Abstract
A multi-residue method was developed to identify and quantify pharmaceutical drug residues in full-fat milk, using a modified QuEChERS extraction procedure and sonication combined with Ultra-High-Performance Liquid Chromatography–High-Resolution Orbitrap Mass Spectrometry (UHPLC-HR-Orbitrap-MS). Sample preparation involves three different QuEChERS extraction procedures and sorbents for the purification step. The optimized modified extraction method, combined with the clean-up approaches using C18 and the EMR-Lipid sorbent, has been validated in terms of linearity, recovery, precision, LOD and LOQ, matrix effects (ME) and expanded uncertainty. The optimized method showed a linearity >0.9903, recoveries within the range 65.1–120.1%, precision (expressed as %RSD) <17.5%, medium (<39.9%) to low (<16.7%) matrix effects and acceptable expanded uncertainty (<33.1%). Finally, the proposed method was applied to representative real samples of milk (by local markets), revealing the existence of one pharmaceutical drug (imidocarb) in one sample.
1. Introduction
Pharmaceutical drugs are widely used in livestock production either for disease treatment, prophylaxis, or growth enhancement [1]. The incorrect use of these chemical compounds or improper withdrawal time may lead to the presence of residues in milk, and the consumption of contaminated milk or dairy products may expose humans to health hazards [2]. The European Union (EU) has set maximum residue limits (MRLs) for active pharmaceutical substances allowed in products of animals intended for human consumption to limit the risks related to the consumption of food containing residues of pharmaceutical drugs [3]. In addition, pharmaceuticals for human use can also be detected in food matrices as a result of their spread in environmental matrices mainly due to the disposal of biosolids in soils, the irrigation of recycled water, or from surface waters receiving wastewater treatment plant effluents. The detection of pharmaceutical drug residues in milk has been reported in previous studies [4,5,6,7,8]. Therefore, the study of pharmaceutical drug residues in food commodities, as well as the monitoring of compliance with MRLs, requires sensitive and rapid analytical methods capable of determining trace levels of a large number of pharmaceuticals in milk in a single analytical procedure.
Milk is a complex matrix containing various components, such as fat, proteins, and other components that can interfere with or prevent proper pharmaceutical drug analysis. Analytical methods for the determination of pharmaceutical drugs in milk have undergone considerable development in recent years. Milk sample preparation techniques based on liquid–liquid extraction (LLE) [9] have been replaced by other approaches, such as solid-phase extraction (SPE) [10,11], solid–liquid extraction (SLE) [5], matrix solid phase dispersion (MSPD) extraction [12,13], solid-phase microextraction (SPME) [14], and more recently by QuEChERS extraction [7,8,15,16,17,18,19].
QuEChERS is a quick, easy, cheap, effective, robust, and safe extraction compared to the above methods, which may present some difficulties related to time consumption and labor intensity, the low recovery of some target compounds, poor reproducibility, and method improvement [20]. Various modifications of QuEChERS extraction have been reported in the literature for the analysis of pharmaceutical drug residues in milk, using different sorbents for the clean-up step or a combination of them, e.g., octadecylsilane, (C18) as the usual sorbent to improve extraction on high-fat food matrices, primary secondary amine (PSA), and sodium acetate (NaOAc) [6,16,21,22]. Moreover, the Enhanced Matrix Removal-Lipid (EMR-Lipid) is a new product used as a dispersive SPE (dSPE), promising the highly selective removal of lipids, without the retention of the analyte. The EMR-Lipid sorbent was initially proposed for the analysis of pesticides in fatty matrices of plant origin [23]. It has subsequently been evaluated for the determination of several target analytes in other fatty matrices, such as bovine tissues (kidney, liver, and muscle) [24], milk [25], kale, salmon, avocado, and pork [26].
Regarding sonication, it is a technique that can effectively reduce the size of milk fat globules (MFG) via shearing, pressure fluctuations, and turbulence [27,28]. The vibrations generated by sonication can improve the extraction efficiency of the target compounds. The combination of extraction techniques can improve analyte recovery and can be efficiently applied to complex matrices such as milk. The application of the QuEChERS method in combination with ultrasound-assisted extraction for the analysis of pharmaceutical drugs in milk has been reported in some studies [29,30].
Until now, various analytical techniques have been used for the analysis of pharmaceutical drug residues in milk, mainly based on liquid chromatography (LC) systems combined with low-resolution (LR) mass spectrometry, such as LC ESI-MS or triple quadrupole LC-MS/MS (QqQ) [21,25,31,32,33,34]. The low-resolution (RL)-MS instruments mentioned above have several limitations, including a lower number of compounds that can be monitored in a single analysis, the limited ability to screen for unknown compounds, and a dependence on reference standards. LRMS requires a targeted approach involving a priori knowledge of the fragmentation pattern of the analytes (MS-MS product transitions along with optimized collision energies). On the other hand, high-resolution (HR) MS offers unique advantages, including the ability to screen samples with little or no knowledge of what is present and to create a digital data archive suitable for retrospective analysis. An LRMS measurement provides information on the nominal mass of the analyte, whereas HRMS measures the exact mass; i.e., the m/z for each ion is measured to four to six decimal places. Interferences can be discarded on the basis of accurate mass in HRMS instruments, and sample preparation procedures can be simplified, resulting in faster analytical methods. The high selectivity and mass accuracy of HRMS instrumentation can overcome false positives and false negatives when screening complex food samples [35,36].
In the last few years, high-resolution acquisition techniques have gained momentum, providing accurate mass measurements for both precursor and product ions. Liquid chromatography (LC)–high-resolution Orbitrap MS instruments offer a range of benefits for analytical applications. The use of the Orbitrap mass analyzer provides high-resolution, high-mass-accuracy, and high-quality MS/MS fragmentation, allowing the determination of an unlimited number of analytes in a single analytical run, even in complex matrices. A variety of screening methodologies can be used for residue analysis in food by LC-HRMS based on different objectives including targeted, suspected, and non-targeted or retrospective analysis. A non-targeted LC–HRMS approach for the analysis of unknown or unexpected sulfonamide residues in honey samples was investigated and optimized by Kırkan et al. [37], whereas an integrated nano LC-HR Orbitrap MS system employing a multiwalled carbon nanotube (MWCNT)-based monolithic stationary phase was applied for the analysis of antibiotics and pesticides in milk and honey by Aydogan et al. [38]. A full MS/all-ion fragmentation acquisition mode was applied in both cases. In addition, Decheng et al. [19] referred to the development of a QuEChERS extraction method for the target analysis of eight carbapenems in milk using LC–Q Exactive (QE) Orbitrap mass spectrometry. The analysis was performed in positive mode in a heated electrospray interface (HEI+) with parallel reaction monitoring (PRM).
Accordingly, the target of the present study was to establish a sensitive and rapid analytical method for the determination of multiclass pharmaceutical drugs in milk using a modified QuEChERS procedure and sonication, followed by liquid chromatography coupled to a high-resolution Orbitrap MS instrument. Different sorbents were used as dispersive SPE (dSPE) agents for the clean-up step, and the modified QuEChERS method (“AOAC 2007.01”) [39] was evaluated and fully validated. To the best of our knowledge, the optimization of the QuEChERS method combined with LC-HR Orbitrap MS in a fully validated study has not been reported so far. Finally, the optimized method was applied to 10 milk samples, which were commercially available in Greek markets for the monitoring of pharmaceutical drug residues.
