Natural Products from Singapore Soil-Derived Streptomycetaceae Family and Evaluation of Their Biological Activities
Abstract
1. Introduction
2. Results and Discussion
2.1. Phylogenetic Analysis and Molecular Identification of Actinobacteria Isolates
2.2. Preliminary Screening of Actinobacteria Isolates
2.3. Isolation and Structural Elucidation of Bioactive Compounds
2.4. Chemical Structural Data of Tetronomycin A (1)
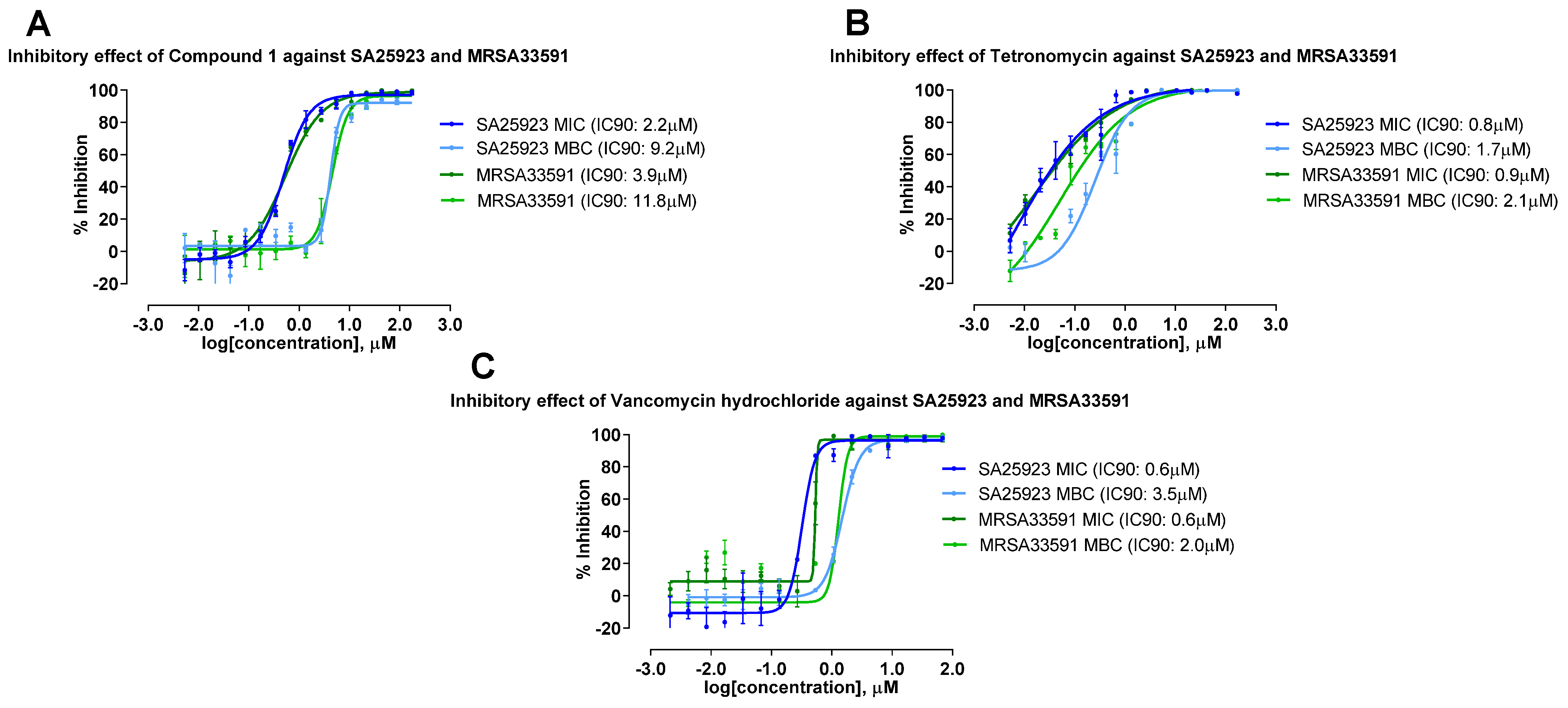
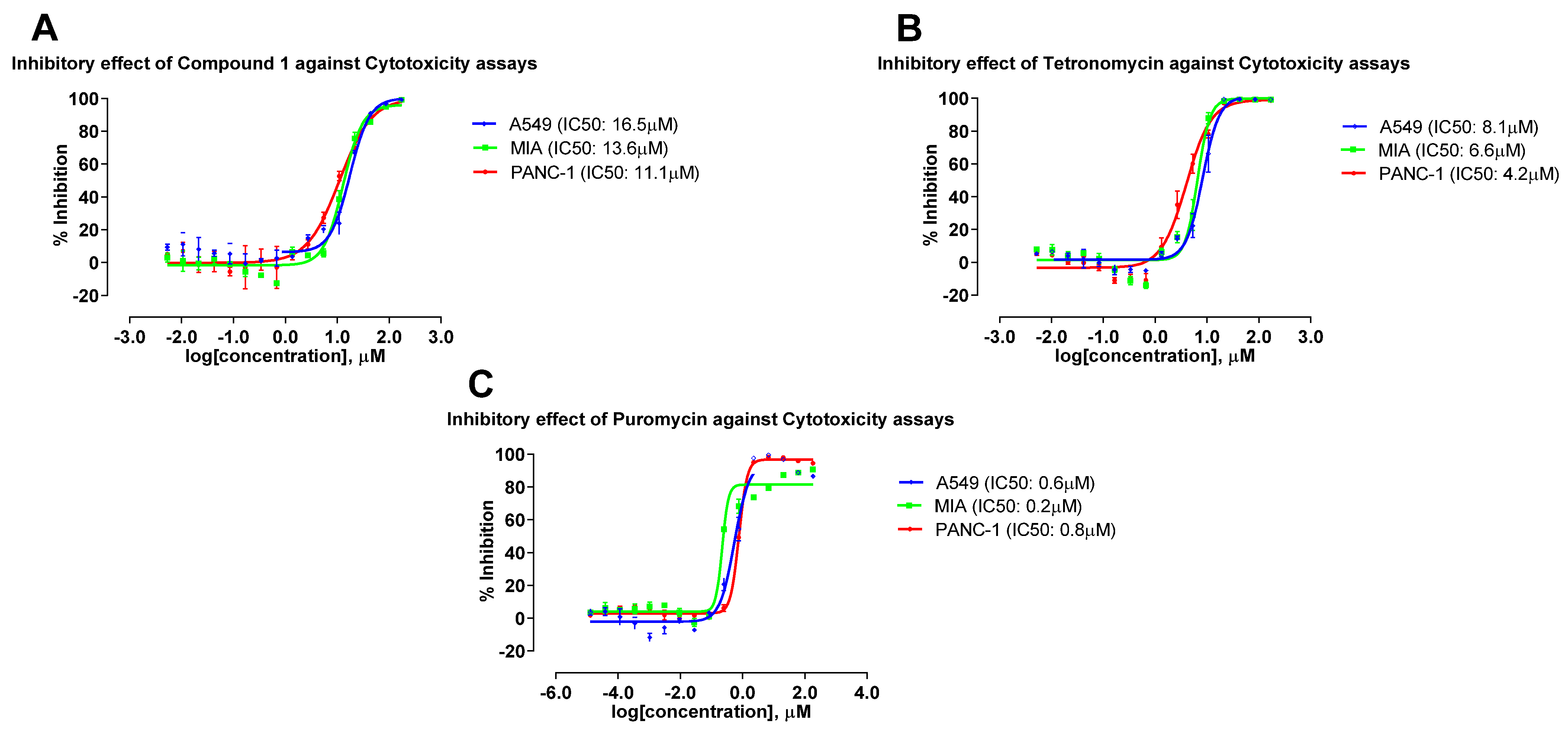
2.5. Antimicrobial and Cytotoxic Activities of Compounds Isolated from the 4 Actinobacterial Strains
2.6. Effects of Growth Media on Production of Bioactive Compounds
3. Materials and Methods
3.1. Molecular Identification and Phylogenetic Analysis of Actinobacteria Isolates
3.2. Fermentation and Extraction of Actinobacterial Crude Extracts
3.3. Biological Assays
3.4. Natural Product Extraction, Compound Isolation, and Structure Elucidation
3.5. General Chemistry Experimental Procedures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Elmaidomy, A.H.; Shady, N.H.; Abdeljawad, K.M.; Elzamkan, M.B.; Helmy, H.H.; Tarshan, E.A.; Adly, A.N.; Hussien, Y.H.; Sayed, N.G.; Zayed, A. Antimicrobial Potentials of Natural Products Against Multidrug Resistance Pathogens: A Comprehensive Review. RSC Adv. 2022, 12, 29078–29102. [Google Scholar] [CrossRef]
