Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins
Abstract
1. Introduction
2. An Overview of the Laccase Structure
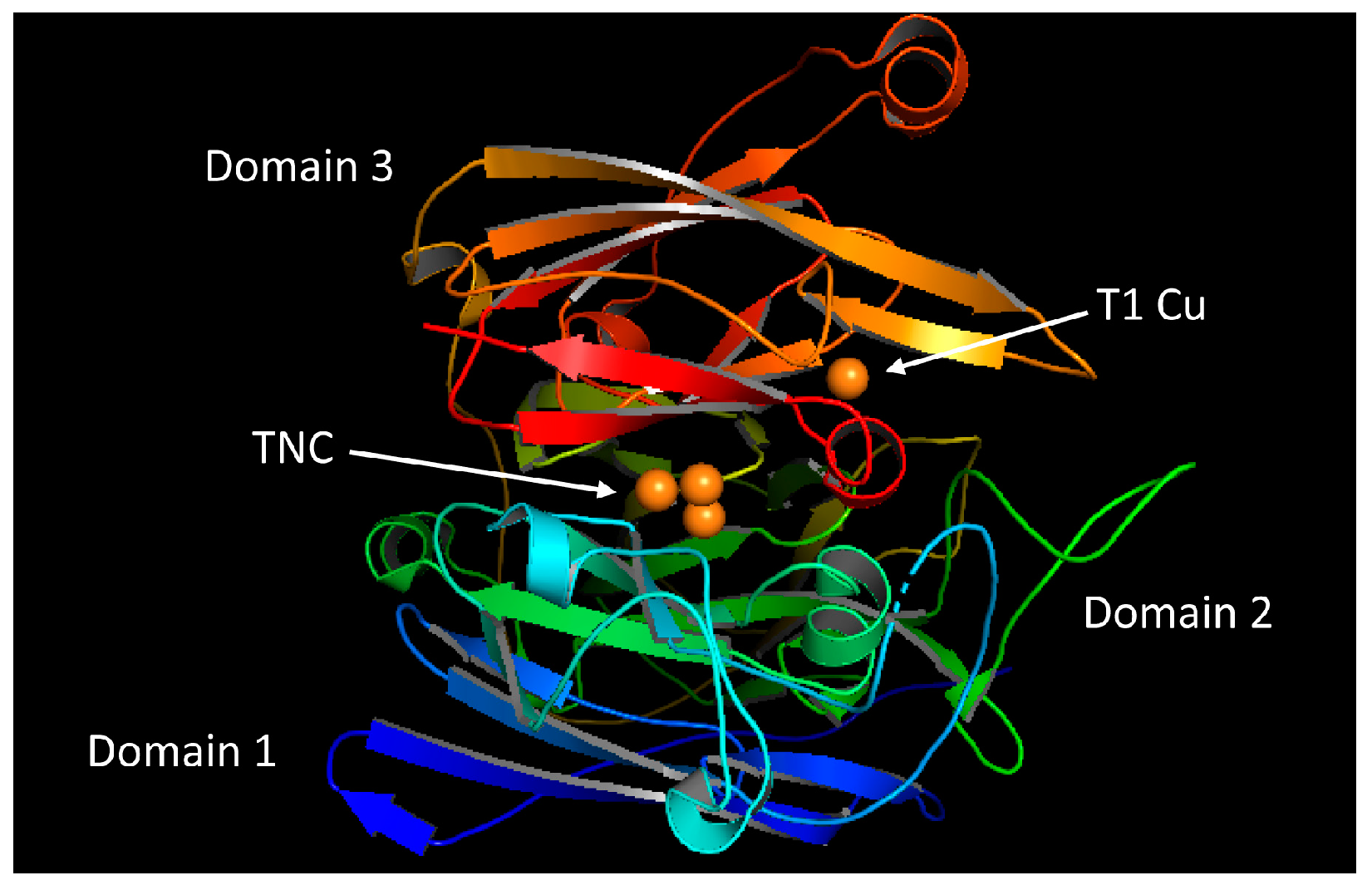
2.1. Overall Structure of the Laccase
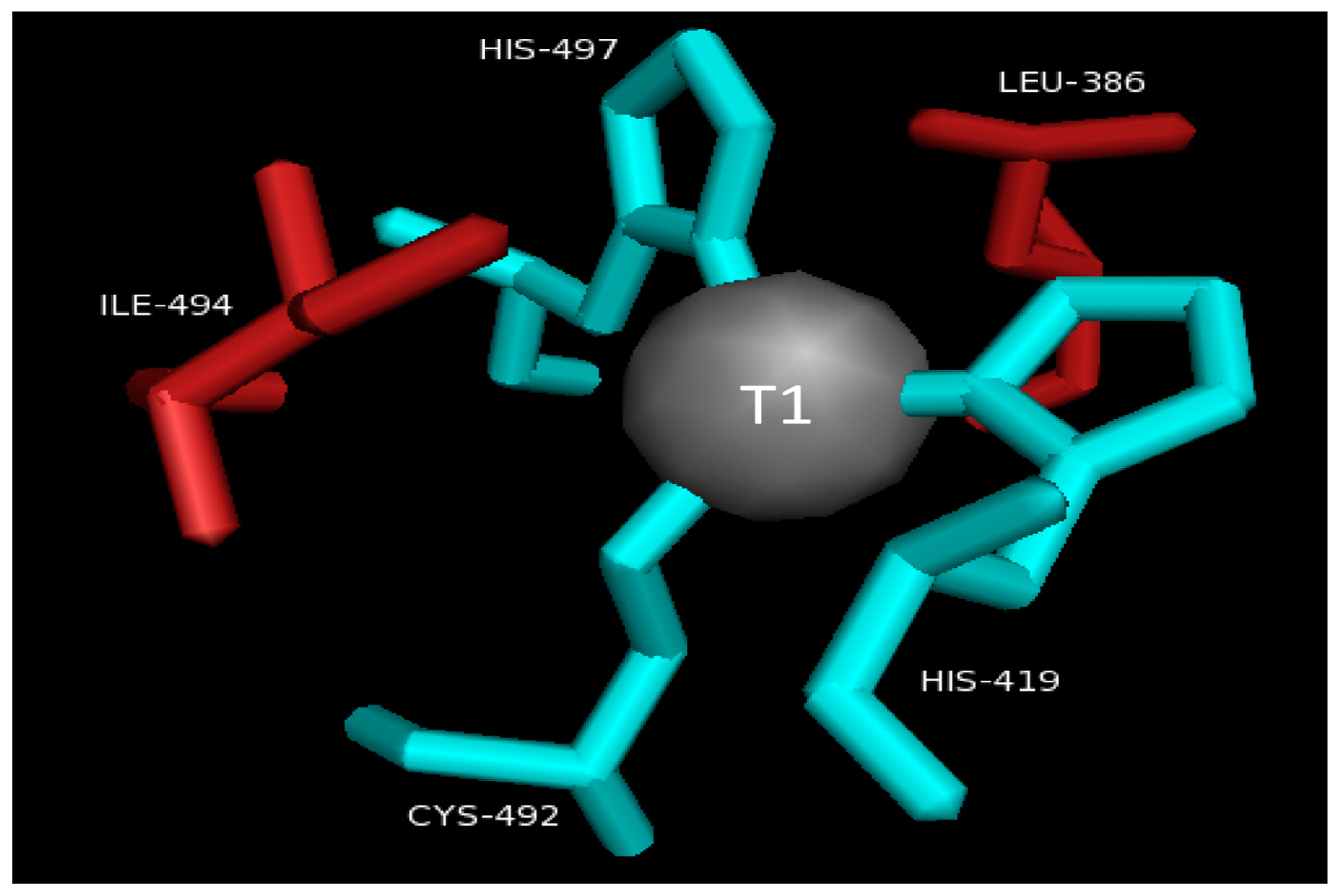
2.2. T1 Site
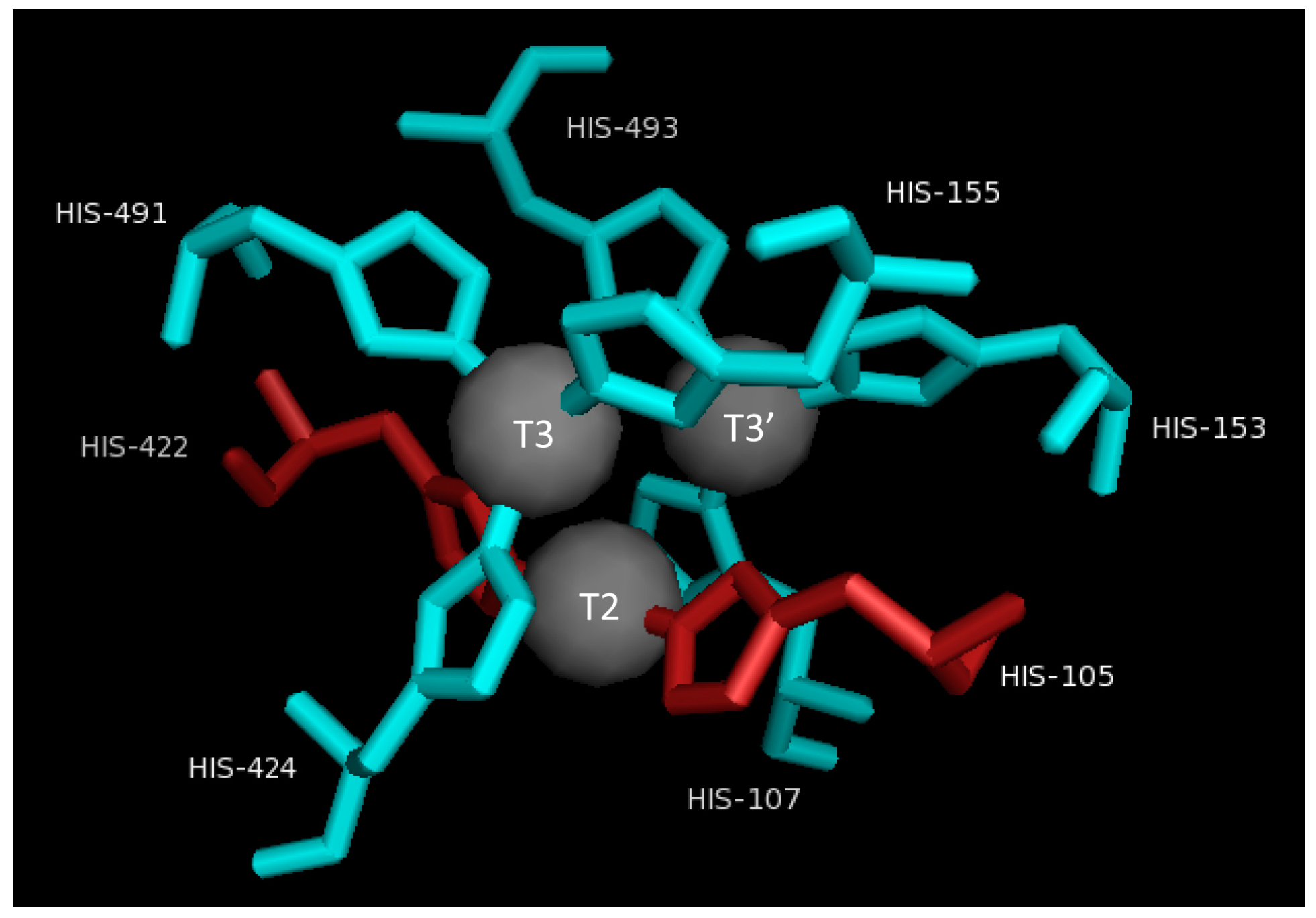
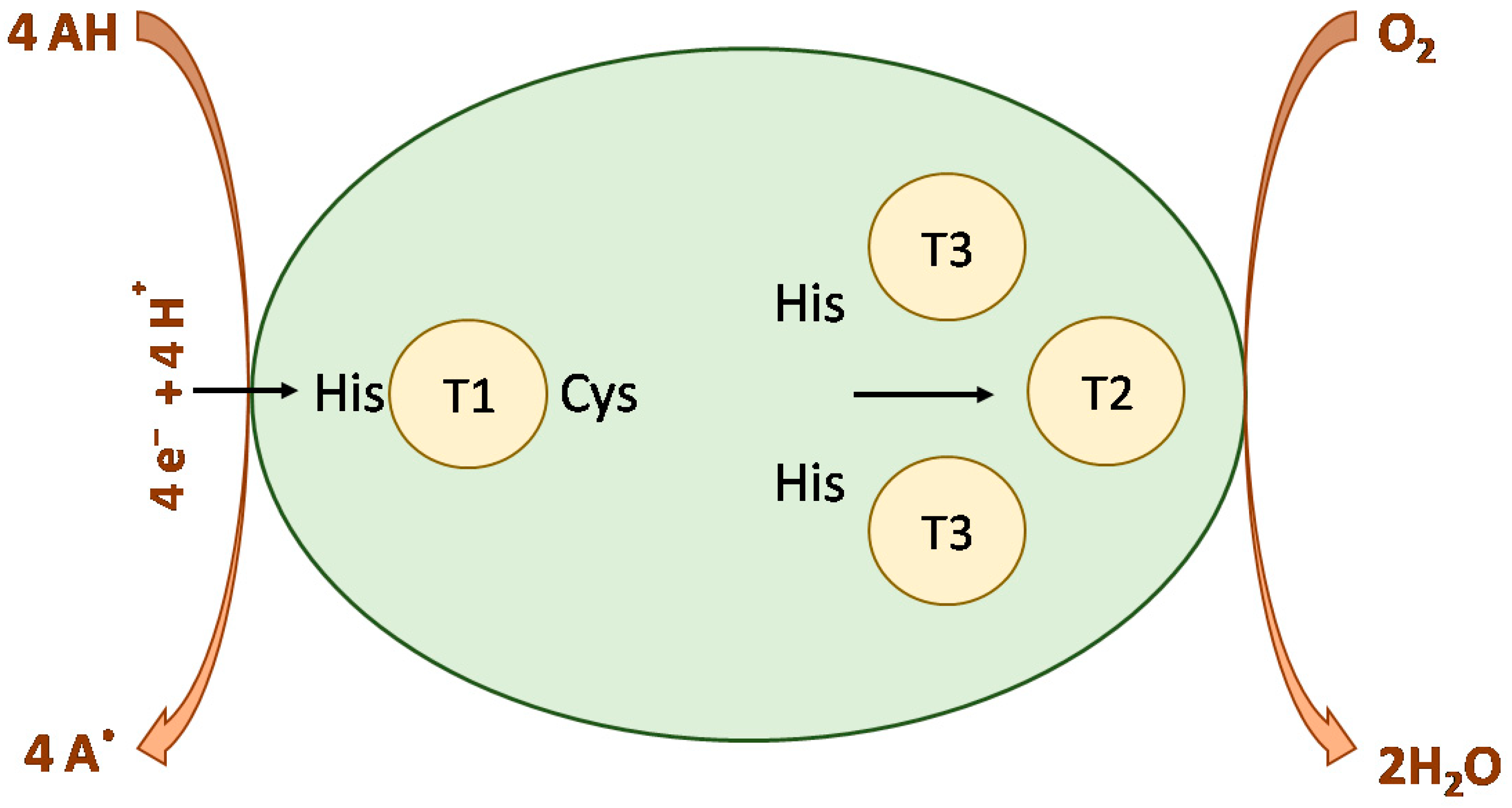
2.3. T2/T3 Site
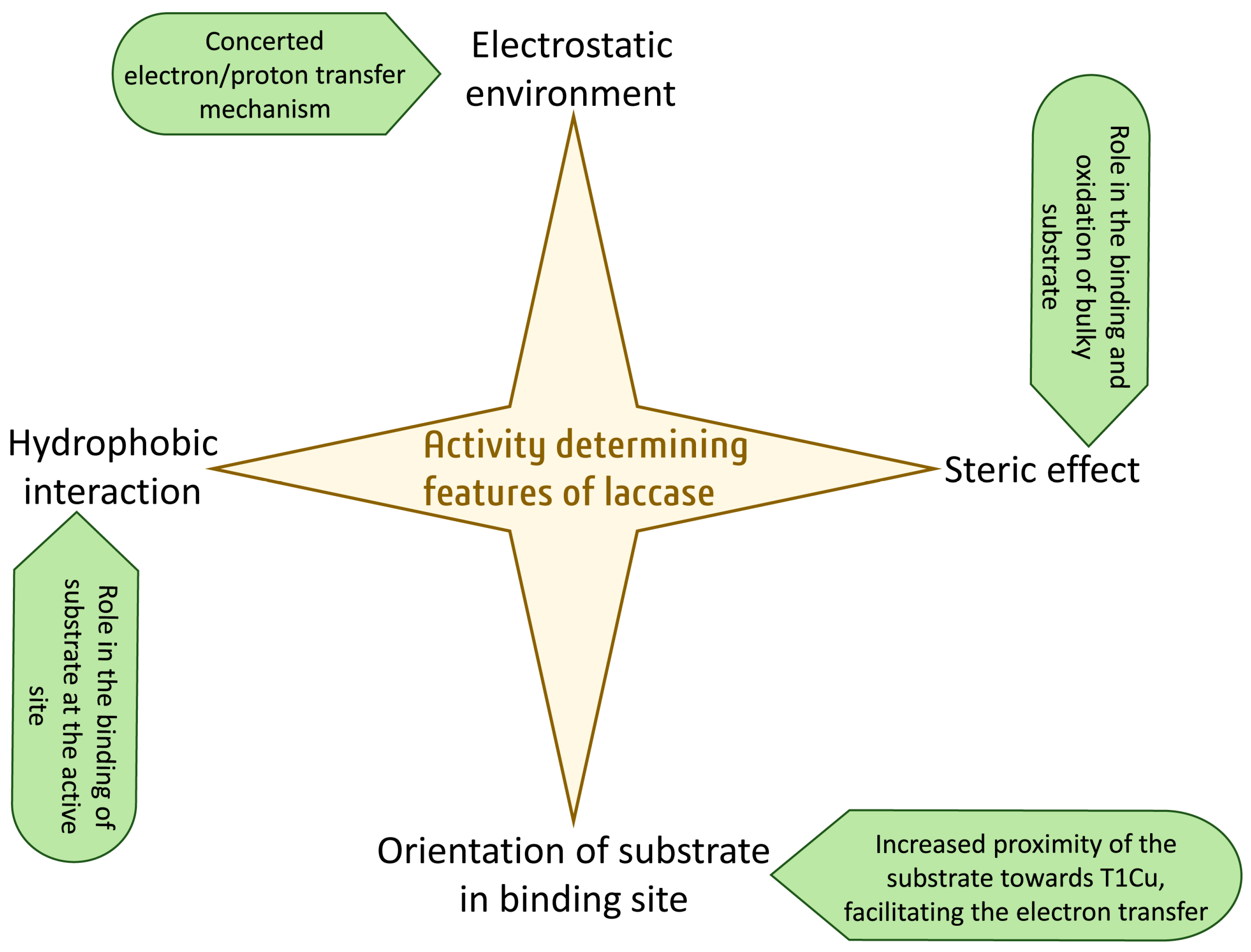
3. Characteristics That Determine Activity Other Than the Redox Potential
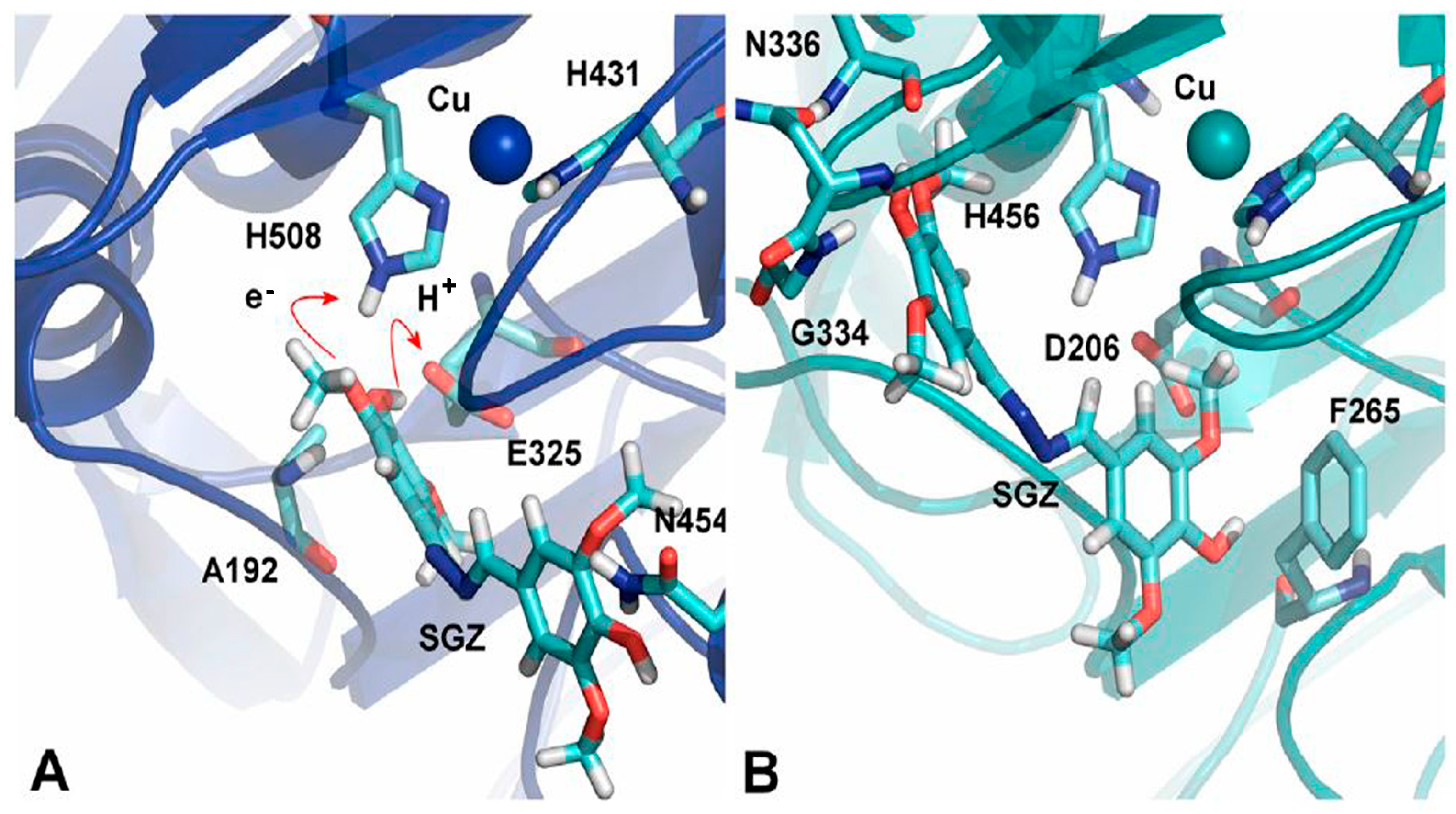
3.1. Electrostatic Environment of the Enzyme Pocket
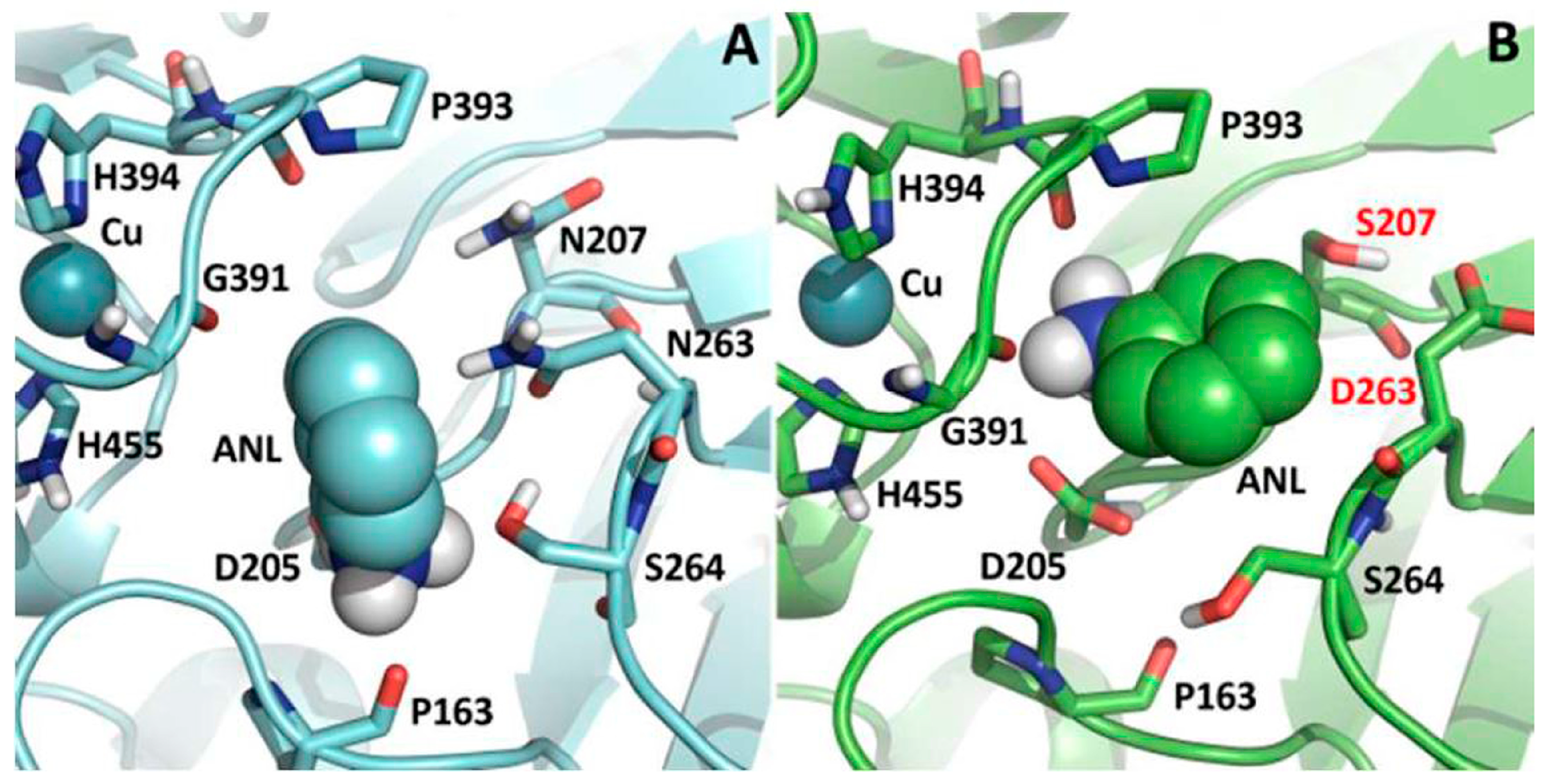
3.2. Steric Hindrance Due to Bulky Structures
3.3. Orientation of Substrate in Binding Site
3.4. Hydrophobic Environment of the Enzyme Pocket
4. Conclusions and Future Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Janusz, G.; Pawlik, A.; Swiderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkolazka, A.; Paszczynski, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper Oxidases and Oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef]
- Kumar, A.; Ahlawat, S.; Mohan, H.; Sharma, K.K. Stabilization-destabilization and redox properties of laccases from medicinal mushroom Ganoderma lucidum and human pathogen Yersinia enterocolitica. Int. J. Biol. Macromol. 2021, 167, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.I.; Chen, P.; Metz, M.; Lee, S.K.; Palmer, A.E. Oxygen Binding, Activation, and Reduction to Water by Copper Proteins. Angew. Chem. Int. Ed. 2001, 40, 4570–4590. [Google Scholar] [CrossRef]
- Bassanini, I.; Ferrandi, E.E.; Riva, S.; Monti, D. Biocatalysis with laccases: An updated overview. Catalysts 2020, 11, 26. [Google Scholar] [CrossRef]
- Ayodeji, F.D.; Shava, B.; Iqbal, H.M.N.; Ashraf, S.S.; Cui, J.D.; Franco, M.; Bilal, M. Biocatalytic Versatilities and Biotechnological Prospects of Laccase for a Sustainable Industry. Catal. Lett. 2023, 153, 1932–1956. [Google Scholar] [CrossRef]
- Cambria, M.T.; Gullotto, D.; Garavaglia, S.; Cambria, A. In silico study of structural determinants modulating the redox potential of Rigidoporus lignosus and other fungal laccases. J. Biomol. Struct. Dyn. 2012, 30, 89–101. [Google Scholar] [CrossRef]
- Chappell, H.A.; Milliken, A.; Farmer, C.; Wendland, N.; Coward, L.; Gregory, D.J.; Johnson, C.M. Efficient remediation of 17α-ethinylestradiol by Lentinula edodes (shiitake) laccase. Biocatal. Agric. Biotechnol. 2017, 10, 64–68. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Tringali, C. Laccase-mediated synthesis of bioactive natural products and their analogues. RSC Chem. Biol. 2022, 3, 614–647. [Google Scholar] [CrossRef]