2. Results and Discussion
2.1. Optimization of QuEChERS Procedure
Firstly, three primary QuEChERS extraction procedures were tested. The acetate method (method B) yielded more target compounds with recoveries ranging from 70 to 120% compared to the buffered (method C) and the original method (method A), as shown in Figure 1. More specifically, the acetate method revealed two compounds with recoveries <60%, the recoveries of five compounds ranged between 60 and 70%, and the rest (eleven compounds) exhibited recoveries from 70 to 120% (Figure 1). The next step was to operate some modifications of the acetate method to achieve higher analyte extractions together with the lower extraction of undesirable interfering compounds.

Figure 1.
Percentages of pharmaceutical drugs exhibiting recoveries within different ranges (70–120%, 60–70%, and <60%) using different QuEChERS methods. Method A (“Original”), method B (“AOAC 2007.01”) [39], and method C (“Buffered CEN 15662”) [40].
Acetonitrile was used as the extraction solvent since it is the most widely used organic solvent in the QuEChERS method. The addition of a chelating agent—EDTA 0.1 M—improves the extraction recovery of some pharmaceutical drugs by preventing their rapid chelation with metal ions [41,42]. Furthermore, the addition of EDTA could improve the adsorption of casein onto milk fat globules (MFG). EDTA is a calcium-chelating agent that can dissociate micellar calcium phosphates, resulting in a partial disintegration of casein micelles [43]. Another important parameter of the QuEChERS extraction procedure is the acidity of the extractant; therefore, the volume of formic acid in acetonitrile was tested by adding 1%, 2%, and 3.35% in the solvent. The best results were obtained after the addition of 3.35% in acetonitrile. According to Zhou et al. [16], the addition of 3.35% formic acid in acetonitrile along with the addition of sodium acetate enhances the salting-out effect and buffers the extract. Next, the contents were placed in a sonication bath (37 kHz, 100 W) for 20 min. The sonication promotes the homogenization of milk and reduces the size of the milk fat globules (MFG) [44].
Another important stage of the QuEChERS procedure concerns the selection of drying salts, which can cause phase separation and affect the distribution of analytes. To date, numerous combinations of QuEChERS salts have been proposed. MgSO4, NaCl, Na2SO4, and NaOAc were the most suitable applicants. The acetate method was performed by applying Na2SO4, NaCl, and NaOAc as drying salts. Since MgSO4 has been shown to promote the chelation of quinolones [21], it was substituted by Na2SO4, NaCl, and NaOAc.
Secondly, the optimization of the clean-up step of the acetate method was performed. The sorbents assayed were PSA/MgSO4, C18, and EMR-Lipid. PSA is a weak anion exchanger sorbent with the ability to remove sugars, organic acids, fatty acids, and polar pigments, while its chemical structure contributes to a high chelating effect [45]. PSA is a sorbent frequently used in the clean-up step of milk extracts for the removal of co-extracted phenolic substances. Anhydrous MgSO4 reduces the volume of the aqueous phase by hydration, whereas non-polar interfering substances such as sterols and long-chain aliphatic compounds could be removed by applying C18 sorbent in the purification step [18,32,46]. However, an excessive amount of C18 can also adsorb lipophilic drugs [47]. C18 sorbents have already been reported in the extraction of veterinary drug residues from milk samples, with and without PSA [6,32,48,49]. EMR-Lipid is a newly invented material applied to difficult matrices [23,24,25,26] that offers the promise of highly selective lipid removal, without analyte retention. The following approaches were evaluated for the purification step: (A) 25 mg of PSA and 150 mg of MgSO4; (B) 50 mg of C18; and (C) 0.5 g of EMR-Lipid. Figure 2 shows the percentage of target compounds obtained with recoveries <60%, between 60 and 70%, and between 70 and 120%, according to the above combinations. As can be seen, both (B) and (C) clean-up approaches exhibited similar results.
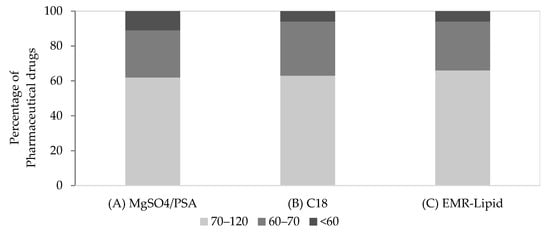
Figure 2.
Percentage of pharmaceutical drugs exhibiting recoveries within different ranges (70–120%, 60–70% and <60%) using different sorbents in the clean-up step. (A) MgSO4/PSA, (B) C18, and (C) EMR-Lipid in milk.
More specifically, in approaches B and C, respectively, 11 and 12 target compounds showed recoveries in the range of 70–120%, and 6 and 5 compounds gave recoveries in the range of 60–70%. In addition, one compound revealed a recovery of <60% in both clean-up approaches. Approach (A) exhibited more target compounds with recoveries below 60%. Thus, approaches (B) and (C) appeared to be more suitable for extract purification. Furthermore, the effectiveness of the purification step was verified by chromatograms with no interfering peaks. Thus, milk samples were further validated according to the previously described and optimized QuEChERS extraction procedure (see Section 3.4.), followed by optimized clean-up approaches (approaches B and C).
2.2. Validation of the Proposed Methods
The validation parameters of the modified QuEChERS procedure for milk with two clean-up approaches are shown in Table 1 and Table 2. As we can see in Table 2, the modified “AOAC 2007.01” QuEChERS method with different clean-up steps (approaches B and C) presented excellent linearity in the tested concentration ranges, with correlation coefficient values ≥ 0.99 in all cases. The extracted ion chromatograms (XIC) of target compounds obtained after the two clean-up approaches, (a) C18 and (b) EMR-Lipid, at a concentration of 50 μg/kg in milk samples are shown in Figure 3. Peak areas did not differ greatly between the two approaches with higher values recorded in most cases (11 analytes) using EMR-Lipid.

Table 1.
Comparison of the methods applying two different clean-up approaches, C18 (approach B) and EMR-Lipid (approach C): intra- and inter-day mean relative recoveries (Rec%), repeatability (expressed as relative standard deviation (%RSDr) and intermediate precision (expressed as relative standard deviation (%RSDWR), at two different spiked levels (n = 6).

Table 2.
Comparison of the methods applying two different clean-up approaches, C18 (approach B) and EMR-Lipid (approach C): linearity, limits of detection (LODs), limits of quantification (LOQs), as well as maximum residue levels (MRLs) for pharmaceutical drug residues in milk.

Figure 3.
Extracted ion chromatogram (XIC) obtained for target compounds for (a) clean-up: C18 (approach B) and (b) clean-up EMR-lipid (approach C) at concentration level of 50 μg/kg of milk.
The trueness and precision of the method were determined in recovery studies with fortified samples at two concentration levels (8 and 50 μg/kg) assayed six times on the same day and six successive days (Table 1). The recoveries at the 8 and 50 μg/kg levels for all the investigated compounds in approach B were within 70–120% with associated RSDs of 19.1%. More specifically, relative recoveries ranged between 75% (sulfathiazole) and 120% (ketoprofen). Intra-day precision in milk ranged from 0.1% for sulfathiazole to 19.0% for fenbendazole, and inter-day precision in milk ranged from 4.3% for diclofenac to 19% for enrofloxacin. In approach C, the recoveries were between 65.1% (diclofenac) and 120.1% (fenbendazole). Intra-day and inter-day precision in milk ranged from 1.9% for albendazole to 9.4% for imidocarb and from 0.4% for enrofloxacin to 17.5% for ketoprofen, respectively.
