2.1. Microbial Community Structure of Coffee Based at the Phylum and Genus Level
Coffee has great microbiological diversity [
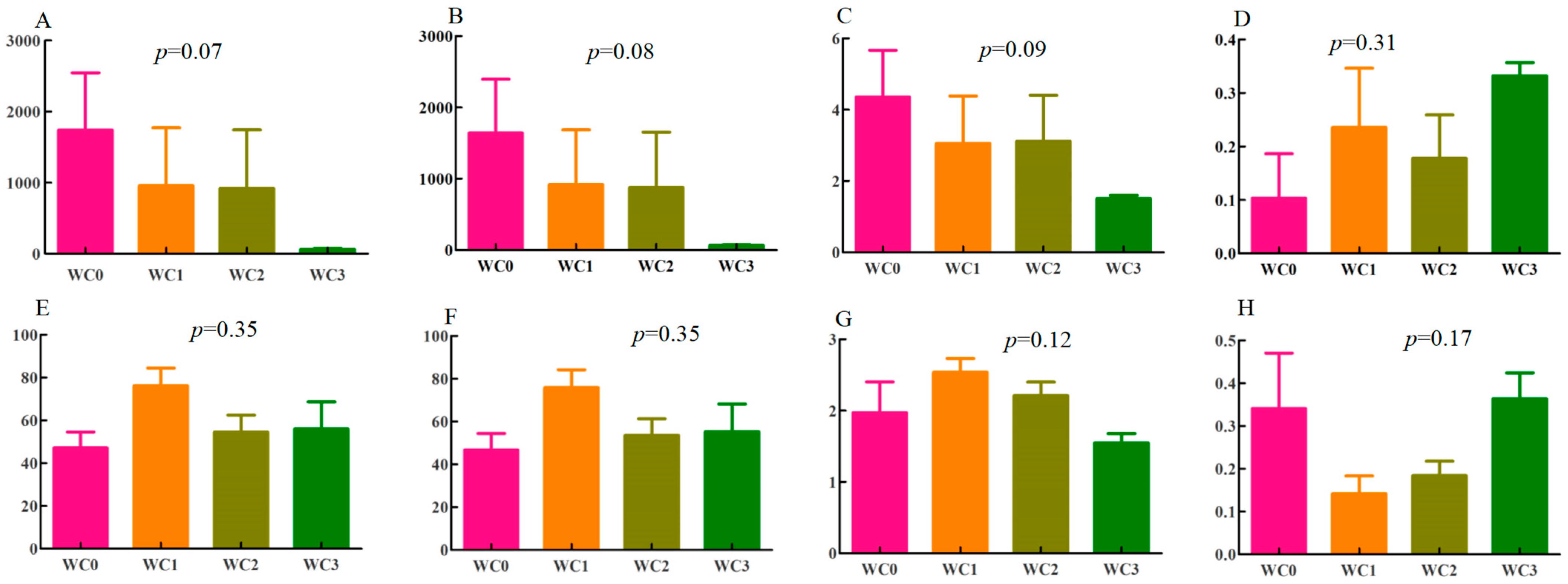
1]. The sequences of bacteria and fungi ranged from 40,037 to 81,509 and from 38,944 to 179,398, respectively, with average lengths of 376 bp for bacteria and 241 bp for fungi. The coverage of coffee samples in each group was higher than 0.99. At the operational-taxonomic-unit (OTU) level, the ace index had
p-values of 0.07 and 0.35 in bacteria and fungi, respectively (
Figure 1A,E). The
p-values of the Chaol index were 0.08 in bacteria and 0.35 in fungi. Among bacteria, WC0 had the highest Chaol index (
Figure 1B), while among fungi, the highest Chaol index was observed in WC1 (
Figure 1F). These results indicated that WC0 and WC1 had the highest species richness in bacteria and fungi, respectively.
The Shannon indices and the Simpon indices can reflect the number, evenness, and diversity of species of coffee samples [
14]. The highest Shannon indices were observed for WC0 for bacteria (
Figure 1C) and WC1 for fungi (
Figure 1G), indicating that they had the highest species evenness for bacteria and fungi, respectively. On the other hand, WC3 showed the highest Simpon indices for bacteria and fungi (
Figure 1D,H), suggesting that it had the highest observed diversity of both bacteria and fungi.