- Jinfeng, E.C.; Mohamad Rafi, M.I.; Chai Hoon, K.; Kok Lian, H.; Yoke Kqueen, C. Analysis of Chemical Constituents, Antimicrobial and Anticancer Activities of Dichloromethane Extracts of Sordariomycetes sp. Endophytic Fungi Isolated from Strobilanthes crispus. World J. Microbiol. Biotechnol. 2017, 33, 5. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Abraham, J. Natural Products from Actinobacteria for Drug Discovery. In Advances in Pharmaceutical Biotechnology; Springer: Singapore, 2020; pp. 333–363. [Google Scholar] [CrossRef]
- Gohain, A.; Manpoong, C.; Saikia, R.; De Mandal, S. Actinobacteria: Diversity and Biotechnological Applications. In Recent Advancements in Microbial Diversity; Academic Press: Cambridge, MA, USA, 2020; pp. 217–231. [Google Scholar] [CrossRef]
- Berdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Mast, Y.; Stegmann, E. Actinomycetes: The Antibiotics Producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Ching, K.-C.; Chin, E.J.; Wibowo, M.; Tan, Z.Y.; Yang, L.-K.; Seow, D.C.; Leong, C.-Y.; Ng, V.W.; Ng, S.-B.; Kanagasundaram, Y. Antibacterial Thiopeptide GE2270-Congeners from Nonomuraea jiangxiensis. Molecules 2023, 28, 101. [Google Scholar] [CrossRef]
- Ching, K.-C.; Chin, E.J.; Wibowo, M.; Tan, Z.Y.; Yang, L.-K.; Seow, D.C.; Leong, C.-Y.; Ng, V.W.; Ng, S.-B.; Kanagasundaram, Y. Antibacterial Spirotetronate Polyketides from an Actinomadura sp. Strain A30804. Molecules 2022, 27, 8196. [Google Scholar] [CrossRef]
- Ibrahim, O.O. Staphylococcus aureus a Gram-positive Coccid Bacterium Causing Microbial Infections, and Toxins Symptoms Including Food Poisoning. EC Microbiol. 2020, 16, 61–76. [Google Scholar]