The LODs and the LOQs of the method are also presented in Table 2. LODs, in approach B, ranged between 0.09 μg/kg for trimethoprim and 15.1 μg/kg for diclofenac, and LOQs ranged between 0.3 μg/kg (trimethoprim) and 50 μg/kg (diclofenac and sulfadiazine), whereas the LODs ranged between 0.09 μg/kg (imidocarb) and 3.1 μg/kg (ketoprofen), and the LOQs were between 0.28 μg/kg (imidocarb) and 10 μg/kg (ketoprofen) in approach C.
The matrix effect values, determined as a signal suppression or enhancement, are illustrated in Figure 4. The ionization efficiency of the analytes may be affected by matrix interferences; thus, calibration curves were established with and without a matrix to evaluate the degree of ion suppression or signal enhancement. In approach B, some of the compounds (six compounds in milk) presented a low matrix effect (between −18.6% and 16.9%), and the rest of the compounds presented a medium matrix effect (twelve compounds between −29.8 to −20.6%). In approach C, twelve compounds presented a low matrix effect, with values between −15.6% and 16.7%, and the rest of the compounds (six compounds) presented a medium matrix effect (between −44.5% and 39.9%).
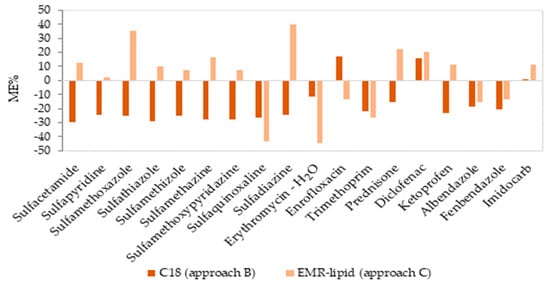
Figure 4.
Matrix effect (%ME) values obtained from the slopes of the solvent and matrix-matched calibration curves in milk for two approaches in clean-up step.
Similar recoveries were found in both approaches at concentration levels of 8 and 50 μg/kg for all the target compounds. More specifically, relative recoveries were within 75–120% with associated RSDs of 19.1% in approach B and between 65.1% and 120.1% with associated RSDs of 17.5% in approach C. Most of the analytes provided similar LOD and LOQ values in both approaches B and C, except two compounds (sulfadiazine and diclofenac) presented higher values in approach B.
The main features of the developed methods proposed in the current study can be compared with those reported in previously published methods, concerning QuEChERS extraction and LC-MS techniques for the evaluation of pharmaceutical drug residues in fatty matrices. Zhao and Lucas [50] assessed the performance of different sorbents (EMR-Lipid, C18 dSPE, and Z-Sep) for screening veterinary drugs in fatty matrices such as bovine liver. The matrix co-extractive removal efficiency, the accuracy, and the precision of the protocol using the EMR-Lipid sorbent for the clean-up step showed better results compared to the other sorbents for the target compounds. On the other hand, Anumol et al. [24] compared two sample preparation approaches using EMR-Lipid and C18 dSPE for the clean-up step for veterinary drugs on fatty samples, including bovine tissues. The results showed that the use of the EMR-Lipid sorbent gave cleaner extracts and improved results for some less polar compounds, such as anthelmintics and tranquilizers, compared to the usage of C18 sorbent. However, the use of the EMR-Lipid sorbent showed much lower recoveries for β-lactam antibiotics and some polar drugs. Furthermore, Tuzimski and Rejczak [49] evaluated nitroimidazole residues in bovine milk by applying different sorbents (Z-Sep, Z-Sep+, PSA, C18, and EMR-Lipid) for sample purification. Similar recoveries were observed by applying both C18 and EMR-Lipid. The application of C18 sorbent obtained recoveries ranging between 56 and 77%, with standard deviation (SD) < 19%, whereas the use of EMR-Lipid provided recoveries of 48 to 77% with SD < 17%. The utilization of PSA sorbent for the purification step revealed higher recoveries than the other sorbents, ranging from 51 to 85%, with SD < 10% (n = 3) for all analytes; thus, the sorbent PSA was used for the purification step. Moreover, according to Jia et al. [8], different sorbents (including PSA, C18, EMR, and Z-Sep, each of them being tested in combination with MgSO4) were evaluated determining veterinary drugs, mycotoxins, and pesticides in bovine milk. The use of both C18 and Z-Sep sorbents to purify the extract performed better than the other two sorbents with more target recoveries falling in the range of 70% to 120%. However, C18 sorbent was preferred for the clean-up step as Z-Sep is not as widely used as C18 in residue analysis. For comparison with low-resolution MS/MS techniques, Jia et al. [8] used QuEChERS extraction with C18 sorbent for the purification step and UHPLC-Qtrap-MS for the determination of multi-class contaminants in milk. This study showed LOD values ranging from 0.01 to 1 µg/kg, and LOQ values were in the range of 0.05 to 5 µg/kg. On the other hand, Castilla-Fernández [25] evaluated two different clean-up steps, (a) EMR-Lipid sorbent and (b) SPE, using UHPLC-QqQ-MS for the analysis of veterinary drugs in milk. The LOQs ranged from 0.01 to 18.25 μg/kg. LC-QqQ-MS and QuEChERS extraction were used for the determination of veterinary drugs in milk by Bang Ye et al. [18]. The method was validated with LOQ values of 5–15 μg/kg. Guo et al. [47] developed a multi-residue method for the determination of 103 veterinary drug residues in milk and dairy products. The method was based on QuEChERS extraction and d-SPE C18/Na2SO4, combined with an LC-QqQ-MS system, and showed LOQ values in the range of 0.1 to 25 μg/kg for milk. In the current study, the LODs ranged between 0.09 and 15.1 μg/kg, and LOQs were in the range of 0.3 to 50 μg/kg in approach B, whereas the LODs ranged between 0.09 and 3.1 μg/kg, and the LOQs were between 0.28 and 10 μg/kg in approach C. LOQ values fulfilled the MRLs set by the EU for milk in both cases. A comparison of sample preparation methods and analytical techniques for the analysis of pharmaceutical compounds in milk are presented in Table 3.
The LR mass analyzers presented much greater sensitivity (lower LOQs) in most cases than the Orbitrap instrument, while the Orbitrap instrument had better selectivity, measuring accurate mass for both parent and fragmented product ions. The current work offers the comparison of a hybrid QuEChERS–sonication extraction combined with different clean-up steps and UHPLC-HR-Orbitrap-MS capability for the determination of drug residues in milk against previous published methods using mainly LC-LR-MS/MS instrumentation.
The expanded MU was estimated individually for each pharmaceutical drug as twice the value of the uncertainty (k = 2, confidence level 95%), and the resulting values are presented in Table 4 and illustrated in Figure 5. In a concentration of 8 μg/kg, the MU values were in the range between 14.80% (albendazole) and 33.04% (fenbendazole and sulfacetamide) in approach B and between 9.94% (sulfadiazine) and 32.78% (diclofenac), in approach C. Both approaches exhibited similar results or better MU values in approach C, except for three compounds (Sulfamethoxypyridazine, diclofenac, and enrofloxacin) that presented higher values, but they were still in compliance with the requirement (50%) of the EU guidance document SANTE/12682/2019 [51].