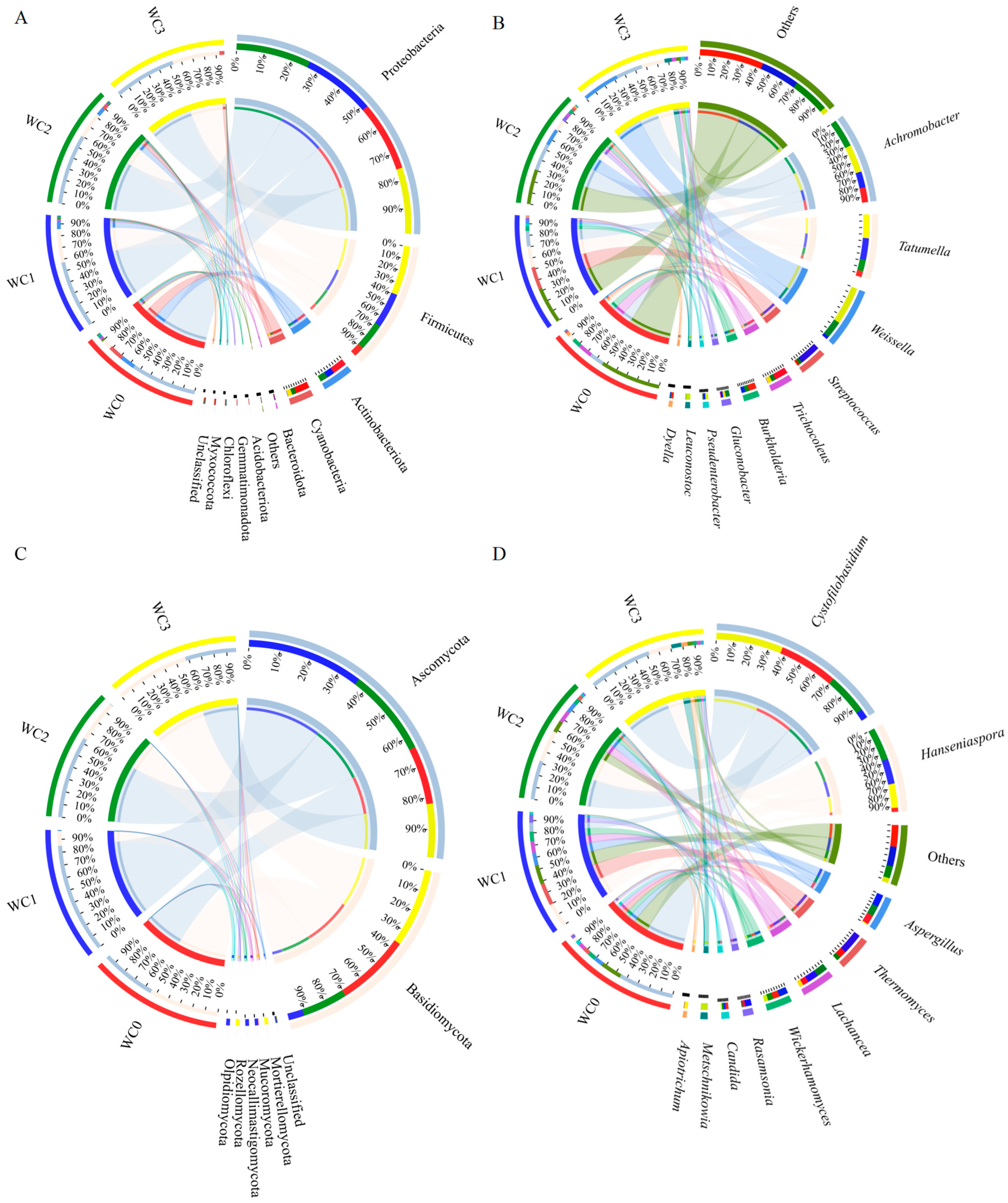
The bacteria in coffee samples from different fermentation times using the washed processing method were classified into 11 phyla by high-throughput sequencing, as shown in
Figure 2A. These phyla included
Proteobacteria,
Firmicutes,
Actinobacteria,
Cyanobacteria,
Bacteroidota,
Acidobacteriota,
Gemmatimonadota,
Chloroflexi, and
Myxococcota. Among them, the dominant phyla were
Proteobacteria (comprising 54.36–61.60% of the community abundance at the phyla level),
Firmicutes (8.57–41.25%),
Actinobacteria (0.27–13.13%),
Cyanobacteria (0.34–13.00%), and
Bacteroidota (0.00–0.16%). Notably, the relative percentage of community abundance of
Proteobacteria in all the samples was more than 50.00%. The relative percent of
Proteobacteria community abundance initially increased from 57.52% in WC0 to 61.60% in WC2, then decreased to 54.36% in WC3. Moreover,
Firmicutes,
Actinobacteria, and
Cyanobacteria exhibited significant changes with fermentation time lengthening. At the start of fermentation, WC0 showed a rich bacterial phyla composition, while at the end of fermentation, WC3 showed a more consistent bacterial phyla composition.
Furthermore, these bacteria were confirmed to 10 genera, as shown in
Figure 2B. These genera included
Achromobacter,
Tatumella,
Weissella,
Streptococcus,
Trichocoleus,
Burkholderia,
Gluconobacter, Pseudenterobacter,
Leuconostoc, and
Dyella. The top five genera were
Achromobacter, Tatumella, Weissella, Streptococcus, and Trichocoleus. At the beginning of the fermentation,
Achromobacter and
Trichocoleus were the dominant bacteria, with 12.95 and 12.88 relative percentages of community abundance at the genus level, respectively.
Achromobacte remained relatively stable during the first 12 h, but its abundance increased over the fermentation process to 24.69% at 24 h and 24.18% at 36 h. In contrast,
Trichocoleus significantly declined as the fermentation processed, reaching its lowest abundance of 0.28% at 12 h.
Burkholderia and
Dyella showed changes similar to those of
Trichocoleus. On the other hand,
Weissella and
Leuconostoc continuously increased throughout the fermentation process, from 0.15% to 33.74% and 0.00% to 6.39%, respectively. The population of
Tatumella followed a similar pattern to that of
Streptococcus, increasing during the first 12 h and then decreasing. The microbial community was highly diverse in coffee fermentation, especially in wet coffee fermentation [
4]. Lactic acid bacteria species, especially
Leuconostoc, were commonly involved in fermentation [
4]. Low
Weissella sp. and
Streptococcus faecalis populations were also identified [
4].
Streptococcus was the perdominant species during the fermentation in the wet process [
4].
