–596. [Google Scholar] [CrossRef]
- Dass, A.V.; Georgelin, T.; Westall, F.; Foucher, F.; De Los Rios, P.; Busiello, D.M.; Liang, S.; Piazza, F. Equilibrium and non-equilibrium furanose selection in the ribose isomerisation network. Nat. Commun. 2021, 12, 2749. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. A multi-faceted approach to elucidate the crystal structure of D-ribose: Similarities to protein structure determination. Angew. Chem. Int. Ed. Engl. 2010, 49, 6487–6489. [Google Scholar] [CrossRef]
- Sisak, D.; McCusker, L.B.; Zandomeneghi, G.; Meier, B.H.; Blaser, D.; Boese, R.; Schweizer, W.B.; Gilmour, R.; Dunitz, J.D. The crystal structure of Dribose at last! Angew. Chem. Int. Ed. Engl. 2010, 49, 4503–4505. [Google Scholar] [CrossRef]
- Arad-Yellin, R.; Green, B.S. Photochemical closing and opening of the guest-binding cavity of cyclodextrins. Nature 1994, 371, 320–322. [Google Scholar] [CrossRef]
- Wójcik, J.; Ejchart, A.; Nowakowski, M. Shape adaptation of quinine in cyclodextrin cavities: NMR studies. Phys. Chem. Chem. Phys. 2019, 21, 6925–6934. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, G.; Sun, K.; Huang, Q. beta-Cyclodextrin coated SiO2@Au@Ag core-shell nanoparticles for SERS detection of PCBs. Phys. Chem. Chem. Phys. 2015, 17, 21149–21157. [Google Scholar] [CrossRef]
