Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein
Abstract
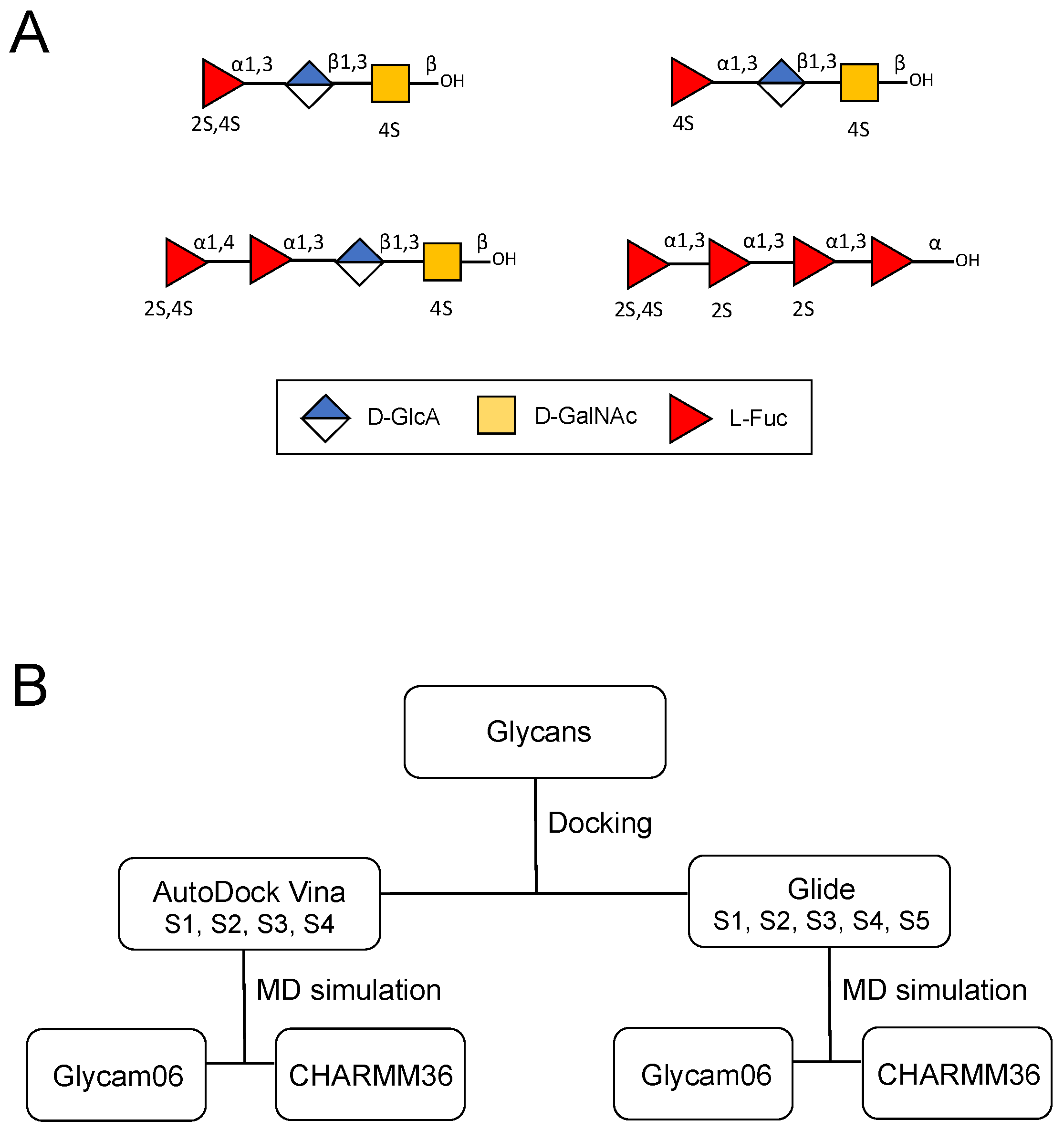
:1. Introduction
2. Results
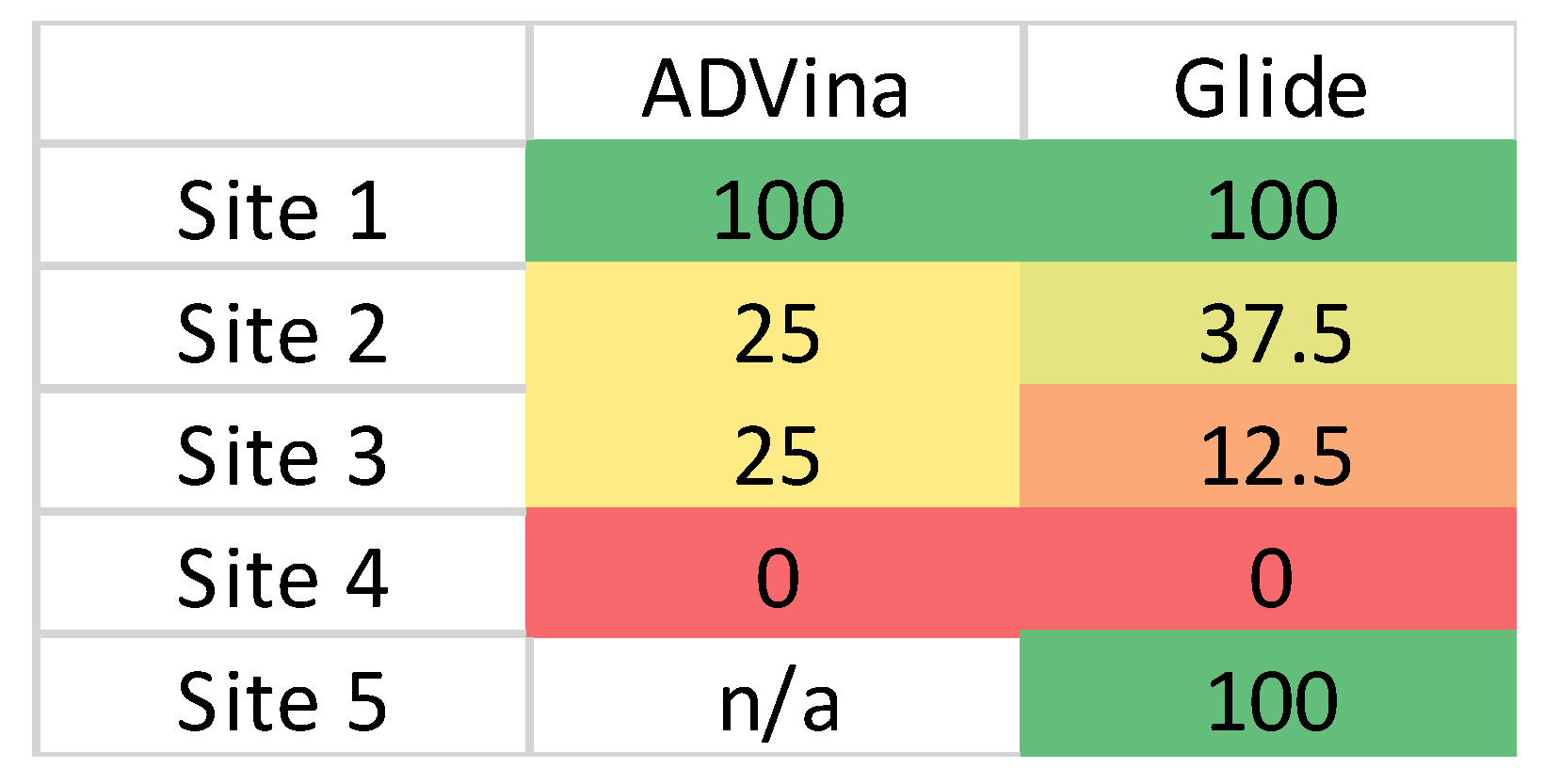
2.1. Identification of Potential Binding Sites in SGP RBD
2.1.1. Site 1 (S1)
2.1.2. Site 2 (S2)
2.1.3. Site 3 (S3)
2.1.4. Site 4 (S4)
2.1.5. Site 5 (S5)
2.2. ADMET Prediction
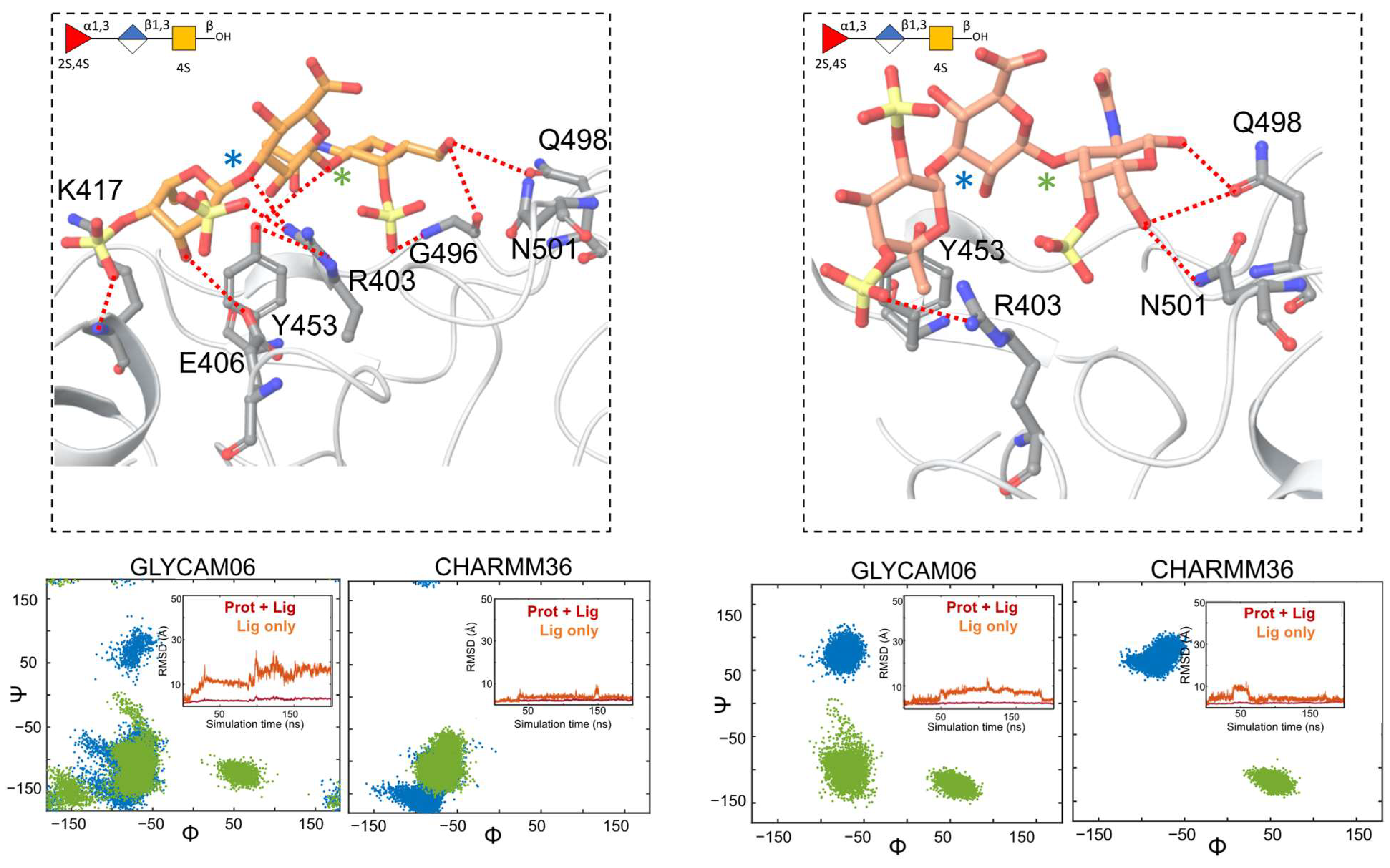
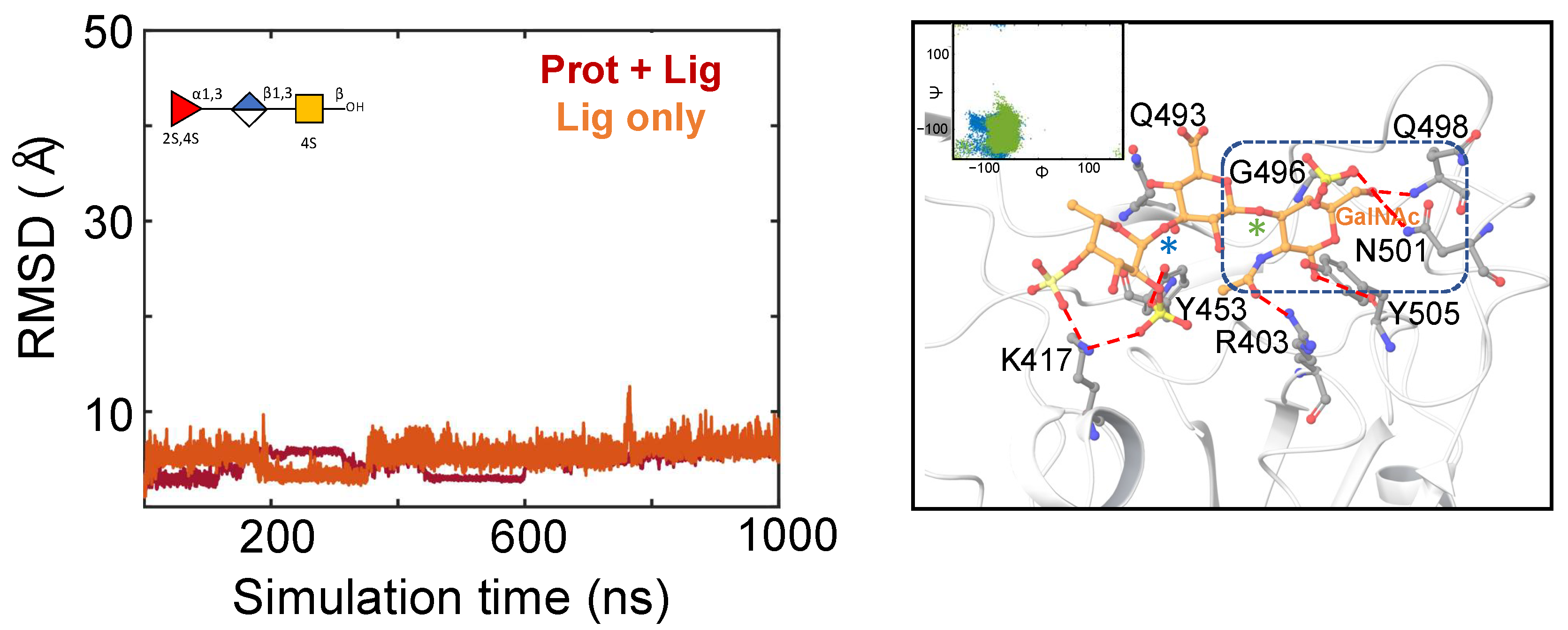
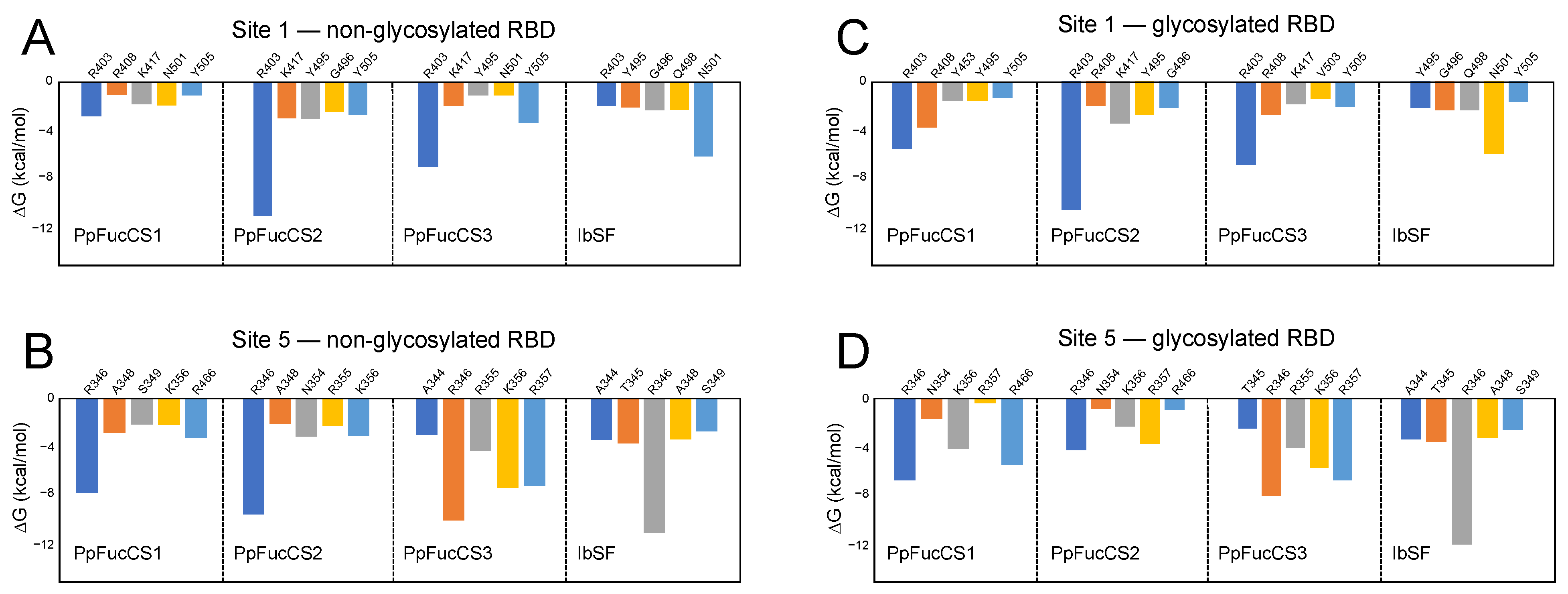
2.3. Characterization of the Holothurian Sulfated Glycan Binding Interaction with SGP RBD
3. Discussion
4. Materials and Methods
4.1. Preparation of 3D Structures of the Holothurian Sulfated Glycan-Derived Composing Oligosaccharides
4.2. Protein Structure Preparation
4.3. Molecular Docking
4.4. SGP–glycan Complex Structure Preparation
4.5. MD Simulations
4.6. Binding-Free Energy Calculations
4.7. ADMET Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Xie, Y.; Du, D.; Karki, C.B.; Guo, W.; Lopez-Hernandez, A.E.; Sun, S.; Juarez, B.Y.; Li, H.; Wang, J.; Li, L.; et al. Revealing the mechanism of SARS-CoV-2 spike protein binding with ACE2. Comput. Sci. Eng. 2020, 22, 21–29. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Nguyen, L.; McCord, K.A.; Bui, D.T.; Bouwman, K.M.; Kitova, E.N.; Elaish, M.; Kumawat, D.; Daskhan, G.C.; Tomris, I.; Han, L.; et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat. Chem. Biol. 2022, 18, 81–90. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Zoepfl, M.; Dwivedi, R.; Taylor, M.C.; Pomin, V.H.; McVoy, M.A. Antiviral activities of four marine sulfated glycans against adenovirus and human cytomegalovirus. Antiviral. Res. 2021, 190, 105077. [Google Scholar] [CrossRef]