The fungi in different coffee samples were of lower diversity than bacteria, which could be classified into seven phyla by high-throughput sequencing, as shown in
Figure 2C. These phyla were
Ascomycota,
Basidiomycota,
Mortierellomycota,
Mucoromycota,
Neocallimastigomycota,
Olpidiomycota, and
Rozellomycota. Among them, the dominant phyla were
Ascomycota (comprising 40.51–87.47% of the community abundance at phyla level) and
Basidiomycota (11.80–59.30%). Notably, the relative abundance of
Ascomycota in different samples was consistently higher than 40.00%. As the fermentation time increased, the relative abundance of
Ascomycota first increased, reaching a maximum of 87.47% at 12 h (WC1), the decreased. On the other hand, the relative abundance of
Basidomycota initially decreased, then increased, reaching a maximum of 59.30% at 36 h (WC3).
Ascomycota is the most diverse and richest phylum in the kingdom of fungi, with 110,000 known species [
15]. Due to their significant metabolic flexibility,
Ascomycota members are widely used in various biotechnological applications, such as the production of fatty alcohols, fatty acids, and biofuels and the reduction and degradation of chemicals and solvents [
15,
16].
Basidiomycota is also a major phylum in the kingdom of fungi, with more than 31,000 known species, which is second only to
Ascomycota in species numbers [
17].
Furthermore, these fungi in coffee samples were confirmed to belong to 10 genera, as shown in
Figure 2D. These genera were
Cystofilobasidum,
Hanseniaspora,
Aspergillus,
Thermomyces,
Lachances,
Wickerhamomyces,
Rasamsonia,
Candida,
Metschnikowia, and
Apiotrichum. The predominant genera were
Cystofilobasidu, Hanseniaspora, Lachancea, Wickerhamomyces, and
Aspergillus. The percentage of community abundance of
Cystofilobasidum at the genus level ranged from 7.54% to 53.41%, with the maximum value observed at 36 h (WC3) and the minimum value at 12 h (WC2). On the other hand, the maximum value for
Aspergillus was 10.76% at 12 h (WC1), and the minimum value was 0.90% at 36 h (WC3).
Wickerhamomyces exhibited a similar pattern to that of
Aspergillus, with a maximum of 7.93% at 12 h (WC1) and a minimum of 3.46% at 36 h (WC3). The percentages of community abundance of
Hanseniaspora were 3.45% at 0 h (WC0) and 26.80% at 24 h (WC2). For
Lachancea, the values of WC1 and WC2 were nearly equivalent at about 9%, and the minimum value was 3.38% at 36 h (WC3). The diversity of fungi in wet coffee processing is often lower than in dry and semi-dry processing, mainly because of the shorter fermentation time and the submerged environment [
4]. The main filamentous fungi are
Aspergillus,
Penicillium,
Fusarium,
Rhizopus,
Mucor, and
Cladosporium [
4,
18]. Yeast species such as
Hanseniaspora and
Candida were frequently isolated from different locations worldwide [
4].
Although many microorganism species are common in coffee fermentation, some are specific to certain regions. For example, in Australia,
Citrobacter and
Leuconostoc are predominant genera, along with
Pichia and
Hanseniaspora [
3]. In Brazil,
Pichia,
Candida, and
Hanseniaspora are the most frequent isolated genera [
5]. In India,
Saccharomyces,
Shizosaccharomyces,
Bacillus,
Lactobacillus,
Leuconostoc,
Pseudomonas, and
Flavobacterium are dominant genera during initial fermentation stages [
18]. In China,
Enterobacter,
Bacillus,
Pseudomonas,
Gluconobacter,
Kluyvera, and
Candida are dominant in the wet processing of
C. arabica [
13]. Based on the results of this study,
Achromobacter, Tatumella, Weissella, Streptococcus, and