- Witayakran, S.; Ragauskas, A.J. Synthetic Applications of Laccase in Green Chemistry. Adv.Synth. Catal. 2009, 351, 1187–1209. [Google Scholar] [CrossRef]
- Chen, Z.; Oh, W.D.; Yap, P.S. Recent advances in the utilization of immobilized laccase for the degradation of phenolic compounds in aqueous solutions: A review. Chemosphere 2022, 307, 135824. [Google Scholar] [CrossRef]
- Mayolo-Deloisa, K.; Gonzalez-Gonzalez, M.; Rito-Palomares, M. Laccases in Food Industry: Bioprocessing, Potential Industrial and Biotechnological Applications. Front. Bioeng. Biotechnol. 2020, 8, 222. [Google Scholar] [CrossRef]
- Stanzione, I.; Pezzella, C.; Giardina, P.; Sannia, G.; Piscitelli, A. Beyond natural laccases: Extension of their potential applications by protein engineering. Appl. Microbiol. Biotechnol. 2020, 104, 915–924. [Google Scholar] [CrossRef]
- Backes, E.; Kato, C.G.; Corre, R.C.G.; Moreira, R.D.P.M.; Peralta, R.A.; Barros, L.; Ferreira, I.C.F.R.; Zanin, G.M.; Bracht, A.; Peralta, R.M. Laccases in food processing: Current status, bottlenecks and perspectives. Trends Food Sci. Technol. 2021, 115, 445–460. [Google Scholar] [CrossRef]
- Minussi, R.C.; Pastore, G.M.; Duran, N. Potential applications of laccase in the food industry. Trends Food Sci. Technol. 2002, 13, 205–216. [Google Scholar] [CrossRef]
- Hussain, A.; Bilal, M.; Rafeeq, H.; Jabeen, Z.; Afsheen, N.; Sher, F.; Kumar, V.; Bharagava, R.N.; Ferreira, L.F.R.; Iqbal, H.M. Role of laccase in the pulp and paper industry. In Nanotechnology in Paper and Wood Engineering; Elsevier: Faisalabad, Pakistan, 2022; pp. 35–60. [Google Scholar]
- Unuofin, J.O.; Falade, A.O.; Aladekoyi, O.J. Applications of microbial laccases in bioremediation of environmental pollutants: Potential issues, challenges, and prospects. Bioremediat. Environ. Sustain. 2021, 519–540. [Google Scholar] [CrossRef]
- Christenson, A.; Dimcheva, N.; Ferapontova, E.E.; Gorton, L.; Ruzgas, T.; Stoica, L.; Shleev, S.; Yaropolov, A.L.; Haltrich, D.; Thorneley, R.N.F.; et al. Direct electron transfer between ligninolytic redox enzymes and electrodes. Electroanalysis 2004, 16, 1074–1092. [Google Scholar] [CrossRef]
- Reinhammar, B.R. Oxidation-reduction potentials of the electron acceptors in laccases and stellacyanin. Biochim. Biophys. Acta 1972, 275, 245–259. [Google Scholar] [CrossRef]