- Vasconcelos, A.A.; Pomin, V.H. Marine Carbohydrate-Based Compounds with Medicinal Properties. Mar. Drugs 2018, 16, 233. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H. Marine Non-Glycosaminoglycan Sulfated Glycans as Potential Pharmaceuticals. Pharmaceuticals 2015, 8, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H. Antimicrobial Sulfated Glycans: Structure and Function. Curr. Top Med. Chem. 2017, 17, 319–330. [Google Scholar] [CrossRef]
- Zong, A.; Cao, H.; Wang, F. Anticancer polysaccharides from natural resources: A review of recent research. Carbohydr. Polym. 2012, 90, 1395–1410. [Google Scholar] [CrossRef] [PubMed]
- Mourão, P.A. Perspective on the use of sulfated polysaccharides from marine organisms as a source of new antithrombotic drugs. Mar. Drugs 2015, 13, 2770–2784. [Google Scholar] [CrossRef]
- Pomin, V.H. A Dilemma in the Glycosaminoglycan-Based Therapy: Synthetic or Naturally Unique Molecules? Med. Res. Rev. 2015, 35, 1195–1219. [Google Scholar] [CrossRef]
- Bose, S.; Banerjee, A.K. Role of heparan sulfate in human parainfluenza virus type 3 infection. Virology 2002, 298, 73–83. [Google Scholar] [CrossRef]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef]