Trichocoleus were found to be predominant in the bacterial community, while
Cystofilobasidu, Hanseniaspora, Lachancea, Wickerhamomyces, and Aspergillus were predominant in the fungal community during complete washed processing.
2.2. Differentially Changed Matabolities (DCMs) Analysis
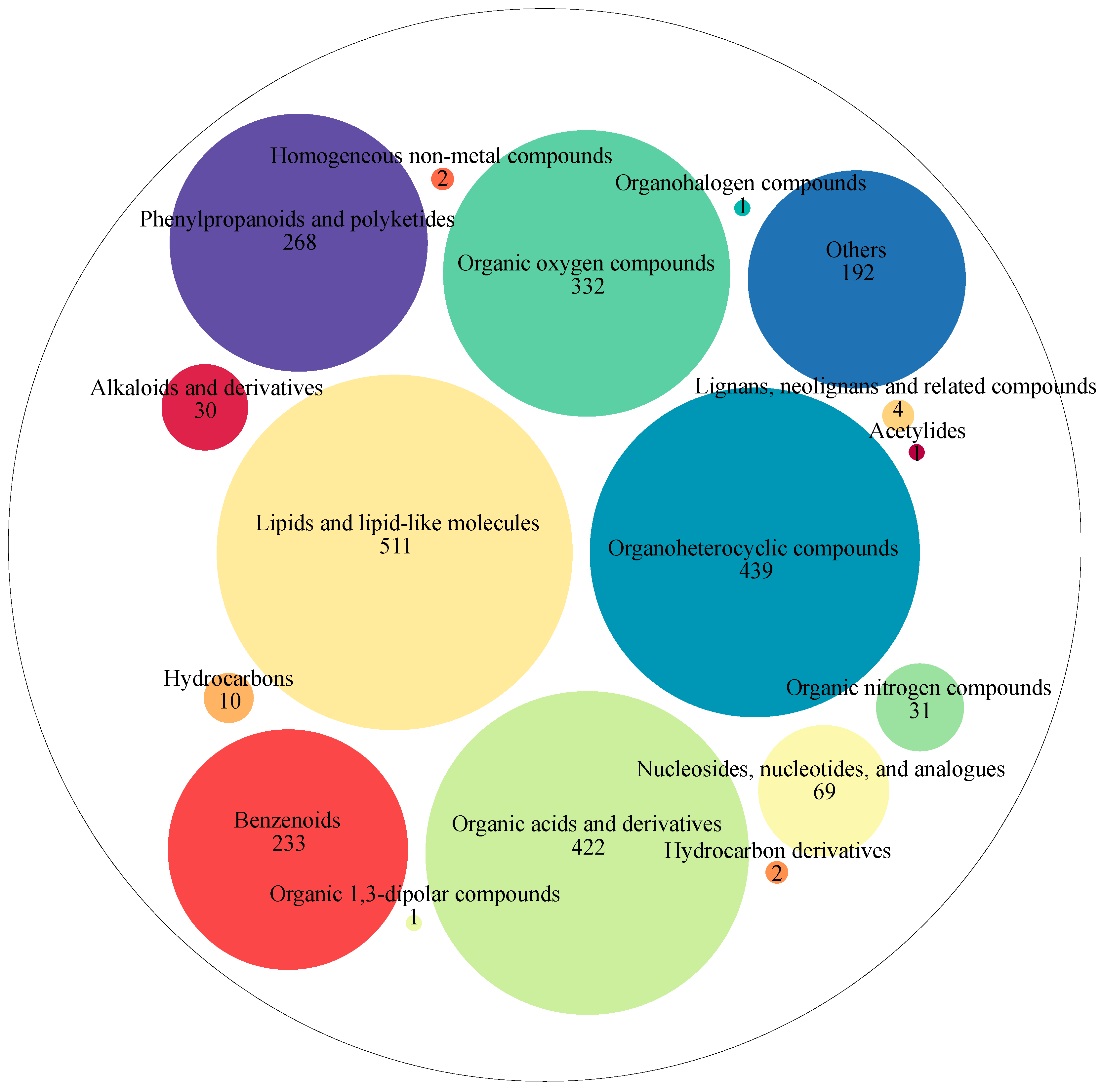
A total of 2548 metabolites were detected in coffee samples from different fermentation times during washed coffee processing. These metabolites were classified into 17 super-classes, as shown in
Figure 3. These super classes included lipids and lipid-like molecules (511 metabolites), organoheterocyclic compounds (439 metabolites), organic acids and derivatives (422 metabolites), organic oxygen compounds (332 metabolites), phenylpropanoids and polyketides (268 metabolites), benzenoids (233 metabolites), nucleosides, nucleotides, and analogs (69 metabolites), organic nitrogen compounds (31 metabolites), alkaloids and derivatives (30 metabolites), hydrocarbons (10 metabolites), lignans, neolignans, and related compounds (4 metabolites), hydrocarbon derivatives (2 metabolites), homogeneous non-metal compounds (2 metabolites), organic 1,3-dipolar compounds (1 metabolite), organohalogen compounds (1 metabolite), acetylides (1 metabolite), and others (192 metabolites).
Then, these metabolites were further grouped into 154 classes. These classes mainly included carboxylic acids and derivatives (352 metabolites), organooxygen compounds (331 metabolites), fatty acyls (203 metabolites), prenol lipids (157 metabolites), benzene and substituted derivatives (128 metabolites), flavonoids (88 metabolites), steroids and steroid derivatives (84 metabolites), phenols (60 metabolites), indoles and derivatives (54 metabolites), glycerophospholipids (52 metabolites), cinnamic acids and derivatives (49 metabolites), pyridines and derivatives (45 metabolites), coumarins and derivatives (41 metabolites), benzopyrans (37 metabolites), organonitrogen compounds (31 metabolites), isoflavonoids (27 metabolites), keto acids and derivatives (27 metabolites), imidazopyrimidines (26 metabolites), quinolines and derivatives (25 metabolites), lactones (19 metabolites), purine nucleosides (19 metabolites), hydroxy acids and derivatives (18 metabolites), naphthalenes (17 metabolites), phenylpropanoic acids (17 metabolites), pyrans (16 metabolites), pyrimidine nucleoside (16 metabolites), dihydrofurans (15 metabolites), piperidines (15 metabolites), peptidomimetics (13 metabolites), azoles (12 metabolites), diazines (12 metabolites), glycerolipids (10 metabolites), heteroaromatic compounds (10 metabolites), pyrrolidines (10 metabolites), and others.
The metabolites detected in different coffee samples were 2441 in WC0, 2391 in WC1, 2409 in WC2, and 2384 in WC3, respectively. There were 47 metabolites found exclusively in WC0, such as isomaltol, cimifugin, 4-methylumbelliferyl acetate, PE (15:0/22:1(13Z)), atalantoflavone, and luteoforol, which might be consumed during fermentation. There were 20 metabolites exclusively found in WC3, including salsoline-1-carboxylate, (S)-2,3-dihydro-7-hydroxy-2-methyl-4-oxo-4H-1-benzopyran-5-acetic acid, indole-3-acetaldehyde, capecitabine, sterigmatocystin, 8-methoxykynurenate, versetamide, calystegin A3, and amphetamine, which might be products of the fermentation process. Additionally, some metabolites, such as oleuropein, prosaikogenin A, and 2-amino-4-[(2-hydroxy-1-oxopropyl)amino]butanoic acid, were found in WC1, WC2, and WC3, indicating that they may have been produced during the initial stages and throughout the fermentation process.