- ul Hassan, S.S.; Anjum, K.; Abbas, S.Q.; Akhter, N.; Shagufta, B.I.; Shah, S.A.A.; Tasneem, U. Emerging Biopharmaceuticals From Marine Actinobacteria. Environ. Toxicol. Pharmacol. 2017, 49, 34–47. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Li, W.-J. Diversity of Actinobacteria in Various Habitats. In Actinobacteria: Microbiology to Synthetic Biology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 37–58. [Google Scholar]
- Ng, S.B.; Kanagasundaram, Y.; Fan, H.; Arumugam, P.; Eisenhaber, B.; Eisenhaber, F. The 160K Natural Organism Library, A Unique Resource for Natural Products Research. Nat. Biotechnol. 2018, 36, 570–573. [Google Scholar] [CrossRef]
- Pepper, I.L.; Gentry, T.J. Earth Environments. In Environmental Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 59–88. [Google Scholar]
- Feeney, M.A.; Newitt, J.T.; Addington, E.; Algora-Gallardo, L.; Allan, C.; Balis, L.; Birke, A.S.; Castaño-Espriu, L.; Charkoudian, L.K.; Devine, R. ActinoBase: Tools and Protocols for Researchers Working on Streptomyces and Other Filamentous Cctinobacteria. Microb. Genom. 2022, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Al-Shaibani, M.M.; Radin Mohamed, R.M.S.; Sidik, N.M.; Enshasy, H.A.E.; Al-Gheethi, A.; Noman, E.; Al-Mekhlafi, N.A.; Zin, N.M. Biodiversity of Secondary Metabolites Compounds Isolated From Phylum Actinobacteria and its Therapeutic Applications. Molecules 2021, 26, 4504. [Google Scholar] [CrossRef] [PubMed]