Table 4.
Measurement uncertainty MU (%) calculated for milk at a concentration level of 8 μg/kg (k = 2, confidence level 95%) and HorRat values for different clean-up approaches.

Figure 5.
Measurement uncertainty (MU%) for milk in a concentration level of 8 μg/kg (n = 6) for two clean-up approaches.

Table 3.
Comparison of recent studies regarding extraction and LC-MS techniques for the analysis of pharmaceutical drugs in milk with the current study.
Table 3.
Comparison of recent studies regarding extraction and LC-MS techniques for the analysis of pharmaceutical drugs in milk with the current study.
| Food Matrix | Compounds | Extraction Method | Clean-Up Method | LC-MS Technique | Acquisition Mode | Linearity | Recovery (%) | LOQs (μg/kg) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Milk | 18 pharmaceutical drugs | Sonication/modified QuEChERS | EMR-Lipid/C18 | LC-hybrid LTQ/Orbitrap-MS | Full MS/dd-MS2 Resolution 60,000/15,000 FWHM | 0.9903 | 65.1–120.1 | 0.28–10 | Current study |
| Bovine milk | 209 veterinary drugs, mycotoxins, and pesticides | Modified QuEChERS | C18 | LC-QTRAP-MS | MRM-IDA-EPI mode | 0.99 | 51.20–129.76 | 0.05–5 | [8] |
| Milk/dairy products | 103 veterinary drugs | Modified QuEChERS | C18 and Na2SO4 | LC-QqQ-MS | MRM mode | 0.9902 | 31.1–120.7 | 0.5–50 | [47] |
| Goat milk | 19 quinolones | “Buffered CEN 15662” QuEChERS | C18 and Na2SO4 | LC–QqQ-MS | MRM mode | 0.9853 | 73.4–114.2 | 5–15 | [18] |
| Cow milk | 66 veterinary drugs | Solvent extraction | EMR-Lipid/SPE | LC-QqQ-MS | MRM mode | 0.998 | 70–120 | 0.02–18.25 | [25] |
| Milk, cheese, and whey | 36 antibiotics | SLE | C18 | LC-Q Exactive Orbitrap-MS | Full scan: resolution at 50,000 FWHM | 0.995 | 70–120 | 1–50 (CCβ) | [52] |
The Horwitz equation for the 8 μg/kg fortification level exhibited an acceptable PRSDWR = 33.1%. Table 4 shows the HorRat value, which was estimated for each one of the pharmaceutical drugs. The HorRat ratio value in all cases was <1, varying between 0.42 (prednisone) and 0.59 (trimethoprim and fenbendazole) in approach B and between 0.42 (erythromycin—H2O) and 0.60 (enrofloxacin) in approach C. To conclude, the applied method has better precision than the maximum allowed.
2.3. Preliminary Application Study
The optimized method was applied to 10 samples of cow’s milk purchased from Greek markets as a preliminary application case study. The analysis revealed that one of the samples contained a pharmaceutical drug. More specifically, imidocarb was detected only in one sample in the concentration of 18 μg/kg, but the concentration found was far below the MRLs (50 μg/kg) set by the EU for milk (Figure 6). MS2 data are depicted in Figure 6c. Two fragment ions were obtained for imidocarb: the first one with an elemental composition of C12H12O2, an m/z of 188.08313, and a mass error of −0.272 ppm and the second one with an elemental composition of C11H14O, an m/z of 162.10373, and a mass error of −1.151 ppm. Imidocarb is an antimicrobial agent that is widely used for the treatment of many diseases in cattle, such as babesiosis and anaplasmosis. Significant residues of imidocarb remain in bovine edible tissues [53] and milk [54] after dosing in cattle. If the recommended withdrawal periods for the drug are not properly implemented, it can lead to the occurrence of imidocarb in edible tissues or milk and eventually in humans through the consumption of contaminated foodstuff.
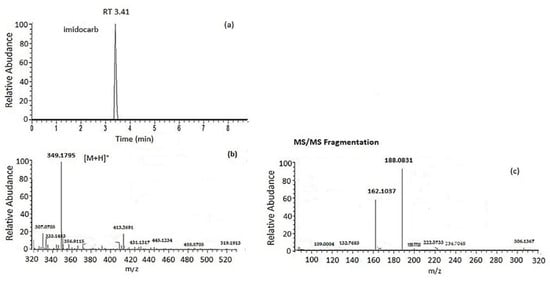
Figure 6.
(a) Extracted ion chromatogram (XIC), (b) full-scan accurate mass parent ion spectrum, and (c) MS/MS data obtained for imidocarb in a milk sample.
3. Materials and Methods
3.1. Chemicals, Reagents, and Samples
Analytical standards (high purity, >98%) of pharmaceutical compounds (sulfacetamide, sulfapyridine, sulfamethoxazole, sulfathiazole, sulfamethizole, sulfamethazine, sulfamethoxypyridazine, sulfaquinoxaline, sulfadiazine, enrofloxacin, trimethoprim, albendazole, erythromycin—H2O, prednisone, diclofenac, fenbendazole, and imidocarb) were purchased from Sigma-Aldrich (Darmstadt, Germany), while ketoprofen was acquired from Tokyo Chemical Industry (Oxford, UK). Individual stock solutions of each compound were prepared in methanol and kept in amber glass bottles at −20 °C.
Methanol, acetonitrile, and water (LC–MS grade) were received from Fisher Scientific (Leicestershire, UK). Acetic acid and formic acid (purity, 98–100%) were obtained from Merck KGaA (Darmstadt, Germany). Ultrapure water was produced in the lab by a Milli-Q water purification system (Millipore, Temecula, CA, USA).
The salts/sorbents used in the QuEChERS extraction were purchased as follows: anhydrous magnesium sulfate (MgSO4), sodium sulfate (Na2SO4), C18 (LiChroprep RP-18, 40–64 μm), and trisodium citrate dehydrate (C6H5Na3O7 · H2O) were purchased from Merck (Darmstadt, Germany); sodium acetate (NaOAc) and sodium chloride (NaCl) were purchased from Riedel-de Haën (Hannover, Germany); primary secondary amine (PSA; 40 μm) and Enhanced Matrix Removal-Lipid (EMR-Lipid) sorbent were purchased from Agilent Technologies (Waldbronn, Germany); sodium citrate dibasic sesquihydrate (C6H6Na2O7 · 1.5H2O) and ethylenediaminetetraacetic acid tetrasodium salt dihydrate (EDTA) were purchased from Sigma-Aldrich (Steinheim, Germany); and syringe filters (polytetrafluoroethylene, 0.22 μm) were purchased from Millipore (Cork, Ireland). Additionally, 50 mL and 15 mL propylene centrifuge tubes were used.
All milk samples for analysis were of Greek origin. Full-fat (3.5%) milk was used to optimize and validate the analytical method. Cow’s milk samples (a total of 10 samples, all full-fat) were purchased from various local supermarkets and markets in Ioannina, Epirus region, NW Greece. Once transferred to the laboratory for analysis, the samples were kept at 4 °C in amber glass bottles until analysis.
3.2. Preliminary Experiments
Initially, three primary QuEChERS methods were investigated. These experiments were aimed to achieve the best extraction efficiency for the target compounds. Ten grams of milk sample were weighed into a 50 mL polypropylene centrifuge tube, spiked at 20 μg/kg fortification level, shaken for 1 min, and extracted as follows below:
- (a)
- Extraction method A (“Original” QuEChERS): Solvent: 10 mL of acetonitrile; extract salts: 4 g of MgSO4 and 1 g of NaCl.