Coffee contains a variety of chemical compounds, including nucleotides and derivatives, tannins, flavonoids, alkaloids, benzene and derivatives, phenylpropanoids, amino acid and derivatives, lipids, heterocyclic compounds, carboxylic acids and derivatives, saccharides, and others [
19]. Members of these compounds have bioactive functions for health. Some of these compounds, such as sugar, proteins, amino acid, and phenolic compounds, are coffee flavor precursors, which form coffee aroma through Maillard reactions, Strecker degradation, caramelization reaction, and fragmentation reactions. [
20]. For example, furanones, such as 4-hydroxy-2,5-dimethyl-3(2H)-furanone and 2(5)-ethyl-4-hydroxy-5(2)-methyl- 3(2H)-furan, generated by the Maillard reaction and subsequent aldol condensation, significantly influence the sweet caramel aroma of roasted coffee [
21].
Caffeine, trigonelline, chlorogenic acids, carboxylic acids, carbohydrates and polymeric polysaccharides, lipids, protein, melanoidins, and minerals are crucial to coffee flavor [
21,
22]. Caffeine can contribute to coffee bitterness [
21,
22], while trigonelline contributes to its overall aroma by forming aromatic compounds [
21,
22]. Furthermore, four chlorogenic acids (chlorogenic acid, isochlorogenic acid, cryptochlorogenic acid, and trans-chlorogenic acid), four feruloylquinic acids (feruloylquinic acid, 3-feruloylquinic acid, 3-
O-feruloylquinic acid, and 4,5-diferuloylquinic acid), and 14 caffeoylquinic acids (1-caffeoylquinic acid, 1-
O-caffeoylquinic acid, 5-caffeoylquinic acid, cis-5-caffeoylquinic acid, dicaffeoylquinic acid, 1,5-dicaffeoylquinic acid, 3,5-di-
O-caffeoylquinic acid, 3,4-di-O-caffeoylquinic acid, 1,3-dicaffeoylquinic acid, 4,5-dicaffeoylquinic acid, 3-caffeoyl-4-feruloylquinic acid, 4-
O-caffeoyl-3-
O-feruloylquinic acid, 3-
O-caffeoyl-4-
O-methylquinic acid, and 3,4,5-tricaffeoylquinic acid) were detected during wet processing, which undergo thermal degradation during calcination to form phenolic compounds and phenolic aromatic compounds that cause bitterness [
22]. Citric acid and derivatives (methylisocitric acid, isocitric acid, and (1R,2R)-isocitric acid), malic acid and derivatives (2-isopropylmalic acid, and 3-isoproylmalic acid), chlorogenic acids and derivatives, quinic acid and derivatives (5-dehydroquinic acids, 3-dehydroquinic acid, and 5 coumaroylquinic acids (p-coumaroylquinic acid, 4-
O-p-coumaroylquinic acid, 4-p-coumaroylquinic acid, 5-p-coumaroylquinic acid, and 3-
O-p-coumaroylquinic acid, and 5-sinapoylquinic acid) were also detected. Citric, malic, chlorogenic, and quinic acids are the primary acidic compounds, occupying about 11% of green coffee beans, which form lactones and volatile phenols during the roasting of coffee beans, influencing coffee aroma [
20,
21].
When mature coffee cherries are harvested, a primary processing is necessary to obtain green coffee beans for the storage, transportation, and roasting of coffee [
1]. There are various methods of obtaining green coffee beans in postharvest processing, including wet, dry, and semi-dry processing methods [
1]. Wet processing is often used for
Coffee arabica and involves submerged fermentation for 12–36 h after removing the mesocarp, resulting in coffee with a higher quality [