- Delbari, Y.; Mohassel, Y.; Bahrami, Y.; Kakaie, E.; Mostafaie, A. A Review on Isolation and Identification of Endophytic Actinobacteria, Their Chemical Structure, Bioactive Compounds, and Potential Medical-Pharmaceutical Applications. J. Maz. Univ. Med. 2020, 30, 195–217. [Google Scholar]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects From Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Allikian, K.; Edgar, R.; Syed, R.; Zhang, S. Fundamentals of Fermentation Media. In Essentials in Fermentation Technology; Springer: Cham, Switzerland, 2019; pp. 41–84. [Google Scholar] [CrossRef]
- VanderMolen, K.M.; Raja, H.A.; El-Elimat, T.; Oberlies, N.H. Evaluation of Culture Media for the Production of Secondary Metabolites in a Natural Products Screening Program. AMB Express 2013, 3, 71. [Google Scholar] [CrossRef]
- Gunsalus, I.; Horecker, B.; Wood, W. Pathways of Carbohydrate Metabolism in Microorganisms. Bacteriol. Rev. 1955, 19, 79–128. [Google Scholar] [CrossRef]
- Keller-Juslen, C.; King, H.D.; Kuhn, M.; Loosli, H.-R.; Pache, W.; Petcher, T.J.; Weber, H.P.; Von Wartburg, A. Tetronomycin, a Novel Polyether of Unusual Structure. J. Antibiot. 1982, 35, 142–150. [Google Scholar] [CrossRef]
- Kimishima, A.; Tsuruoka, I.; Kanto, H.; Tsutsumi, H.; Arima, N.; Sakai, K.; Sugamata, M.; Matsui, H.; Watanabe, Y.; Iwatsuki, M. Rediscovery of Tetronomycin as a Broad-Spectrum and Potent Antibiotic Against Drug-Resistant Gram-Positive Bacteria. ACS Omega 2023, 8, 11556–11563. [Google Scholar] [CrossRef]