- Xu, F.; Shin, W.S.; Brown, S.H.; Wahleithner, J.A.; Sundaram, U.M.; Solomon, E.I. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta 1996, 1292, 303–311. [Google Scholar] [CrossRef]
- Klonowska, A.; Gaudin, C.; Fournel, A.; Asso, M.; Le Petit, J.; Giorgi, M.; Tron, T. Characterization of a low redox potential laccase from the basidiomycete C30. Eur. J. Biochem. 2002, 269, 6119–6125. [Google Scholar] [CrossRef]
- Shleev, S.V.; Morozova, O.V.; Nikitina, O.V.; Gorshina, E.S.; Rusinova, T.V.; Serezhenkov, V.A.; Burbaev, D.S.; Gazaryan, I.G.; Yaropolov, A.I. Comparison of physico-chemical characteristics of four laccases from different basidiomycetes. Biochimie 2004, 86, 693–703. [Google Scholar] [CrossRef]
- Olbrich, A.C.; Schild, J.N.; Urlacher, V.B. Correlation between the T1 copper reduction potential and catalytic activity of a small laccase. J. Inorg. Biochem. 2019, 201, 110843. [Google Scholar] [CrossRef]
- Mateljak, I.; Monza, E.; Lucas, M.F.; Guallar, V.; Aleksejeva, O.; Ludwig, R.; Leech, D.; Shleev, S.; Alcalde, M. Increasing Redox Potential, Redox Mediator Activity, and Stability in a Fungal Laccase by Computer-Guided Mutagenesis and Directed Evolution. ACS Catal. 2019, 9, 4561–4572. [Google Scholar] [CrossRef]
- Barber-Zucker, S.; Mateljak, I.; Goldsmith, M.; Kupervaser, M.; Alcalde, M.; Fleishman, S.J. Designed High-Redox Potential Laccases Exhibit High Functional Diversity. ACS Catal. 2022, 12, 13164–13173. [Google Scholar] [CrossRef]
- Jiang, Q.; Cui, Z.; Wei, R.; Nie, K.; Xu, H.; Liu, L. Feasible Cluster Model Method for Simulating the Redox Potentials of Laccase CueO and Its Variant. Front. Bioeng. Biotechnol. 2022, 10, 957694. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ouyang, X.; Li, J.; Zhao, Y.L. Natural Syringyl Mediators Accelerate Laccase-Catalyzed beta-O-4 Cleavage and Calpha-Oxidation of a Guaiacyl Model Substrate via an Aggregation Mechanism. ACS Omega 2021, 6, 22578–22588. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Pardo, I.; Canas, A.I.; Molina, P.; Record, E.; Martinez, A.T.; Martinez, M.J.; Alcalde, M. Engineering platforms for directed evolution of Laccase from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 2012, 78, 1370–1384. [Google Scholar] [CrossRef]
- Camarero, S.; Ibarra, D.; Martinez, A.T.; Romero, J.; Gutierrez, A.; del Rio, J.C. Paper pulp delignification using laccase and natural mediators. Enzym. Microb. Technol. 2007, 40, 1264–1271. [Google Scholar] [CrossRef]