23]. Fermentation is a natural and critical process for removing the mucilage and reducing water content [
4,
24] through enzymes that naturally occur in the coffee fruit and microflora acquired from the environment. During fermentation, microbial metabolites can migrate into the coffee and change various physiological parameters, such as water content, simple sugars, aroma, and other flavor precursors [
4,
25].
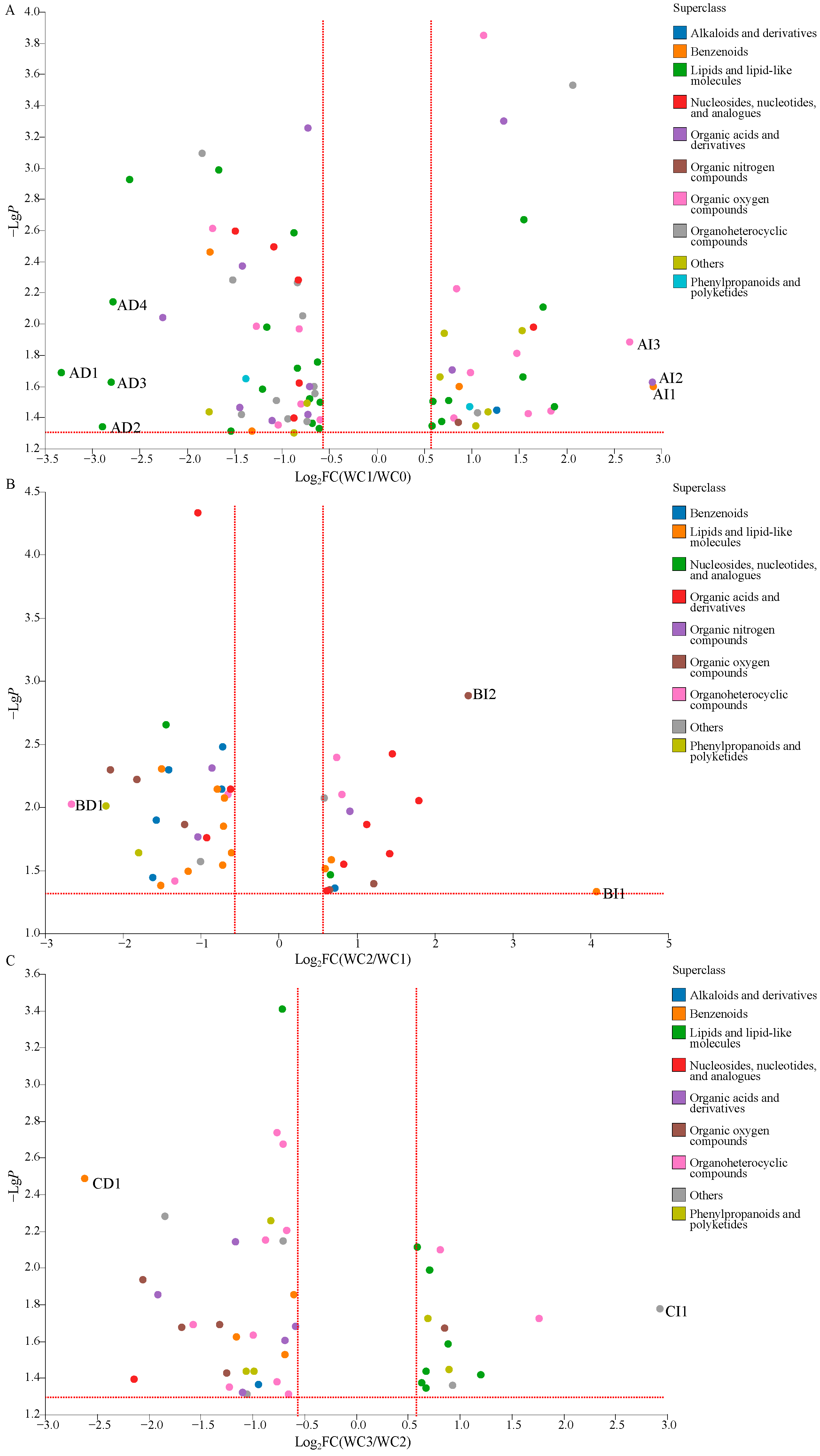
To gain further information about wet coffee processing, an overall comparison of the DCMs of WC0, WC1, WC2, and WC3 was carried out. As shown in
Figure 4A, 82 DCMs were detected when comparing WC1 to WC0. These DCMs related to alkaloids and derivatives (one metabolite), benzenoids (four metabolites), lipids and lipid-like molecules (24 metabolites), nucleosides, nucleotides, and analogues (six metabolites), organic acids and derivatives (10 metabolites), organic nitrogen compounds (one metabolite), organic oxygen compounds (14 metabolites), organoheterocyclic compounds (12 metabolites), phenylpropanoids and polyketides (2 metabolites), and others (eight metabolites). Among them, the relative levels of 50 metabolites were decreased significantly (VIP > 1.0,
p < 0.05, and FC < 0.67), included benzenoids (two metabolites, 4-hydroxy-3-methoxycinnmaldehyde, and diethyl phthalic acid), lipids and lipid-like molecules (16 metabolites, e.g., pimelic acid and carboprost methyl), nucleosides, nucleotides, and analogues (five metabolites, e.g., inosine, 5′-guanylic acid, and 3′-amino-3′-deoxythimidine glucuronide), organic acids and derivatives (seven metabolites, e.g., 4-fluoro-L-phenylalanine and γ-glutamylphenylalanine), organic oxygen compounds (six metabolites, e.g., hygromycin B and glucoheptonic acid), organoheterocyclic compounds (10 metabolites, e.g., isoeugenitol and guanine), phenylpropanoids and polyketides (one metabolite, 7-ethoxycoumarin), and others (three metabolites, securinine, sinapoyl aldehyde, and SM (d18:0/20:3(8Z,11Z,14Z)-2OH(5,6))). Among these DCMs, four metabolites, BULLEYACONITINE A, PS(15:0/24:1(15Z)), PC(18:2(9Z,12Z)/18:1(11Z)),and PC(14:1(9Z)/20:1(11Z)), were significantly decreased, with the value of FC less than 0.20. Meanwhile, the relative levels of 32 metabolites increased significantly (VIP > 1.0,
p < 0.05, and FC > 1.5), including alkaloids and derivatives (one metabolite, harmalol), benzenoids (two metabolites, 4-(phenylamino)benzoic acid, and 3,4-dihydroxymandelic acid), lipids and lipid-like molecules (eight metabolites, e.g., 3-hexaprenyl-4-hydroxybenzoic acid, and pelanin), nucleosides, nucleotides, and analogues (one metabolite, uridine diphosphate glucose), organic acids and derivatives (three metabolites, 2-aminoisobutyric acid, portulacaxanthinⅡ, and 10-EdAM), organic nitrogen compounds (one metabolite, agmatine), organic oxygen compounds (eight metabolites, e.g., 3-hydroxynevirapine glucuronide and panthenol), organoheterocyclic compounds (two metabolites, niacinamide and N,Nitrosopiperidine), phenylpropanoids and polyketides (one metabolite, 4-feruloyl-1,5-quinolactone), and others (five metabolites, e.g., N-docosahexaenoyl lysine and 2,6-dihydroxynaphthalene). Among these DCMs, three metabolites, 3,4-dihydroxymandelic acid, portulacaxanthin II, and mycophenolic acid
O-acyl-glucuronide were important increased with the value of FC over 5.0.