- Henkel, T.; Zeeck, A. Secondary Metabolites by Chemical Screening. 15 Structure and Absolute Configuration of Naphthomevalin, A New Dihydro-Naphthoquinone Antibiotic From Streptomyces sp. J. Antibiot. 1991, 44, 665–669. [Google Scholar] [CrossRef]
- Miles, Z.D.; Diethelm, S.; Pepper, H.P.; Huang, D.M.; George, J.H.; Moore, B.S. A Unifying Paradigm for Naphthoquinone-Based Meroterpenoid (Bio) Synthesis. Nat. Chem. 2017, 9, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Bockholt, H.; Udvarnoki, G.; Rohr, J.; Mocek, U.; Beale, J.M.; Floss, H.G. Biosynthetic Studies on the Xanthone Antibiotics Lysolipins X And I. J. Org. Chem. 1994, 59, 2064–2069. [Google Scholar] [CrossRef]
- Guimarães, L.A.; Jimenez, P.C.; Sousa, T.D.S.; Freitas, H.P.S.; Rocha, D.D.; Wilke, D.V.; Martín, J.; Reyes, F.; Pessoa, O.D.L.; Costa-Lotufo, L.V. Chromomycin A2 Induces Autophagy in Melanoma Cells. Mar. Drugs 2014, 12, 5839–5855. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Y.-P. Synthesis of Nonactin and the Proposed Structure of Trilactone. Org. Lett. 2006, 8, 2831–2834. [Google Scholar] [CrossRef]
- Kitagawa, I.; Yoshikawa, M.; Yosioka, I. Saponin And Sapogenol. XIII. Structures of Three Soybean Saponins: Soyasaponin I, Soyasaponin II, And Soyasaponin III. Chem. Pharm. Bull. 1976, 24, 121–129. [Google Scholar] [CrossRef]
- Haneda, M.; Nawata, Y.; Hayashi, T.; Ando, K. Tetranactin, A New Miticidal Antibiotic. VI Determination of Dinactin, Trinactin and Tetranactin in Their Mixtures by NMR Spectroscopy. J. Antibiot. 1974, 27, 555–557. [Google Scholar] [CrossRef]