- Singh, G.; Pandey, S.P.; Singh, P.K. Guest Binding with Sulfated Cyclodextrins: Does the Size of Cavity Matter? Chemphyschem 2023, 24, e202200421. [Google Scholar] [CrossRef]
- Yao, G.; Zhai, Z.; Zhong, J.; Huang, Q. DFT and SERS Study of N-15 Full-Labeled Adenine Adsorption on Silver and Gold Surfaces. J. Phys. Chem. C 2017, 121, 9869–9878. [Google Scholar] [CrossRef]
- Hamzah, R.; Abu Bakar, M.; Dahham, O.S.; Zulkepli, N.N.; Dahham, S.S. A structural study of epoxidized natural rubber (ENR-50) ring opening under mild acidic condition. J. Appl. Polym. Sci. 2016, 133, 44123. [Google Scholar] [CrossRef]
- Guido, E.; Colleoni, C.; De Clerck, K.; Plutino, M.R.; Rosace, G. Influence of catalyst in the synthesis of a cellulose-based sensor: Kinetic study of 3-glycidoxypropyltrimethoxysilane epoxy ring opening by Lewis acid. Sens. Actuators B Chem. 2014, 203, 213–222. [Google Scholar] [CrossRef]
- Wang, S.Y.; Li, L.; Xiao, Y.; Wang, Y. Recent advances in cyclodextrins-based chiral-recognizing platforms. TrAC Trend. Anal. Chem. 2019, 121, 115691. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Walker, M.; Harvey, A.J.; Sen, A.; Dessent, C.E. Performance of M06, M06-2X, and M06-HF density functionals for conformationally flexible anionic clusters: M06 functionals perform better than B3LYP for a model system with dispersion and ionic hydrogen-bonding interactions. J. Phys. Chem. A 2013, 117, 12590–12600. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. Density functionals for noncovalent interaction energies of biological importance. J. Chem. Theory Comput. 2007, 3, 289–300. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. A prototype for graphene material simulation: Structures and interaction potentials of coronene dimers. J. Phys. Chem. C 2008, 112, 4061–4067. [Google Scholar] [CrossRef]
- Hohenstein, E.G.; Chill, S.T.; Sherrill, C.D. Assessment of the Performance of the M05-2X and M06-2X Exchange-Correlation Functionals for Noncovalent Interactions in Biomolecules. J. Chem. Theory Comput. 2008, 4, 1996–2000. [Google Scholar] [CrossRef]
- Yao, G.; Zhang, J.; Huang, Q. Conformational and vibrational analyses of meta-tyrosine: An experimental and theoretical study. Spectrochim Acta A 2015, 151, 111–123. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.; Millam, J. GaussView 5; Semichem Inc.: Shawnee Mission, KS, USA, 2009. [Google Scholar]

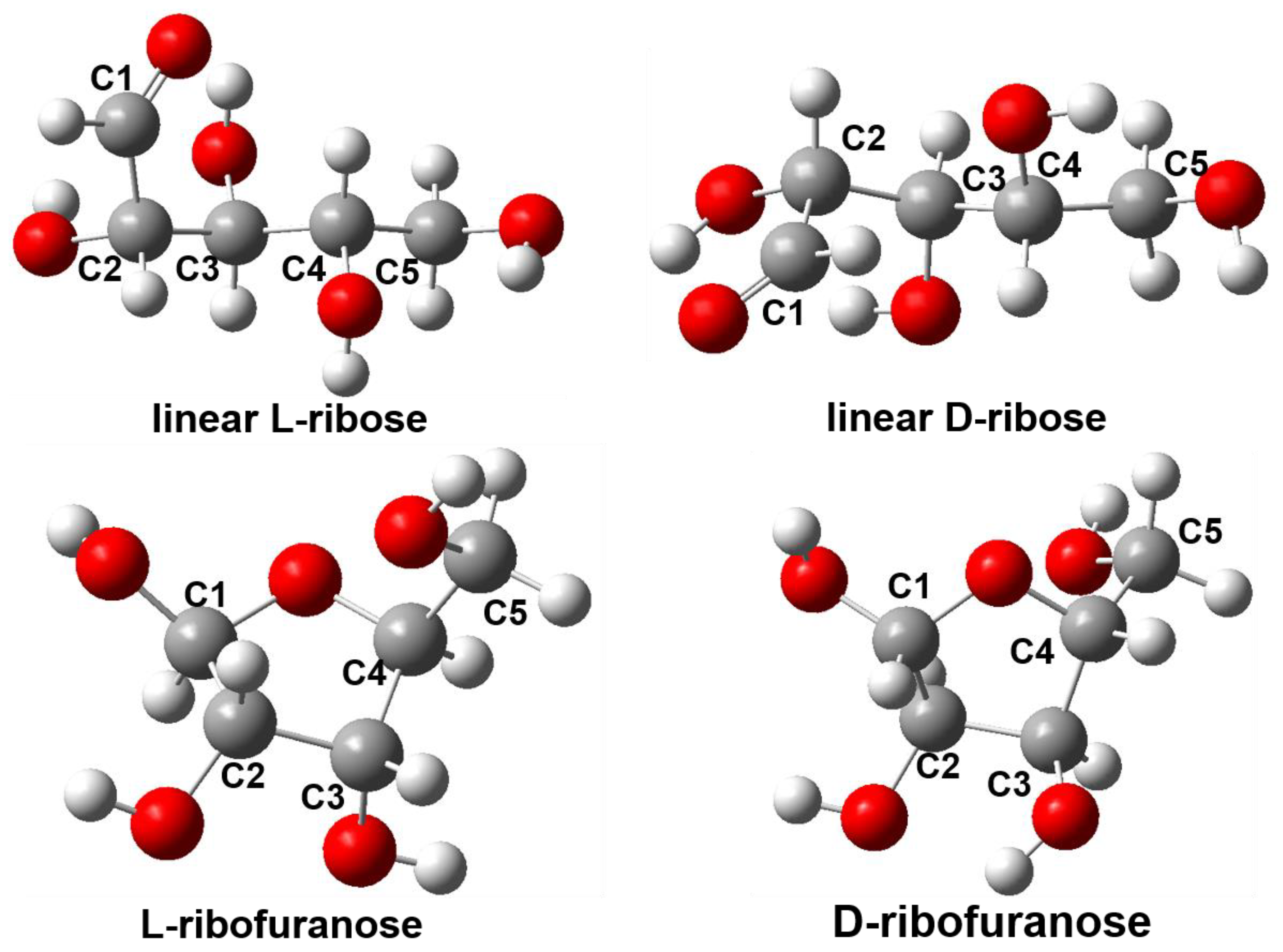
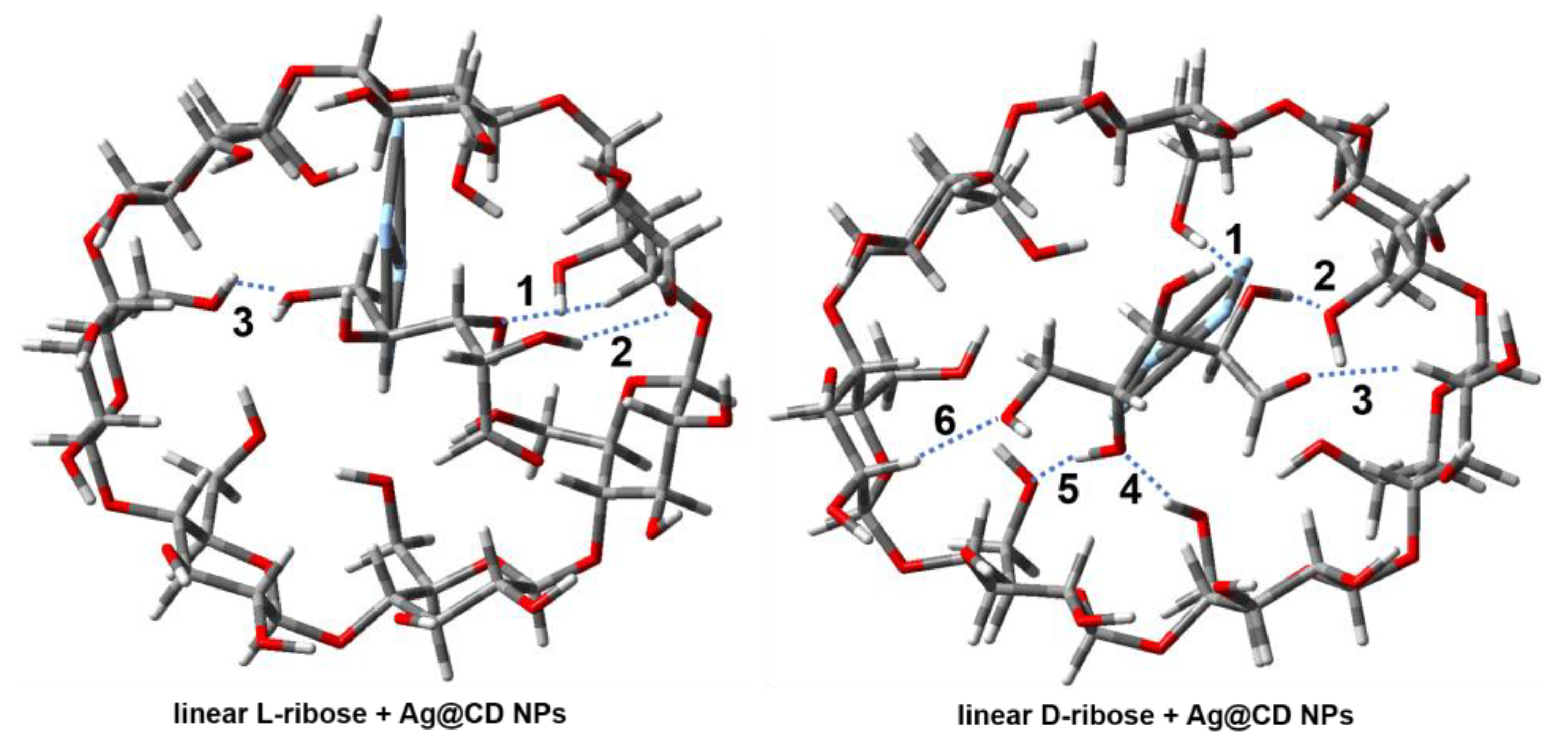
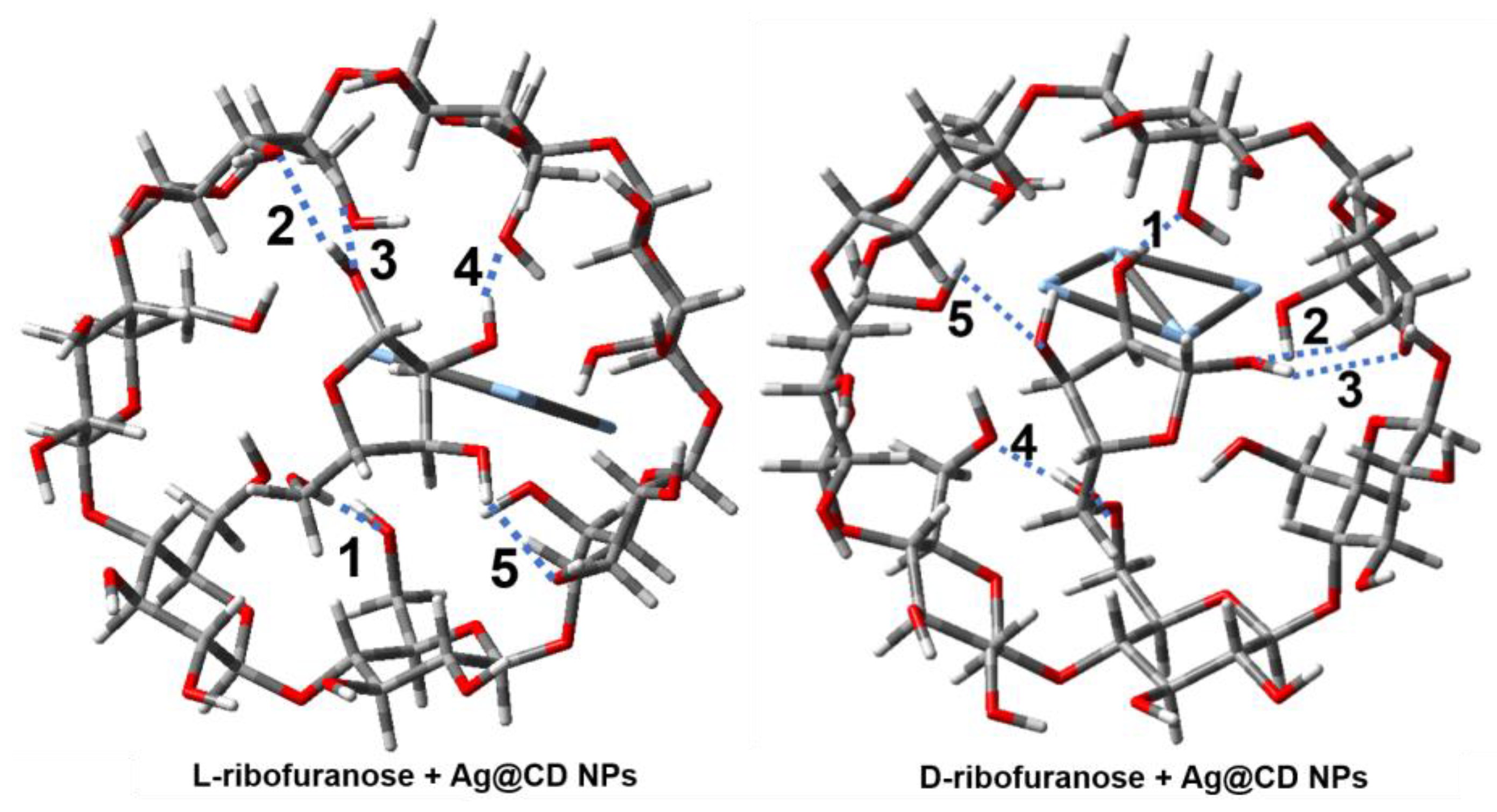
| Complex | E(Bind) (in kJ mol−1) | Size a (in a Å × b Å) |
|---|---|---|
| linear L-ribose and Ag@CD | −73.406 | 14.2 × 13.4 |
| linear D-ribose and Ag@CD | −125.877 | 15.4 × 12.8 |