Similarly, 46 DCMs were detected between WC2 and WC1 (
Figure 4B), including benzenoids (six metabolites), lipids and lipid-like molecules (11 metabolites), nucleosides, nucleotides, and analogues (two metabolites), organic acids and derivatives (nine metabolites), organic nitrogen compounds (three metabolites), organic oxygen compounds (six metabolites), organoheterocyclic compounds (five metabolites), phenylpropanoids and polyketides (two metabolites), and others (two metabolites). The relative levels of 28 DCMs were decreased significantly (VIP > 1.0,
p < 0.05, and FC < 0.67), included benzenoids (five metabolites, e.g., menadione and isoeugenol), lipids and lipid-like molecules (eight metabolites, e.g., dodecanol and PE(16:0/0:0)), nucleosides, nucleotides, and analogs (one metabolite, inosine), organic acids and derivatives (three metabolites, 2-aminoisobutyric acid, 5-keto-D-gluconate, and enalaprilat), organic nitrogen compounds (two metabolites, betaine aldehyde and trolamine), organic oxygen compounds (three metabolites, caffeic acid 3-glucoside, D-gluconic acid, and difructose anhydrideⅢ), organoheterocyclic compounds (three metabolites, cantharidin, 7-methyladenine, and urocanic acid), phenylpropanoids and polyketides (two metabolites, 3-phenyllactic acid and trioxsalen), and others (one metabolite, DG(2:0/20:3(8Z,11Z.14Z)-2OH(5,6)/0:0)). Meanwhile, the relative levels of 18 DCMs were increased significantly (VIP > 1.0,
p < 0.05, and FC > 1.5), including benzenoids (one metabolite, ginnalin B), lipids and lipid-like molecules (3 metabolites, palmitic amide, α-dimorphecolic acid, and stanozolol), nucleosides, nucleotides, and analogues (one metabolite, 5′-N-methylcarboxamidoadenosine), organic acids and derivatives (six metabolites, e.g., N-valylphenylalanine, and 5-hydroxy saxagliptin), organic nitrogen compounds (one metabolite, xestoaminol C), organic oxygen compounds (three metabolites, D-arabitol, α-CEHC glucuronide, and 2-deoxy-D-ribose), organoheterocyclic compounds (two metabolites, dihydrobiopterin and austdiol), and others (one metabolite, 6-bromoquinolin-2(1H)-one). Two DCMs, stanozolol and 2-deoxy-D-ribose, were significantly increased with a value of FC over 5.0, and 1 DCM, cantharidin, was significantly decreased with a value of FC less than 0.20.
A total of 45 DCMs were detected between WC3 and WC2 (
Figure 4C), including alkaloids and derivatives (one metabolite), benzenoids (four metabolites), lipids and lipid-like molecules (eight metabolites), nucleosides, nucleotides, and analogues (1 metabolite), organic acids and derivatives (five metabolites), organic oxygen compounds (5 metabolites), organoheterocyclic compounds (11 metabolites), phenylpropanoids and polyketides (five metabolites), and others (five metabolites). Among them, the relative levels of 31 DCMs were decreased significantly (VIP > 1.0,
p < 0.05, and FC < 0.67), including alkaloids and derivatives (one metabolite, mdo-npa), benzenoids (four metabolites, e.g.,bisnoryangonin, and phenyl acetate), lipids and lipid-like molecules (one metabolite, lysoPI(16:0/0:0)), nucleosides, nucleotides, and analogues (one metabolite, didanosine), organic acids and derivatives (five metabolites, e.g., tetrahydrodipicolinate and D-dopachrome), organic oxygen compounds (four metabolites, e.g., melilotoside and uridine 2′-phosphate), organoheterocyclic compounds (nine metabolites, e.g., indoleacetic acid and khellin), phenylpropanoids and polyketides (three metabolites, biochanin A, cis-
p-coumaric acid, and biochanin A), and others (three metabolites, 13Z-docosenamide, citrinin hydrate, and PA(PGE2/i-16:0)). Meanwhile, the relative levels of 14 DCMs were increased significantly (VIP > 1.0,
p < 0.05, and FC > 1.5), including lipids and lipid-like molecules (seven metabolites, e.g., 15-demethyl plumieride and digalactosyldiacylglycerol), organic oxygen compounds (one metabolite, (
S)-Nerolidol 3-O-[a-L-Rhamnopyranosyl-(1->4)-a-L-rhamnopyranosyl-(1->2)-[4-(4-hydroxy-3-methoxycinnamoyl)-(E)-a-L-rhamnopyranosyl-(1->6)]-b-D-glucopyranoside]), organoheterocyclic compounds (two metabolites, lettucenin A, and 2,3-dimethylmaleic anhydride), phenylpropanoids and polyketides (two metabolites, 3″,6″-di-O-p-coumaroyltrifolin, and Fevicordin B 2-[rhamnosyl-(1->4)-glucosyl-(1->6)-glucoside]), and others (two metabolites, L-quinate, and DG(2:0/20:3(8Z,11Z,14Z)-2OH(5,6)/0:0)). Among them, L-quinate was a significantly increased DCM with a value of FC over 5.0 and bisnoryangonin was a significantly increased DCMs with a value of FC less than 0.20.