- Vivès, R.R.; Imberty, A.; Sattentau, Q.J.; Lortat-Jacob, H. Heparan sulfate targets the HIV-1 envelope glycoprotein gp120 coreceptor binding site. J. Biol. Chem. 2005, 280, 21353–21357. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Spear, P.G. Herpesviruses and heparan sulfate: An intimate relationship in aid of viral entry. J. Clin. Investig. 2001, 108, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Sharp, J.S.; Zhang, F.; Pomin, V.H.; Ashpole, N.M.; Mitra, D.; McCandless, M.G.; Jin, W.; Liu, H.; Sharma, P.; et al. Effective Inhibition of SARS-CoV-2 Entry by Heparin and Enoxaparin Derivatives. J. Virol. 2021, 95, e01987-20. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jin, W.; Sood, A.; Montgomery, D.W.; Grant, O.C.; Fuster, M.M.; Fu, L.; Dordick, J.S.; Woods, R.J.; Zhang, F.; et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral. Res. 2020, 181, 104873. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.; Maciorowski, D.; Zak, S.E.; Kulkarni, C.V.; Herbert, A.S.; Durvasula, R.; Fareed, J.; Dye, J.M.; Kempaiah, P. Heparin: A simplistic repurposing to prevent SARS-CoV-2 transmission in light of its in-vitro nanomolar efficacy. Int. J. Biol. Macromol. 2021, 183, 203–212. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Gandhi, N.S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, R.; Samanta, P.; Sharma, P.; Zhang, F.; Mishra, S.K.; Kucheryavy, P.; Kim, S.B.; Aderibigbe, A.O.; Linhardt, R.J.; Tandon, R.; et al. Structural and kinetic analyses of holothurian sulfated glycans suggest potential treatment for SARS-CoV-2 infection. J. Biol. Chem. 2021, 297, 101207. [Google Scholar] [CrossRef] [PubMed]