- Ayala, M. Redox Potential of Peroxidases. In Biocatalysis Based on Heme Peroxidases; Torres, E., Ayala, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 61–77. [Google Scholar]
- Gu, Y.; Yuan, L.; Jia, L.; Xue, P.; Yao, H. Recent developments of a co-immobilized laccase-mediator system: A review. RSC Adv. 2021, 11, 29498–29506. [Google Scholar] [CrossRef]
- Aza, P.; de Salas, F.; Molpeceres, G.; Rodriguez-Escribano, D.; de la Fuente, I.; Camarero, S. Protein Engineering Approaches to Enhance Fungal Laccase Production in S. cerevisiae. Int. J. Mol. Sci. 2021, 22, 1157. [Google Scholar] [CrossRef]
- Rivera-Hoyos, C.M.; Morales-Alvarez, E.D.; Poutou-Pinales, R.A.; Pedroza-Rodriguez, A.M.; Rodriguez-Vazquez, R.; Delgado-Boada, J.M. Fungal laccases. Fungal Biol. Rev. 2013, 27, 67–82. [Google Scholar] [CrossRef]
- Pardo, I.; Camarero, S. Laccase engineering by rational and evolutionary design. Cell. Mol. Life Sci. 2015, 72, 897–910. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gomez-Gil, X.; Gutierrez-Soto, G.; Hernandez-Luna, C.E.; Herrera de Los Santos, M.; Levin, L.; Rojo-Dominguez, A.; Romero-Martinez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Factories 2019, 18, 200. [Google Scholar] [CrossRef] [PubMed]
- Glazunova, O.A.; Trushkin, N.A.; Moiseenko, K.V.; Filimonov, I.S.; Fedorova, T.V. Catalytic Efficiency of Basidiomycete Laccases: Redox Potential versus Substrate-Binding Pocket Structure. Catalysts 2018, 8, 152. [Google Scholar] [CrossRef]
- Leontievsky, A.A.; Vares, T.; Lankinen, P.; Shergill, J.K.; Pozdnyakova, N.N.; Myasoedova, N.M.; Kalkkinen, N.; Golovleva, L.A.; Cammack, R.; Thurston, C.F.; et al. Blue and yellow laccases of ligninolytic fungi. FEMS Microbiol. Lett. 1997, 156, 9–14. [Google Scholar] [CrossRef]
- Koroljova-Skorobogat’ko, O.V.; Stepanova, E.V.; Gavrilova, V.P.; Morozova, O.V.; Lubimova, N.V.; Dzchafarova, A.N.; Jaropolov, A.I.; Makower, A. Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus and effect of inducers on laccase synthesis. Biotechnol. Appl. Biochem. 1998, 28, 47–54. [Google Scholar]
- Shin, K.S.; Lee, Y.J. Purification and characterization of a new member of the laccase family from the white-rot basidiomycete Coriolus hirsutus. Arch. Biochem. Biophys. 2000, 384, 109–115. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef]
- Yaropolov, A.I.; Skorobogatko, O.V.; Vartanov, S.S.; Varfolomeyev, S.D. Laccase—Properties, Catalytic Mechanism, and Applicability. Appl. Biochem. 1994, 49, 257–280. [Google Scholar] [CrossRef]