- (b)
- Extraction method B (“AOAC 2007.01” QuEChERS) [39]: Solvent: 10 mL of acetonitrile containing 1% acetic acid; extract salts: 6 g of MgSO4 and 1.5 g of NaOAc.
- (c)
- Extraction method C (“Buffered CEN 15662” QuEChERS) [40]: Solvent: 10 mL of acetonitrile; extract salts: 4 g of MgSO4, 1 g of NaCl, 1 g of C6H5Na3O7⋅H2O. and 0.5 g of C6H6Na2O7⋅1.5H2O.
In all cases, after the addition of the salts, the tubes were shaken for 1 min and centrifuged for 5 min at 4000 rpm. Then, 1 mL of the supernatant was transferred into a 15 mL centrifuge tube containing 25 mg of PSA and 150 mg of MgSO4 for the clean-up step (clean-up approach A). The tubes were shaken for 1 min and centrifuged for 5 min at 4000 rpm. The supernatant was transferred to a glass testing tube, evaporated to dryness under a gentle stream of nitrogen, and then reconstituted in 1 mL of H2O:MeOH (90:10 v/v) acidified with 0.1% formic acid. The sample was filtered using syringe membrane filters (polytetrafluoroethylene, 0.22 μm) before injection into UHPLC-LTQ/Orbitrap MS.
3.3. UHPLC-Orbitrap MS Parameters
The UHPLC system (Accela, Thermo Fisher Scientific, Bremen, Germany) was coupled to a hybrid LTQ Orbitrap XL Fourier transform mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) fitted with an Ion Max electrospray ionization probe. The full scan was applied in positive ionization mode, with a mass range of 120–1000 Da and a mass resolving power of 60,000 FWHM. Extracted ion chromatograms were used for initial identification followed by data-dependent MS/MS (resolution set at 15,000 FWHM), using collision-induced dissociation (CID) with normalized collision energy (NCE) at 35% for all analytes. The mass accuracy tolerance window was set to 5 ppm. Target compounds, retention times, and detection parameters for the data-dependent acquisition (full MS/dd-MS2) analysis are presented in Table 5. The main parameters of the mass spectrometer were as follows: spray voltage, 4 kV; tube lens, 90 V; sheath gas flow rate, 35 arbitrary units (au); auxiliary gas flow rate, 10 au, capillary temperature, 320 °C. The scans were applied by targeting the automatic gain control (AGC) at 4 × 105 ions. To process the data. Thermo Xcalibur 2.1 software (Thermo Electron, San Jose, CA, USA) was used.

Table 5.
UHPLC–LTQ Orbitrap MS analysis data. Target pharmaceutical drugs, retention times (tR), and detection parameters for full MS/dd-MS2 analysis.
UHPLC was equipped with an Accela AS autosampler (model 2.1.1) and an Accela quaternary gradient pump. A reversed-phase Fortis C18 (Fortis Technologies, Neston, UK) analytical column (50 × 2.1 mm, 1.7 μm particle size) was used to achieve separation, and column oven temperature was maintained at 35 °C. Mobile phase was a mixture of solvent A, H2O acidified with 0.1% formic acid, and solvent B, MeOH acidified with 0.1% formic acid. The elution gradient started at 95% A; remained at 95% for 1 min; changed to 30% after 1 more min; changed to 0% after 3 more min, where it remained for 2 min; and finally returned to the initial conditions. The total run time was 10 min. The injection volume was 5 μL, and the flow rate was 250 μL/min.
3.4. QuEChERS Extraction Procedure
The sample extraction and clean-up procedures followed in method B (“AOAC 2007.01” QuEChERS) [39] were selected as the most efficient method, and some further modifications have been studied for optimizing the method. Ten grams of the milk sample were weighed into a 50 mL polypropylene centrifuge tube and spiked at 8 and 50 μg/kg fortification levels. A total of 10 mL of EDTA 0.1 M was added, and the tubes were immediately shaken for 1 min. Then, 10 mL of acetonitrile containing 3.35% formic acid were added, and the tubes were shaken again for 1 min. The mixture was placed in a sonication bath (37 kHz, 100 W, Elmasonic P, Singen, Germany) for 20 min. Afterwards, 4 g of Na2SO4, 1.2 of NaCl, and 0.7 g of NaOAc were added; shaken again for 1 min; and centrifuged for 5 min at 4000 rpm. Two alternative clean-up approaches were evaluated: 50 mg of C18 (clean-up approach B) and 0.5 g of EMR-Lipid (clean-up approach C). A total of 1 mL of the organic upper phase was transferred into a 15 mL polypropylene centrifuge tube containing 50 mg of C18. On the other hand, 5 mL of the organic upper phase was transferred into a 15 mL polypropylene centrifuge tube containing 0.5 g of EMR-Lipid, which was firstly conditioned with 2.5 mL of Milli-Q water and shaken for 30 s. Then, it was transferred into a 15 mL polypropylene centrifuge tube containing 1.6 g of MgSO4 and 0.4 g of NaCl. For both the above clean-up approaches, after the addition of the sorbents, the tubes were vigorously shaken for 1 min and then centrifuged for 5 min at 4000 rpm. Finally, 1 mL of the supernatant was transferred to a glass testing tube, evaporated to dryness under a gentle stream of nitrogen at 40 °C, and reconstituted into 1 mL of H2O:MeOH (90:10, v/v) acidified with 0.1% formic acid. Finally, the sample was filtered using syringe membrane filters (polytetrafluoroethylene, 0.22 μm) before being injected into the UHPLC-LTQ/Orbitrap MS instrument.
3.5. Method Validation
Matrix-matched standard calibration curves prepared in H2O:MeOH (90:10, v/v) acidified with 0.1% formic acid were used for the quantification of drug residues. Blank milk samples were used for the construction of matrix-matched calibration curves. The method was validated by determining analytical parameters such as sensitivity/linearity, recovery, precision, limit of detection (LOD) and limit of quantification (LOQ), matrix effects (ME), and measurement uncertainty (MU), following the European Commission (EC) documents 2002/657/EC and N° SANTE/12682/2019 [51,55].
The linearity of the method was checked in the range of 5 to 100 µg/kg, using weighted least-squares regression and expressed as a determination coefficient (R2). Blank samples of biologically produced milk (previously analyzed for drug residues) were spiked with a pharmaceutical drug mixture at 8 and 50 μg/kg fortification levels to determine recovery rates. In all cases, six replicates (n = 6) were prepared for each spike level. The method’s trueness and precision were studied via the mean recoveries (Rec%) and the relative standard deviation (RSD%) at 8 and 50 μg/kg. Repeatability (%RSDr) and intermediate precision (%RSDWR) were tested on the same day (n = 6) and for six consecutive days, respectively. According to SANTE guidance documents, mean recoveries should range between 70 and 120%, with an associated RSD ≤ 20%, for all analytes. In some cases, mean recovery rates can be accepted outside the range of 70–120% if RSD ≤ 20%, but the mean recovery must not be lower than 30% or higher than 140%. The sensitivity of the method was evaluated by determining limits of quantification (LOQs) and limits of detection (LODs) of the target compounds using a signal-to-noise ratio (S/N) of 3 and 10, respectively. According to N° SANTE/12682/2019, LOQ values should be ≤ MRLs. The matrix effect (ME) values for the target compounds were studied by comparing a calibration curve prepared in milk extract and a calibration curve prepared in the solvent, at the same concentration range, according to the formula below:
%ME is a useful parameter to assess the effectiveness of the method since co-extractants could increase or reduce the analytical signal. %ME values <±20%, <±20–±50%, and >±50% are considered low, medium, and high, respectively [56].