- Kearns, F.L.; Sandoval, D.R.; Casalino, L.; Clausen, T.M.; Rosenfeld, M.A.; Spliid, C.B.; Amaro, R.E.; Esko, J.D. Spike-heparan sulfate interactions in SARS-CoV-2 infection. Curr. Opin. Struct. Biol. 2022, 76, 102439. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chopra, P.; Tomris, I.; van der Woude, R.; Liu, L.; de Vries, R.P.; Boons, G.J. Well-Defined Heparin Mimetics Can Inhibit Binding of the Trimeric Spike of SARS-CoV-2 in a Length-Dependent Manner. JACS Au 2023, 3, 1185–1195. [Google Scholar] [CrossRef]
- Kim, S.H.; Kearns, F.L.; Rosenfeld, M.A.; Votapka, L.; Casalino, L.; Papanikolas, M.; Amaro, R.E.; Freeman, R. SARS-CoV-2 evolved variants optimize binding to cellular glycocalyx. Cell Rep. Phys. Sci. 2023, 4, 101346. [Google Scholar] [CrossRef] [PubMed]
- Hirsh, J.; Anand, S.S.; Halperin, J.L.; Fuster, V.; Association, A.H. AHA Scientific Statement: Guide to anticoagulant therapy: Heparin: A statement for healthcare professionals from the American Heart Association. Arterioscler. Thromb. Vasc. Biol. 2001, 21, E9. [Google Scholar] [CrossRef]
- Hirsh, J.; Anand, S.S.; Halperin, J.L.; Fuster, V.; Association, A.H. Guide to anticoagulant therapy: Heparin: A statement for healthcare professionals from the American Heart Association. Circulation 2001, 103, 2994–3018. [Google Scholar] [CrossRef]
- Warkentin, T.E.; Levine, M.N.; Hirsh, J.; Horsewood, P.; Roberts, R.S.; Gent, M.; Kelton, J.G. Heparin-induced thrombocytopenia in patients treated with low-molecular-weight heparin or unfractionated heparin. N. Engl. J. Med. 1995, 332, 1330–1335. [Google Scholar] [CrossRef]