| L-ribofuranose and Ag@CD | −53.342 | 14.1 × 13.4 |
| D-ribofuranose and Ag@CD | −68.250 | 14.3 × 13.5 |
| Complex | 1 a | 2 | 3 | 4 | 5 | 6 | Avg. b |
|---|---|---|---|---|---|---|---|
| linear L-ribose and Ag@CD | 2.198 | 2.098 | 2.069 | 2.122 | |||
| linear D-ribose and Ag@CD | 1.774 | 1.644 | 2.269 | 1.949 | 1.738 | 2.055 | 1.905 |
| L-ribofuranose and Ag@CD | 1.989 | 2.142 | 2.121 | 1.923 | 2.106 | 2.056 | |
| D-ribofuranose and Ag@CD | 1.679 | 2.169 | 2.288 | 1.830 | 2.228 | 2.039 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, G.; Liu, C.; Elsherbiny, S.M.; Huang, Q. Chiral Recognition of D/L-Ribose by Visual and SERS Assessments. Molecules 2023, 28, 6480. https://doi.org/10.3390/molecules28186480
Yao G, Liu C, Elsherbiny SM, Huang Q. Chiral Recognition of D/L-Ribose by Visual and SERS Assessments. Molecules. 2023; 28(18):6480. https://doi.org/10.3390/molecules28186480
Chicago/Turabian StyleYao, Guohua, Chao Liu, Shereen M. Elsherbiny, and Qing Huang. 2023. "Chiral Recognition of D/L-Ribose by Visual and SERS Assessments" Molecules 28, no. 18: 6480. https://doi.org/10.3390/molecules28186480
APA StyleYao, G., Liu, C., Elsherbiny, S. M., & Huang, Q. (2023). Chiral Recognition of D/L-Ribose by Visual and SERS Assessments. Molecules, 28(18), 6480. https://doi.org/10.3390/molecules28186480