- Gabdulkhakov, A.; Kolyadenko, I.; Kostareva, O.; Mikhaylina, A.; Oliveira, P.; Tamagnini, P.; Lisov, A.; Tishchenko, S. Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces griseoflavus Ac-993. Int. J. Mol. Sci. 2019, 20, 3184. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Solomon, E.I. Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. 2015, 72, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Andberg, M.; Kallio, J.; Koivula, A.; Kruus, K.; Rouvinen, J. A near atomic resolution structure of a Melanocar pusalbomyces laccase. J. Struct. Biol. 2008, 162, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Osipov, E.; Polyakov, K.; Kittl, R.; Shleev, S.; Dorovatovsky, P.; Tikhonova, T.; Hann, S.; Ludwig, R.; Popov, V. Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2913–2923. [Google Scholar] [CrossRef] [PubMed]
- Zhukhlistova, N.E.; Zhukova, Y.N.; Lyashenko, A.V.; Zaitsev, V.N.; Mikhailov, A.M. Three-dimensional organization of three-domain copper oxidases: A review. Crystallogr. Rep. 2008, 53, 92–109. [Google Scholar] [CrossRef]
- Serrano-Posada, H.; Centeno-Leija, S.; Rojas-Trejo, S.P.; Rodríguez-Almazán, C.; Stojanoff, V.; Rudiño-Piñera, E. X-ray-induced catalytic active-site reduction of a multicopper oxidase: Structural insights into the proton-relay mechanism and O2-reduction states. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2396–2411. [Google Scholar] [CrossRef]
- Sekretaryova, A.; Jones, S.M.; Solomon, E.I. O2 Reduction to Water by High Potential Multicopper Oxidases: Contributions of the T1 Copper Site Potential and the Local Environment of the Trinuclear Copper Cluster. J. Am. Chem. Soc. 2019, 141, 11304–11314. [Google Scholar] [CrossRef]
- Bento, I.; Silva, C.S.; Chen, Z.; Martins, L.O.; Lindley, P.F.; Soares, C.M. Mechanisms underlying dioxygen reduction in laccases. Structural and modelling studies focusing on proton transfer. BMC Struct. Biol. 2010, 10, 28. [Google Scholar] [CrossRef]
- Xu, F.; Berka, R.M.; Wahleithner, J.A.; Nelson, B.A.; Shuster, J.R.; Brown, S.H.; Palmer, A.E.; Solomon, E.I. Site-directed mutations in fungal laccase: Effect on redox potential, activity and pH profile. Biochem. J. 1998, 334 Pt 1, 63–70. [Google Scholar] [CrossRef]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry 2002, 41, 7325–7333. [Google Scholar] [CrossRef]