- Pomin, V.H. Holothurian fucosylated chondroitin sulfate. Mar. Drugs 2014, 12, 232–254. [Google Scholar] [CrossRef]
- Soares, P.A.G.; Ribeiro, K.A.; Valente, A.P.; Capillé, N.V.; Oliveira, S.M.C.G.; Tovar, A.M.F.; Pereira, M.S.; Vilanova, E.; Mourão, P.A.S. A unique fucosylated chondroitin sulfate type II with strikingly homogeneous and neatly distributed α-fucose branches. Glycobiology 2018, 28, 565–579. [Google Scholar] [CrossRef]
- Chen, S.; Hu, Y.; Ye, X.; Li, G.; Yu, G.; Xue, C.; Chai, W. Sequence determination and anticoagulant and antithrombotic activities of a novel sulfated fucan isolated from the sea cucumber Isostichopus badionotus. Biochim. Biophys. Acta 2012, 1820, 989–1000. [Google Scholar] [CrossRef]
- Nagle, V.; Gaikwad, M.; Pawar, Y.; Dasgupta, S. Marine red alga Porphyridium sp. as a source of sulfated polysaccharides (SPs) for combating against COVID-19. Preprints 2020, 2020040168. [Google Scholar]
- Mottarella, S.E.; Beglov, D.; Beglova, N.; Nugent, M.A.; Kozakov, D.; Vajda, S. Docking server for the identification of heparin binding sites on proteins. J. Chem. Inf. Model. 2014, 54, 2068–2078. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target Ther. 2021, 6, 396. [Google Scholar] [CrossRef]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef]