Six replicate analyses performed on different days at a level of 8 μg/kg were used to evaluate the expanded MU for the two clean-up approaches. A default expanded MU of 50% should not be overstepped (equivalent to a confidence level of 95% and a coverage factor of 2). Furthermore, another practice for evaluating acceptable measurement precision is to use the Horwitz equation and the Horwitz ratio (HorRat) [57].
4. Conclusions
Different approaches have been effectively applied in the extraction and clean-up steps for the analysis of pharmaceutical drugs in milk using modified QuEChERS extraction combined with sonication and UHPLC-Orbitrap MS. The acidified acetate method was selected as more efficient, and the optimization of the purification revealed C18 and EMR-Lipid as the best dispersive SPE (dSPE) agents. The optimized methods were further validated and the parameters of linearity, trueness, precision, LOD, and LOQ and expanded uncertainty fulfilled the requirements according to the SANTE guidelines, while the HorRat values revealed a better method precision than allowed. The EMR-Lipid approach provides better validation parameter values for most of the analytes. The method was applied to real samples, revealing the existence of one pharmaceutical drug (imidocarb) in one milk sample. The developed method can serve as an effective and easy approach for the determination of a wide range of pharmaceutical drug residues in milk.
Author Contributions
Conceptualization, I.K. and D.H.; methodology, O.K., D.H. and I.K.; validation, O.K. and M.K.; formal analysis, O.K. and M.K.; investigation, O.K. and M.K.; resources, I.K. and T.A.; data curation, O.K. and M.K.; writing—original draft preparation, O.K. and I.K.; writing—review and editing, O.K., D.H., and I.K.; visualization, O.K.; supervision, I.K. and D.H.; funding acquisition, T.A. and I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the project “Development of research infrastructure for the design, production, development of quality characteristics and safety of agrofoods and functional foods (RI-Agrofoods)” (MIS 5047235), which is implemented under the action “Reinforcement of the Research and Innovation Infrastructure”, funded by the operational program “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are included in the manuscript.
Acknowledgments
The authors would like to thank the Unit of Environmental, Organic and Biochemical High-Resolution Analysis–Orbitrap-LC–MS of the University of Ioannina for providing access to the facilities.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Not applicable.
References
- He, Z.; Zhang, H. (Eds.) Applied Manure and Nutrient Chemistry for Sustainable Agriculture and Environment; Springer: Dordrecht, The Netherlands, 2014; ISBN 978-94-017-8806-9. [Google Scholar]
- Beyene, T. Veterinary Drug Residues in Food-Animal Products: Its Risk Factors and Potential Effects on Public Health. J. Vet. Sci. Technol. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- European Commission. Regulation (EU) No 37/2010 of the Commission of 22 December 2009 on pharmacologically active substances and their classification as regards the maximum residue limits in foodstuffs of animal origin. Off. J. Eur. Union 2010, L15, 1–72. [Google Scholar]
- Tsiboukis, D.; Sazakli, E.; Jelastopulu, E.; Leotsinidis, M. Anthelmintics Residues in Raw Milk. Assessing Intake by a Children Population. Pol. J. Vet. Sci. 2013, 16, 85–91. [Google Scholar] [CrossRef]
- Dasenaki, M.E.; Thomaidis, N.S. Multi-Residue Determination of 115 Veterinary Drugs and Pharmaceutical Residues in Milk Powder, Butter, Fish Tissue and Eggs Using Liquid Chromatography–Tandem Mass Spectrometry. Anal. Chim. Acta 2015, 880, 103–121. [Google Scholar] [CrossRef]
- Grabsk, A.; de Souza, J.; De Marchi, F.; do Prado, R.; dos Santos, G.; Porto, C.; Pilau, E. Determination of Antibiotics Residues in Milk Using a QuEChERS Method Using Full Factorial Design and Liquid Chromatography-Tandem Mass Spectrometry. J. Braz. Chem. Soc. 2019, 35, 1498–1505. [Google Scholar] [CrossRef]
- Souza, R.; Fernández, P.; Muela, A.; Cesio, M.V.; Heinzen, H.; Pareja, L. Development of a Methodology for the Simultaneous Analysis of Multiclass Contaminants in Milk. Food Anal. Methods 2021, 14, 1075–1086. [Google Scholar] [CrossRef]
- Jia, Q.; Qiu, J.; Zhang, L.; Liao, G.; Jia, Y.; Qian, Y. Multiclass Comparative Analysis of Veterinary Drugs, Mycotoxins, and Pesticides in Bovine Milk by Ultrahigh-Performance Liquid Chromatography–Hybrid Quadrupole–Linear Ion Trap Mass Spectrometry. Foods 2022, 11, 331. [Google Scholar] [CrossRef]
- Unsal, I.A.; Tasan, M.; Gokcen, T.; Goren, A.C. Determination of Sulfonamides in Milk by ID-LC-MS/MS. J. Chem. Metrol. 2018, 12, 70–78. [Google Scholar] [CrossRef]
- Yue, Z.; Qiu, Y.; Liu, X.; Ji, C. Determination of Multi-Residues of Tetracyclines and Their Metabolites in Milk by High Performance Liquid Chromatography-Tandem Positive-Ion Electrospray Ionization Mass Spectrometry. Chin. J. Anal. Chem. 2006, 34, 1255–1259. [Google Scholar] [CrossRef]
- Xie, J.; Peng, T.; Zhu, A.; He, J.; Chang, Q.; Hu, X.; Chen, H.; Fan, C.; Jiang, W.; Chen, M.; et al. Multi-Residue Analysis of Veterinary Drugs, Pesticides and Mycotoxins in Dairy Products by Liquid Chromatography–Tandem Mass Spectrometry Using Low-Temperature Cleanup and Solid Phase Extraction. J. Chromatogr. B 2015, 1002, 19–29. [Google Scholar] [CrossRef]
- Bogialli, S.; Curini, R.; Di Corcia, A.; Laganà, A.; Mele, M.; Nazzari, M. Simple Confirmatory Assay for Analyzing Residues of Aminoglycoside Antibiotics in Bovine Milk: Hot Water Extraction Followed by Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. A 2005, 1067, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bogialli, S.; D’Ascenzo, G.; Di Corcia, A.; Laganà, A.; Nicolardi, S. A Simple and Rapid Assay Based on Hot Water Extraction and Liquid Chromatography–Tandem Mass Spectrometry for Monitoring Quinolone Residues in Bovine Milk. Food Chem. 2008, 108, 354–360. [Google Scholar] [CrossRef]
- Lu, Y.; Lü, L.; He, J.; Zhao, T. Preparation of Hydrophilic Molecularly Imprinted Solid-phase Microextraction Fiber for the Selective Removal and Extraction of Trace Tetracyclines Residues in Animal Derived Foods. J. Sep. Sci. 2020, 43, 2172–2179. [Google Scholar] [CrossRef]
- de Oliveira Arias, J.L.; Schneider, A.; Batista-Andrade, J.A.; Vieira, A.A.; Caldas, S.S.; Primel, E.G. Chitosan from Shrimp Shells: A Renewable Sorbent Applied to the Clean-up Step of the QuEChERS Method in Order to Determine Multi-Residues of Veterinary Drugs in Different Types of Milk. Food Chem. 2018, 240, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xu, J.-J.; Cong, J.-M.; Cai, Z.-X.; Zhang, J.-S.; Wang, J.-L.; Ren, Y.-P. Optimization for Quick, Easy, Cheap, Effective, Rugged and Safe Extraction of Mycotoxins and Veterinary Drugs by Response Surface Methodology for Application to Egg and Milk. J. Chromatogr. A 2018, 1532, 20–29. [Google Scholar] [CrossRef]