- Autore, F.; Del Vecchio, C.; Fraternali, F.; Giardina, P.; Sannia, G.; Faraco, V. Molecular determinants of peculiar properties of a Pleurotus ostreatus laccase: Analysis by site-directed mutagenesis. Enzym. Microb. Technol. 2009, 45, 507–513. [Google Scholar] [CrossRef]
- Galli, C.; Madzak, C.; Vadala, R.; Jolivalt, C.; Gentili, P. Concerted electron/proton transfer mechanism in the oxidation of phenols by laccase. Chembiochem 2013, 14, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Monza, E.; Lucas, M.F.; Camarero, S.; Alejaldre, L.C.; Martinez, A.T.; Guallar, V. Insights into Laccase Engineering from Molecular Simulations: Toward a Binding-Focused Strategy. J. Phys. Chem. Lett. 2015, 6, 1447–1453. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.; de Salas, F.; Lucas, M.F.; Monza, E.; Acebes, S.; Martinez, A.T.; Camarero, S.; Guallar, V. Computer-Aided Laccase Engineering: Toward Biological Oxidation of Arylamines. ACS Catal. 2016, 6, 5415–5423. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, M.; Zheng, Y.L.; Zhao, X.X.; Li, B.; Huan, W.W. Atomic-scale investigation of the interaction between coniferyl alcohol and laccase for lignin degradation using molecular dynamics simulations and spectroscopy. J. Dispers. Sci. Technol. 2019, 40, 686–694. [Google Scholar] [CrossRef]
- Loi, M.; Glazunova, O.; Fedorova, T.; Logrieco, A.F.; Mule, G. Fungal Laccases: The Forefront of Enzymes for Sustainability. J. Fungi 2021, 7, 1048. [Google Scholar] [CrossRef]
- Rochefort, D.; Leech, D.; Bourbonnais, R. Electron transfer mediator systems for bleaching of paper pulp. Green Chem. 2004, 6, 14–24. [Google Scholar] [CrossRef]
- Tadesse, M.A.; D’Annibale, A.; Galli, C.; Gentili, P.; Sergi, F. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 2008, 6, 868–878. [Google Scholar] [CrossRef]
- Moldes, D.; Diaz, M.; Tzanov, T.; Vidal, T. Comparative study of the efficiency of synthetic and natural mediators in laccase-assisted bleaching of eucalyptus kraft pulp. Bioresour. Technol. 2008, 99, 7959–7965. [Google Scholar] [CrossRef]
- Galli, C.; Gentili, P.; Jolivalt, C.; Madzak, C.; Vadalà, R. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 2011, 91, 123–131. [Google Scholar] [CrossRef]