- Sanda, M.; Morrison, L.; Goldman, R. N- and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 2021, 93, 2003–2009. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, W.; Mao, Y.; Chen, Y.; Wang, S.; Zhong, Y.; Su, T.; Gong, M.; Du, D.; Lu, X.; et al. Site-specific N-glycosylation Characterization of Recombinant SARS-CoV-2 Spike Proteins. Mol. Cell. Proteom. 2021, 20, 100058. [Google Scholar] [CrossRef]
- Woo, H.; Park, S.J.; Choi, Y.K.; Park, T.; Tanveer, M.; Cao, Y.; Kern, N.R.; Lee, J.; Yeom, M.S.; Croll, T.I.; et al. Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J. Phys. Chem. B 2020, 124, 7128–7137. [Google Scholar] [CrossRef]
- Fiorentini, S.; Messali, S.; Zani, A.; Caccuri, F.; Giovanetti, M.; Ciccozzi, M.; Caruso, A. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect. Dis. 2021, 21, e147. [Google Scholar] [CrossRef]
- Luan, B.; Wang, H.; Huynh, T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: Insights from molecular dynamics simulations. FEBS Lett. 2021, 595, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Uciechowska-Kaczmarzyk, U.; Chauvot de Beauchene, I.; Samsonov, S.A. Docking software performance in protein-glycosaminoglycan systems. J. Mol. Graph. Model. 2019, 90, 42–50. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Pisabarro, M.T. Computational analysis of interactions in structurally available protein–glycosaminoglycan complexes. Glycobiology 2016, 26, 850–861. [Google Scholar] [CrossRef]
- Simulations Plus, Inc. Lancaster, CA, USA. Available online: http://www.simulations-plus.com (accessed on 29 August 2023).