- Rageh, A.H.; Abdel-Rahim, S.A.; Askal, H.F.; Saleh, G.A. Hydrophilic-Interaction Planar Chromatography in Ultra-Sensitive Determination of α-Aminocephalosporin Antibiotics. Application to Analysis of Cefalexin in Goat Milk Samples Using Modified QuEChERS Extraction Technique. J. Pharm. Biomed. Anal. 2019, 166, 421–434. [Google Scholar] [CrossRef]
- Bang Ye, S.; Huang, Y.; Lin, D.-Y. QuEChERS Sample Pre-Processing with UPLC–MS/MS: A Method for Detecting 19 Quinolone-Based Veterinary Drugs in Goat’s Milk. Food Chem. 2022, 373, 131466. [Google Scholar] [CrossRef]
- Decheng, S.; Xia, F.; Zhiming, X.; Liyang; Peilong, W. Simultaneous Determination of Eight Carbapenems in Milk by Modified QuEChERS and Ultra High Performance Liquid Chromatography Coupled with High-Field Quadrupole-Orbitrap High-Resolution Mass Spectrometry. J. Chromatogr. A 2022, 1670, 462979. [Google Scholar] [CrossRef]
- Lambropoulou, D.A.; Albanis, T.A. Methods of Sample Preparation for Determination of Pesticide Residues in Food Matrices by Chromatography–Mass Spectrometry-Based Techniques: A Review. Anal. Bioanal. Chem. 2007, 389, 1663–1683. [Google Scholar] [CrossRef]
- Desmarchelier, A.; Fan, K.; Minh Tien, M.; Savoy, M.-C.; Tarres, A.; Fuger, D.; Goyon, A.; Bessaire, T.; Mottier, P. Determination of 105 Antibiotic, Anti-Inflammatory, Antiparasitic Agents and Tranquilizers by LC-MS/MS Based on an Acidic QuEChERS-like Extraction. Food Addit. Contam. Part A 2018, 35, 647–661. [Google Scholar] [CrossRef]
- Zhang, X.; Li, T.; Zhang, L.; Hu, T.; Fu, Y.; Guo, Z. Simultaneous Determination of Sulfoxaflor in 14 Daily Foods Using LC-MS/MS. Int. J. Environ. Anal. Chem. 2019, 99, 557–567. [Google Scholar] [CrossRef]
- López-Blanco, R.; Nortes-Méndez, R.; Robles-Molina, J.; Moreno-González, D.; Gilbert-López, B.; García-Reyes, J.F.; Molina-Díaz, A. Evaluation of Different Cleanup Sorbents for Multiresidue Pesticide Analysis in Fatty Vegetable Matrices by Liquid Chromatography Tandem Mass Spectrometry. J. Chromatogr. A 2016, 1456, 89–104. [Google Scholar] [CrossRef]
- Anumol, T.; Lehotay, S.J.; Stevens, J.; Zweigenbaum, J. Comparison of Veterinary Drug Residue Results in Animal Tissues by Ultrahigh-Performance Liquid Chromatography Coupled to Triple Quadrupole or Quadrupole–Time-of-Flight Tandem Mass Spectrometry after Different Sample Preparation Methods, Including Use of a Commercial Lipid Removal Product. Anal. Bioanal. Chem. 2017, 409, 2639–2653. [Google Scholar] [CrossRef] [PubMed]
- Castilla-Fernández, D.; Moreno-González, D.; Beneito-Cambra, M.; Molina-Díaz, A. Critical Assessment of Two Sample Treatment Methods for Multiresidue Determination of Veterinary Drugs in Milk by UHPLC-MS/MS. Anal. Bioanal. Chem. 2019, 411, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Matarrita, J.; Sapozhnikova, Y.; Lehotay, S.J. Evaluation of a Recent Product to Remove Lipids and Other Matrix Co-Extractives in the Analysis of Pesticide Residues and Environmental Contaminants in Foods. J. Chromatogr. A 2016, 1449, 17–29. [Google Scholar] [CrossRef]
- Abesinghe, A.M.N.L.; Vidanarachchi, J.K.; Islam, N.; Prakash, S.; Silva, K.F.S.T.; Bhandari, B.; Karim, M.A. Effects of Ultrasonication on the Physicochemical Properties of Milk Fat Globules of Bubalus Bubalis (Water Buffalo) under Processing Conditions: A Comparison with Shear-Homogenization. Innov. Food Sci. Emerg. Technol. 2020, 59, 102237. [Google Scholar] [CrossRef]
- Nguyen, N.H.A.; Anema, S.G. Ultrasonication of Reconstituted Whole Milk and Its Effect on Acid Gelation. Food Chem. 2017, 217, 593–601. [Google Scholar] [CrossRef]
- Karageorgou, E.; Myridakis, A.; Stephanou, E.G.; Samanidou, V. Multiresidue LC-MS/MS Analysis of Cephalosporins and Quinolones in Milk Following Ultrasound-Assisted Matrix Solid-Phase Dispersive Extraction Combined with the Quick, Easy, Cheap, Effective, Rugged, and Safe Methodology: Sample Preparation. J. Sep. Sci. 2013, 36, 2020–2027. [Google Scholar] [CrossRef]
- Qin, Y.; Jatamunua, F.; Zhang, J.; Li, Y.; Han, Y.; Zou, N.; Shan, J.; Jiang, Y.; Pan, C. Analysis of Sulfonamides, Tilmicosin and Avermectins Residues in Typical Animal Matrices with Multi-Plug Filtration Cleanup by Liquid Chromatography–Tandem Mass Spectrometry Detection. J. Chromatogr. B 2017, 1053, 27–33. [Google Scholar] [CrossRef]
- Sun, H.; Yu, Q.-W.; He, H.-B.; Lu, Q.; Shi, Z.-G.; Feng, Y.-Q. Nickel Oxide Nanoparticle-Deposited Silica Composite Solid-Phase Extraction for Benzimidazole Residue Analysis in Milk and Eggs by Liquid Chromatography–Mass Spectrometry. J. Agric. Food Chem. 2016, 64, 356–363. [Google Scholar] [CrossRef]
- Whelan, M.; Kinsella, B.; Furey, A.; Moloney, M.; Cantwell, H.; Lehotay, S.J.; Danaher, M. Determination of Anthelmintic Drug Residues in Milk Using Ultra High Performance Liquid Chromatography–Tandem Mass Spectrometry with Rapid Polarity Switching. J. Chromatogr. A 2010, 1217, 4612–4622. [Google Scholar] [CrossRef] [PubMed]
- van Pamel, E.; Daeseleire, E. A Multiresidue Liquid Chromatographic/Tandem Mass Spectrometric Method for the Detection and Quantitation of 15 Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) in Bovine Meat and Milk. Anal. Bioanal. Chem. 2015, 407, 4485–4494. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ren, X.; Diao, Y.; Chen, Y.; Wang, Q.; Jin, W.; Zhou, P.; Fan, Q.; Zhang, Y.; Liu, H. Multiclass Analysis of 25 Veterinary Drugs in Milk by Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry. Food Chem. 2018, 257, 259–264. [Google Scholar] [CrossRef]
- Aydoğan, C. Recent Advances and Applications in LC-HRMS for Food and Plant Natural Products: A Critical Review. Anal. Bioanal. Chem. 2020, 412, 1973–1991. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A. The Current Role of High-Resolution Mass Spectrometry in Food Analysis. Anal. Bioanal. Chem. 2012, 403, 1233–1249. [Google Scholar] [CrossRef] [PubMed]
- Kırkan, E.; Tahir, A.O.; Bengü, A.Ş.; Aslan, H.; Çiftçi, M.; Aydoğan, C. Rapid Determination of Sulfonamide Residues in Honey Samples Using Non-targeted Liquid Chromatography-high Resolution Mass Spectrometry. Sep. Sci. PLUS 2020, 3, 451–459. [Google Scholar] [CrossRef]
- Aydoğan, C.; El Rassi, Z. MWCNT Based Monolith for the Analysis of Antibiotics and Pesticides in Milk and Honey by Integrated Nano-Liquid Chromatography-High Resolution Orbitrap Mass Spectrometry. Anal. Methods 2019, 11, 21–28. [Google Scholar] [CrossRef]
- AOAC Official Method 2007. 01. Pesticide Residues in Foods by Acetonitrile Extraction and Partitioning with Magnesium Sulfate; AOAC International: Rockville, MD, USA, 2017. [Google Scholar]
- EN 15662; Foods of Plant Origin—Determination of Pesticide Residues Using GC-MS and/or LC-MS/MS Following Acetonitrile Extraction/Partitioning and Clean-Up by Dispersive SPE—QuEChERS Method. European Committee for Standardization: Brussels, Belgium, 2018.