- Prasad, N.K.; Vindal, V.; Narayana, S.L.; Ramakrishna, V.; Kunal, S.P.; Srinivas, M. In silico analysis of Pycnoporus cinnabarinus laccase active site with toxic industrial dyes. J. Mol. Model. 2012, 18, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Chinnathambi, V.; Arumugam, P.; Suresh, P.K. In silico and in vitro physicochemical screening of Rigidoporus sp. crude laccase-assisted decolorization of synthetic dyes--approaches for a cost-effective enzyme-based remediation methodology. Appl. Biochem. Biotechnol. 2013, 169, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, S.; Singh, D.; Virdi, J.S.; Sharma, K.K. Molecular modeling and MD-simulation studies: Fast and reliable tool to study the role of low-redox bacterial laccases in the decolorization of various commercial dyes. Environ. Pollut. 2019, 253, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Sadasivam, S.K.; Gunalan, S.; Shanmugam, G.; Kothandan, G. Application of docking and active site analysis for enzyme linked biodegradation of textile dyes. Environ. Pollut. 2019, 248, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Miele, A.; Giardina, P.; Notomista, E.; Piscitelli, A.; Sannia, G.; Faraco, V. A semi-rational approach to engineering laccase enzymes. Mol. Biotechnol. 2010, 46, 149–156. [Google Scholar] [CrossRef][Green Version]
- Toscano, M.D.; De Maria, L.; Lobedanz, S.; Ostergaard, L.H. Optimization of a small laccase by active-site redesign. Chembiochem 2013, 14, 1209–1211. [Google Scholar] [CrossRef]
- Xie, T.; Liu, Z.C.; Liu, Q.; Wang, G.G. Structural insight into the oxidation of sinapic acid by CotA laccase. J. Struct.Biol. 2015, 190, 155–161. [Google Scholar] [CrossRef]
- Pardo, I.; Santiago, G.; Gentili, P.; Lucas, F.; Monza, E.; Medrano, F.J.; Galli, C.; Martinez, A.T.; Guallar, V.; Camarero, S. Re-designing the substrate binding pocket of laccase for enhanced oxidation of sinapic acid. Catal. Sci. Technol. 2016, 6, 3900–3910. [Google Scholar] [CrossRef]
- Lucas, M.F.; Monza, E.; Jorgensen, L.J.; Ernst, H.A.; Piontek, K.; Bjerrum, M.J.; Martinez, A.T.; Camarero, S.; Guallar, V. Simulating Substrate Recognition and Oxidation in Laccases: From Description to Design. J. Chem. Theory Comput. 2017, 13, 1462–1467. [Google Scholar] [CrossRef]
- Mehra, R.; Meyer, A.S.; Kepp, K.P. Molecular dynamics derived life times of active substrate binding poses explain KM of laccase mutants. RSC Adv. 2018, 8, 36915–36926. [Google Scholar] [CrossRef]
- Conceicao, J.C.S.; Dias, H.J.; Peralva, C.M.S.; Crotti, A.E.M.; da Rocha Pita, S.S.; de Oliveira Silva, E. Phenolic Compound Biotransformation by Trametes versicolor ATCC 200801 and Molecular Docking Studies. Appl. Biochem. Biotechnol. 2020, 190, 1498–1511. [Google Scholar] [CrossRef] [PubMed]