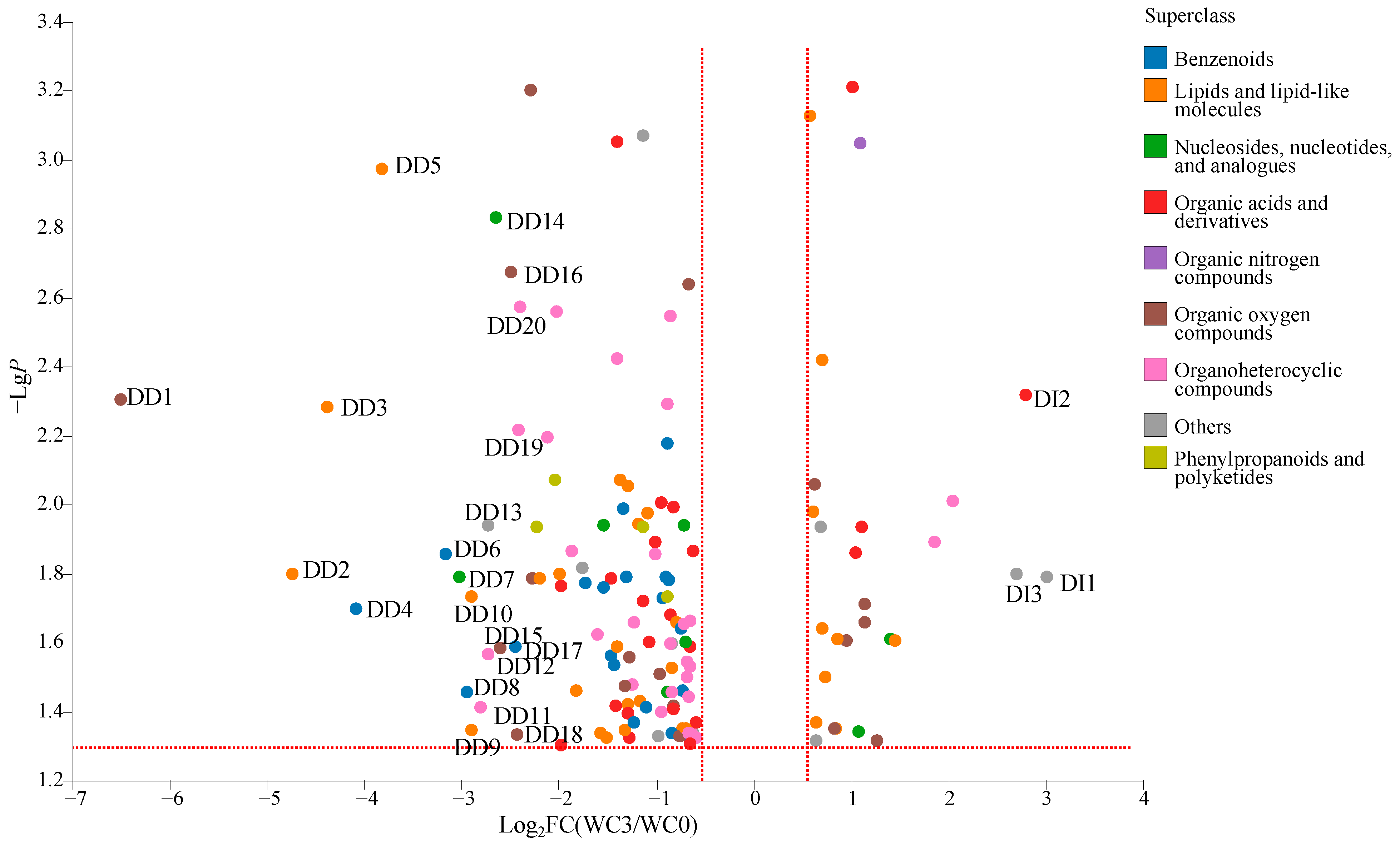
A total of 143 DCMs were detected between WC3 and WC0 (
Figure 5), including benzenoids (20 metabolites), lipids and lipid-like molecules (31 metabolites), nucleosides, nucleotides, and analogues (eight metabolites), organic acids and derivatives (22 metabolites), organic nitrogen compounds (one metabolite), organic oxygen compounds (18 metabolites), organoheterocyclic compounds (29 metabolites), phenylpropanoids and polyketides (4 metabolites), and others (10 metabolites). Among them, the relative levels of 115 DCMs were decreased significantly (VIP > 1.0,
p < 0.05, and FC < 0.67), including benzenoids (20 metabolites, e.g., ethyl benzoate and isoeugenol), lipids and lipid-like molecules (22 metabolites, e.g., 5-hexyl-2-furanoctanoic acid and valtrate), nucleosides, nucleotides, and analogues (six nVCs, e.g., inosine and guanosine), organic acids and derivatives (18 metabolites, e.g., trioxyethylene dimethacrylate and 3-methyl-2-oxovaleric acid), organic oxygen compounds (12 metabolites, e.g., melilotoside and D-gluconic acid), organoheterocyclic compounds (27 metabolites, e.g., isoeugenitol and cabotegravir), phenylpropanoids and polyketides (four metabolites, e.g., trioxsalen and sinapic acid), and others (six metabolites, e.g., citrinin hydrate and 2-aminophenol). Meanwhile, the relative levels of 28 DCMs were increased significantly (VIP > 1.0,
p < 0.05, and FC > 1.5), including lipids and lipid-like molecules (nine metabolites, e.g., arctiol and α-dimorphecolic acid), nucleosides, nucleotides, and analogues (two metabolites, uridine diphosphate-N-acetylglucosamine, and uridine diphosphate glucose), organic acids and derivatives (four metabolites, e.g., sulbactam and portulacaxanthin Ⅱ), organic nitrogen compounds (one metabolite, xestoaminol C), organic oxygen compounds (six metabolites, e.g., aziridyl benzoquinone and panthenol), organoheterocyclic compounds (two metabolites, N-nitrosopiperidine, and niacinamide), and others (four metabolites, e.g., L-quinate and dodemorph). Meanwhile, 23 DCMs were important, with FC > 5.0 (DI1-DI3) or FC < 0.20 (DD1-DD20).