- Gros, M.; Petrović, M.; Barceló, D. Tracing Pharmaceutical Residues of Different Therapeutic Classes in Environmental Waters by Using Liquid Chromatography/Quadrupole-Linear Ion Trap Mass Spectrometry and Automated Library Searching. Anal. Chem. 2009, 81, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Thomaidis, N.S. Multianalyte Method for the Determination of Pharmaceuticals in Wastewater Samples Using Solid-Phase Extraction and Liquid Chromatography–Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2015, 407, 4229–4245. [Google Scholar] [CrossRef]
- Sikand, V.; Tong, P.S.; Vink, S.; Roy, S. Physicochemical Properties of Skim Milk Powders Prepared with the Addition of Mineral Chelators. J. Dairy Sci. 2016, 99, 4146–4153. [Google Scholar] [CrossRef]
- Chávez-Martínez, A.; Reyes-Villagrana, R.A.; Rentería-Monterrubio, A.L.; Sánchez-Vega, R.; Tirado-Gallegos, J.M.; Bolivar-Jacobo, N.A. Low and High-Intensity Ultrasound in Dairy Products: Applications and Effects on Physicochemical and Microbiological Quality. Foods 2020, 9, 1688. [Google Scholar] [CrossRef] [PubMed]
- Tuzimski, T.; Szubartowski, S. Method Development for Selected Bisphenols Analysis in Sweetened Condensed Milk from a Can and Breast Milk Samples by HPLC–DAD and HPLC-QqQ-MS: Comparison of Sorbents (Z-SEP, Z-SEP Plus, PSA, C18, Chitin and EMR-Lipid) for Clean-Up of QuEChERS Extract. Molecules 2019, 24, 2093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Liu, X.; Zhang, J.; Cao, Y.; Shi, Z.; Sun, H. Multi-Class, Multi-Residue Analysis of Trace Veterinary Drugs in Milk by Rapid Screening and Quantification Using Ultra-Performance Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry. J. Dairy Sci. 2015, 98, 8433–8444. [Google Scholar] [CrossRef]
- Guo, X.; Tian, H.; Yang, F.; Fan, S.; Zhang, J.; Ma, J.; Ai, L.; Zhang, Y. Rapid Determination of 103 Common Veterinary Drug Residues in Milk and Dairy Products by Ultra Performance Liquid Chromatography Tandem Mass Spectrometry. Front. Nutr. 2022, 9, 879518. [Google Scholar] [CrossRef]
- Ehling, S.; Reddy, T.M. Liquid Chromatography–Mass Spectrometry Method for the Quantitative Determination of Residues of Selected Veterinary Hormones in Powdered Ingredients Derived from Bovine Milk. J. Agric. Food Chem. 2013, 61, 11782–11791. [Google Scholar] [CrossRef] [PubMed]
- Tuzimski, T.; Rejczak, T. A QuEChERS-Based Sample Preparation Method for the Analysis of 5-Nitroimidazoles in Bovine Milk by HPLC–DAD. J. AOAC Int. 2017, 100, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lucas, D. Multiresidue Analysis of Veterinary Drugs in Bovine Liver by LC/MS/MS: Agilent Bond Elut EMR-Lipid. Appl. Note Agil. Technol. 2015, 5, 5991–6098. [Google Scholar]
- European Commission. Analytical Quality Control and Method Validation for Pesticide Residues Analysis in Food and Feed. SANTE 2019, 12682, 21–22. [Google Scholar]
- Igualada, C.; Giraldo, J.; Font, G.; Yusà, V. Validation of a Multi-Residue UHPLC-HRMS Method for Antibiotics Screening in Milk, Fresh Cheese, and Whey. J. Food Compos. Anal. 2022, 106, 104265. [Google Scholar] [CrossRef]
- Tang, Y.; Yu, N.; Liu, C.; Han, M.; Wang, H.; Chen, X.; Kang, J.; Li, X.; Liu, Y. Residue Depletion of Imidocarb in Bovine Tissues by UPLC-MS/MS. Animals 2022, 13, 104. [Google Scholar] [CrossRef]
- Traynor, I.M.; Thompson, C.S.; Armstrong, L.; Fodey, T.; Danaher, M.; Jordan, K.; Kennedy, D.G.; Crooks, S.R.H. Determination of Imidocarb Residues in Bovine and Ovine Liver and Milk by Immunobiosensor. Food Addit. Contam. Part A 2013, 30, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Decision 96/23/Ec Commission (2002) 96/23/EC COMMISSION DECISION of 12 August 2002 implementing Council Directive 96/23/EC concerning the performance of analytical methods and the interpretation of results (notified under document number C (2022) 3044) (Text withEEA relevance) (2022/657/EC). 96/23/Ec Comm Desis 29. Off. J. Eur. Union 2002, L221, 8. [Google Scholar]
- Nannou, C.I.; Boti, V.I.; Albanis, T.A. Trace Analysis of Pesticide Residues in Sediments Using Liquid Chromatography–High-Resolution Orbitrap Mass Spectrometry. Anal. Bioanal. Chem. 2018, 410, 1977–1989. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, W.; Albert, R. The Horwitz Ratio (HorRat): A Useful Index of Method Performance with Respect to Precision. J. AOAC Int. 2006, 89, 1095–1109. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).