- Mishra, S.K.; Koča, J. Assessing the performance of MM/PBSA, MM/GBSA, and QM–MM/GBSA approaches on protein/carbohydrate complexes: Effect of implicit solvent models, QM methods, and entropic contributions. J. Phys. Chem. B 2018, 122, 8113–8121. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Zoepfl, M.; Samanta, P.; Zhang, F.; Xia, K.; Thara, R.; Linhardt, R.J.; Doerksen, R.J.; McVoy, M.A.; Pomin, V.H. Fractionation of sulfated galactan from the red alga Botryocladia occidentalis separates its anticoagulant and anti-SARS-CoV-2 properties. J. Biol. Chem. 2022, 298, 101856. [Google Scholar] [CrossRef] [PubMed]
- Maurya, A.K.; Sharma, P.; Samanta, P.; Shami, A.A.; Misra, S.K.; Zhang, F.; Thara, R.; Kumar, D.; Shi, D.; Linhardt, R.J. Structure, anti-SARS-CoV-2, and anticoagulant effects of two sulfated galactans from the red alga Botryocladia occidentalis. Int. J. Biol. Macromol. 2023, 238, 124168. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.J.; et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 2020, 370, 426–431. [Google Scholar] [CrossRef]
- Carino, A.; Moraca, F.; Fiorillo, B.; Marchianò, S.; Sepe, V.; Biagioli, M.; Finamore, C.; Bozza, S.; Francisci, D.; Distrutti, E.; et al. Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain. Front. Chem. 2020, 8, 572885. [Google Scholar] [CrossRef]
- Schuurs, Z.P.; Hammond, E.; Elli, S.; Rudd, T.R.; Mycroft-West, C.J.; Lima, M.A.; Skidmore, M.A.; Karlsson, R.; Chen, Y.H.; Bagdonaite, I.; et al. Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Comput. Struct. Biotechnol. J. 2021, 19, 2806–2818. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W.; Mackerell, A.D. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef]
- Singh, A.; Tessier, M.B.; Pederson, K.; Wang, X.; Venot, A.P.; Boons, G.J.; Prestegard, J.H.; Woods, R.J. Extension and validation of the GLYCAM force field parameters for modeling glycosaminoglycans. Can. J. Chem. 2016, 94, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Lee, J.; Qi, Y.; Kern, N.R.; Lee, H.S.; Jo, S.; Joung, I.; Joo, K.; Im, W. CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 2019, 29, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Fadda, E. Molecular simulations of complex carbohydrates and glycoconjugates. Curr. Opin. Chem. Biol. 2022, 69, 102175. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- Nagarajan, B.; Sankaranarayanan, N.V.; Desai, U.R. Rigorous analysis of free solution glycosaminoglycan dynamics using simple, new tools. Glycobiology 2020, 30, 516–527. [Google Scholar] [CrossRef]
- Kim, S.H.; Kearns, F.L.; Rosenfeld, M.A.; Casalino, L.; Papanikolas, M.J.; Simmerling, C.; Amaro, R.E.; Freeman, R. Cell Surface-Inspired Universal Sensor for Betacoronaviruses. ACS Cent. Sci. 2022, 8, 22–42. [Google Scholar] [CrossRef]
- Paiardi, G.; Richter, S.; Oreste, P.; Urbinati, C.; Rusnati, M.; Wade, R.C. The binding of heparin to spike glycoprotein inhibits SARS-CoV-2 infection by three mechanisms. J. Biol. Chem. 2022, 298, 101507. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Pepi, L.E.; Rouhani, D.S.; Heiss, C.; Azadi, P. Glycosylation of SARS-CoV-2: Structural and functional insights. Anal. Bioanal. Chem. 2021, 413, 7179–7193. [Google Scholar] [CrossRef]
- Hsu, Y.P.; Frank, M.; Mukherjee, D.; Shchurik, V.; Makarov, A.; Mann, B.F. Structural remodeling of SARS-CoV-2 spike protein glycans reveals the regulatory roles in receptor-binding affinity. Glycobiology 2023, 33, 126–137. [Google Scholar] [CrossRef]
- Allen, J.D.; Chawla, H.; Samsudin, F.; Zuzic, L.; Shivgan, A.T.; Watanabe, Y.; He, W.T.; Callaghan, S.; Song, G.; Yong, P.; et al. Site-specific steric control of SARS-CoV-2 spike glycosylation. Biochemistry 2021, 60, 2153–2169. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-2: Glide; Schrödinger, LLC: New York, NY, USA, 2021.