- Cambria, M.T.; Di Marino, D.; Falconi, M.; Garavaglia, S.; Cambria, A. Docking Simulation and Competitive Experiments Validate the Interaction Between the 2,5-Xylidine Inhibitor and Rigidoporus lignosus Laccase. J. Biomol. 2010, 27, 501–509. [Google Scholar] [CrossRef]
- Gupta, N.; Lee, F.S.; Farinas, E.T. Laboratory evolution of laccase for substrate specificity. J. Mol. Catal. B Enzym. 2010, 62, 230–234. [Google Scholar] [CrossRef]
- Chen, M.; Zeng, G.M.; Lai, C.; Li, J.; Xu, P.; Wu, H.P. Molecular basis of laccase bound to lignin: Insight from comparative studies on the interaction of Trametes versicolor laccase with various lignin model compounds. RSC Adv. 2015, 5, 52307–52313. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, Q.; Zhou, W.; Xie, Z.; Cai, Y.J.; Liao, X.R.; Guan, Z.B. Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis. Appl. Microbiol. Biotechnol. 2017, 101, 1935–1944. [Google Scholar] [CrossRef]
- Kyomuhimbo, H.D.; Brink, H.G. Applications and immobilization strategies of the copper-centred laccase enzyme; a review. Heliyon 2023, 9, e13156. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Zeng, G.; Yuan, X.; Chen, M.; Hu, L.; Li, H.; Wang, H.; Xu, P.; Lai, C.; Wan, J.; et al. Molecular docking simulation on the interactions of laccase from Trametes versicolor with nonylphenol and octylphenol isomers. Bioprocess Biosyst. Eng. 2018, 41, 331–343. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhan, J.; Zhang, Y.; Lin, Y.; Yang, X. The K428 residue from Thermus thermophilus SG0.5JP17-16 laccase plays the substantial role in substrate binding and oxidation. J. Biomol. Struct. Dyn. 2021, 39, 1312–1320. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.; Bhardwaj, P.; Ishqi, H.M.; Shahid, M.; Islam, A. Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins. Molecules 2023, 28, 6209. https://doi.org/10.3390/molecules28176209
Ali M, Bhardwaj P, Ishqi HM, Shahid M, Islam A. Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins. Molecules. 2023; 28(17):6209. https://doi.org/10.3390/molecules28176209
Chicago/Turabian StyleAli, Misha, Priyanka Bhardwaj, Hassan Mubarak Ishqi, Mohammad Shahid, and Asimul Islam. 2023. "Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins" Molecules 28, no. 17: 6209. https://doi.org/10.3390/molecules28176209
APA StyleAli, M., Bhardwaj, P., Ishqi, H. M., Shahid, M., & Islam, A. (2023). Laccase Engineering: Redox Potential Is Not the Only Activity-Determining Feature in the Metalloproteins. Molecules, 28(17), 6209. https://doi.org/10.3390/molecules28176209