- Hu, Y.; Espindola, A.P.D.; Stewart, N.A.; Wei, S.; Posner, B.A.; MacMillan, J.B. Chromomycin SA Analogs from A Marine-Derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 5183–5189. [Google Scholar] [CrossRef]
- Beck, J.; Gerlach, H.; Prelog, V.; Voser, W. Stoffwechselprodukte von Actinomyceten. 35. Mitteilung. Über die Konstitution der Makrotetrolide Monactin, Dinactin und Trinactin. Helv. Chim. Acta 1962, 45, 620–630. [Google Scholar] [CrossRef]
- Nakahashi, A.; Yaguchi, Y.; Miura, N.; Emura, M.; Monde, K. A Vibrational Circular Dichroism Approach to the Determination of the Absolute Configurations of Flavorous 5-substituted-2 (5 H)-furanones. J. Nat. Prod. 2011, 74, 707–711. [Google Scholar] [CrossRef]
- Yong, K.W.; Barnych, B.; De Voss, J.J.; Vatèle, J.-M.; Garson, M.J. Plakortolide Stereochemistry Revisited: The Checkered History of Plakortolides E and I. J. Nat. Prod. 2012, 75, 1792–1797. [Google Scholar] [CrossRef]
- Cho, E.; Kwon, O.-S.; Chung, B.; Lee, J.; Sun, J.; Shin, J.; Oh, K.-B. Antibacterial Activity of Chromomycins from a Marine-Derived Streptomyces microflavus. Mar. Drugs 2020, 18, 522. [Google Scholar] [CrossRef] [PubMed]
- Guang, C.; Chen, J.; Sang, S.; Cheng, S. Biological Functionality of Soyasaponins and Soyasapogenols. J. Agric. Food Chem. 2014, 62, 8247–8255. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hu, X.; Li, L.; Hu, X.; Wang, J.; Hu, X.; Liu, H.; Yu, L.; You, X.; Jiang, B. 1-hydroxy-7-oxolavanducyanin and Δ7″, 8″-6″-hydroxynaphthomevalin from Streptomyces sp. CPCC 203577. J. Antibiot. 