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Mallajosyula, S.S.; Jo, S.; Im, W.; MacKerell, A.D. Molecular dynamics simulations of glycoproteins using CHARMM. Methods Mol. Biol. 2015, 1273, 407–429. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Cheatham, T.I.; Miller, J.; Fox, T.; Darden, T.; Kollman, P. Molecular dynamics simulations on solvated biomolecular systems: The particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995, 117, 4193–4194. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.v.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2015.
- Schrödinger Release 2021-2: Maestro; Schrödinger, LLC: New York, NY, USA, 2021.
- Nagae, M.; Mishra, S.K.; Neyazaki, M.; Oi, R.; Ikeda, A.; Matsugaki, N.; Akashi, S.; Manya, H.; Mizuno, M.; Yagi, H.; et al. 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy. Genes Cells 2017, 22, 348–359. [Google Scholar] [CrossRef]
- Hawkins, G.D.; Cramer, C.J.; Truhlar, D.G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 1996, 100, 19824–19839. [Google Scholar] [CrossRef]
- Tian, M.; Li, H.; Yan, X.; Gu, J.; Zheng, P.; Luo, S.; Zhangsun, D.; Chen, Q.; Ouyang, Q. Application of per-Residue Energy Decomposition to Design Peptide Inhibitors of PSD95 GK Domain. Front. Mol. Biosci. 2022, 9, 848353. [Google Scholar] [CrossRef] [PubMed]



| Non-Glycosylated RBD | ||||||
|---|---|---|---|---|---|---|
| Ligand | VdWaals | Elec. | EGB | ESurf | ∆Total | |
| S1 | PpFucCS1 | −14.23 | −273.2 | 274.3 | −1.850 | −14.94 |
| PpFucCS2 | −25.54 | −303.5 | 303.2 | −3.454 | −29.30 | |
| PpFucCS3 | −22.51 | −314.3 | 318.2 | −2.789 | −21.43 | |
| IbSF | −34.51 | −267.6 | 270.6 | −3.287 | −34.81 | |
| S5 | PpFucCS1 | −23.27 | −447.9 | 442.9 | −3.012 | −31.25 |
| PpFucCS2 | −27.15 | −367.6 | 369.0 | −3.214 | −28.95 | |
| PpFucCS3 | −36.74 | −501.6 | 491.9 | −4.417 | −50.80 | |
| IbSF | −36.49 | −431.4 | 436.4 | −4.046 | −35.50 | |
| Glycosylated RBD | ||||||
| S1 | PpFucCS1 | −24.92 | −304.7 | 311.1 | −3.124 | −21.63 |
| PpFucCS2 | −24.36 | −326.0 | 322.6 | −3.633 | −31.30 | |
| PpFucCS3 | −25.01 | −322.6 | 325.0 | −3.187 | −25.83 | |
| IbSF | −35.03 | −266.8 | 270.3 | −3.329 | −34.92 | |
| S5 | PpFucCS1 | −29.86 | −474.5 | 469.6 | −3.548 | −38.30 |
| PpFucCS2 | −16.23 | −293.8 | 293.1 | −2.097 | −18.98 | |
| PpFucCS3 | −32.63 | −466.3 | 459.3 | −3.974 | −43.58 | |
| IbSF | −35.77 | −416.5 | 419.3 | −3.844 | −36.82 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, P.; Mishra, S.K.; Pomin, V.H.; Doerksen, R.J. Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein. Molecules 2023, 28, 6413. https://doi.org/10.3390/molecules28176413
Samanta P, Mishra SK, Pomin VH, Doerksen RJ. Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein. Molecules. 2023; 28(17):6413. https://doi.org/10.3390/molecules28176413
Chicago/Turabian StyleSamanta, Priyanka, Sushil K. Mishra, Vitor H. Pomin, and Robert J. Doerksen. 2023. "Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein" Molecules 28, no. 17: 6413. https://doi.org/10.3390/molecules28176413
APA StyleSamanta, P., Mishra, S. K., Pomin, V. H., & Doerksen, R. J. (2023). Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein. Molecules, 28(17), 6413. https://doi.org/10.3390/molecules28176413