2020, 73, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Jizba, J.; Sedmera, P.; Zima, J.; Beran, M.; Blumauerová, M.; Kandybin, N.; Samoukina, G. Macrotetrolide Antibiotics Produced by Streptomyces globisporus. Folia Microbiol. 1991, 36, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.P.; Prova, S.S.; Moraes, L.A.B.; Ifa, D.R. Characterization and Mapping of Secondary Metabolites of Streptomyces sp. from Caatinga by Desorption Electrospray Ionization Mass Spectrometry (DESI–MS). Anal. Bioanal. Chem. Res. 2018, 410, 7135–7144. [Google Scholar] [CrossRef]
- Kuncharoen, N.; Bunbamrung, N.; Intaraudom, C.; Choowong, W.; Thawai, C.; Tanasupawat, S.; Pittayakhajonwut, P. Antimalarial and Antimicrobial Substances Isolated from the Endophytic Actinomycete, Streptomyces aculeolatus MS1-6. Phytochemistry 2023, 207, 113568. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and Evaluation of Useful Bacterium-specific PCR Primers That Amplify Genes Coding for Bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sirota, F.L.; Goh, F.; Low, K.-N.; Yang, L.-K.; Crasta, S.C.; Eisenhaber, B.; Eisenhaber, F.; Kanagasundaram, Y.; Ng, S.B. Isolation and identification of an Anthracimycin analogue from Nocardiopsis kunsanensis, a halophile from a Saltern, by genomic mining strategy. J. Genom. 2018, 6, 63. [Google Scholar] [CrossRef]
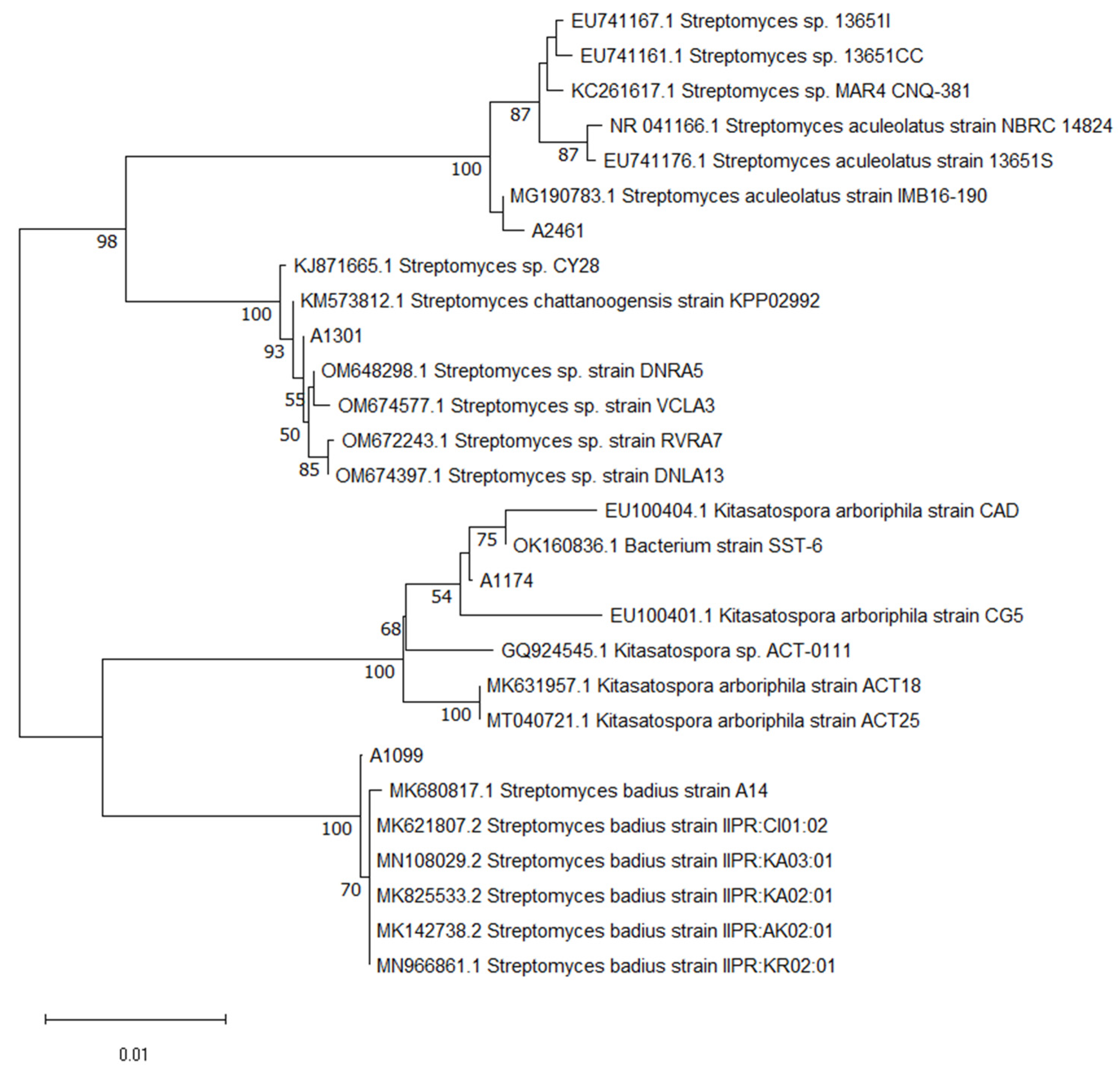

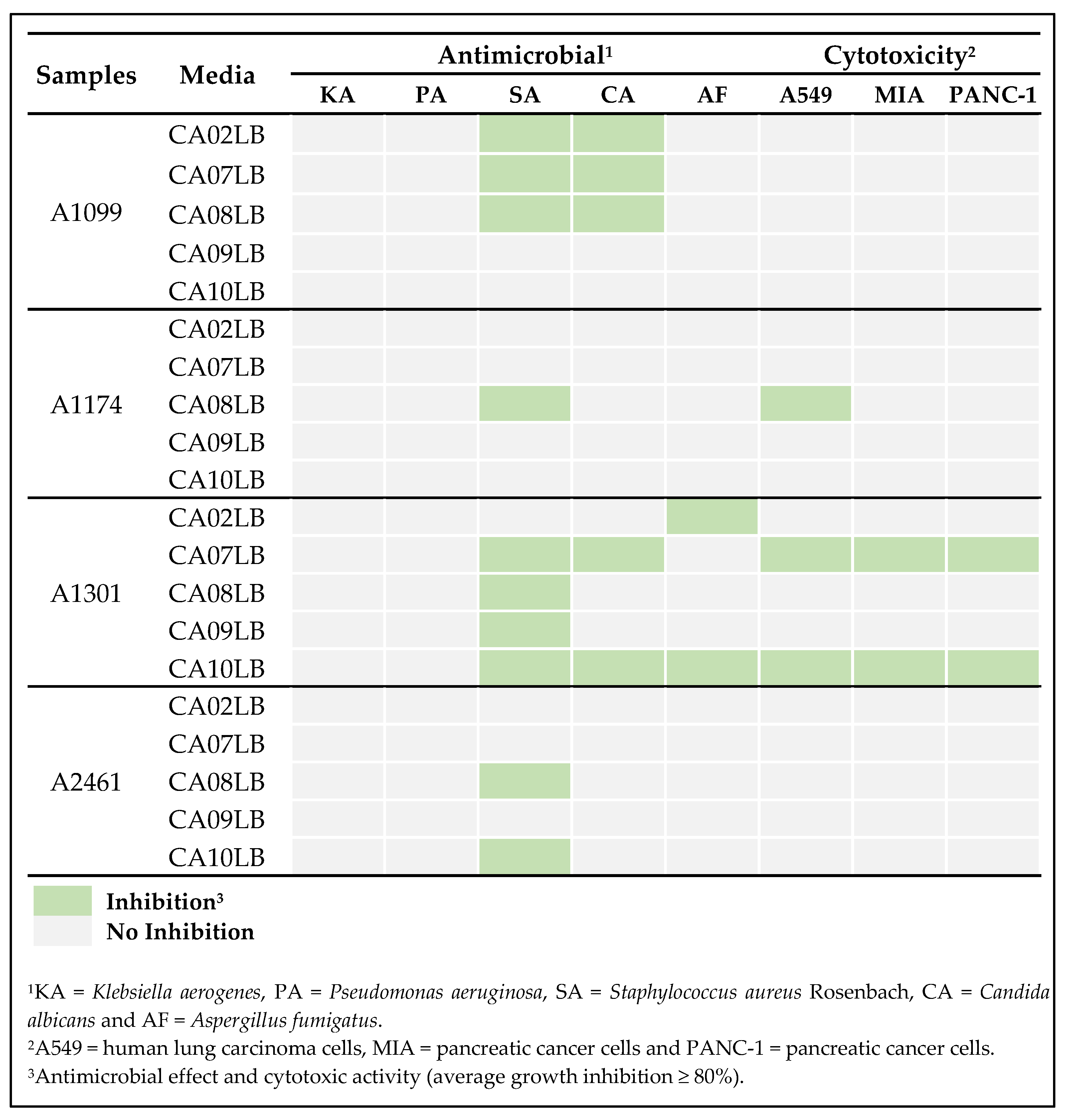

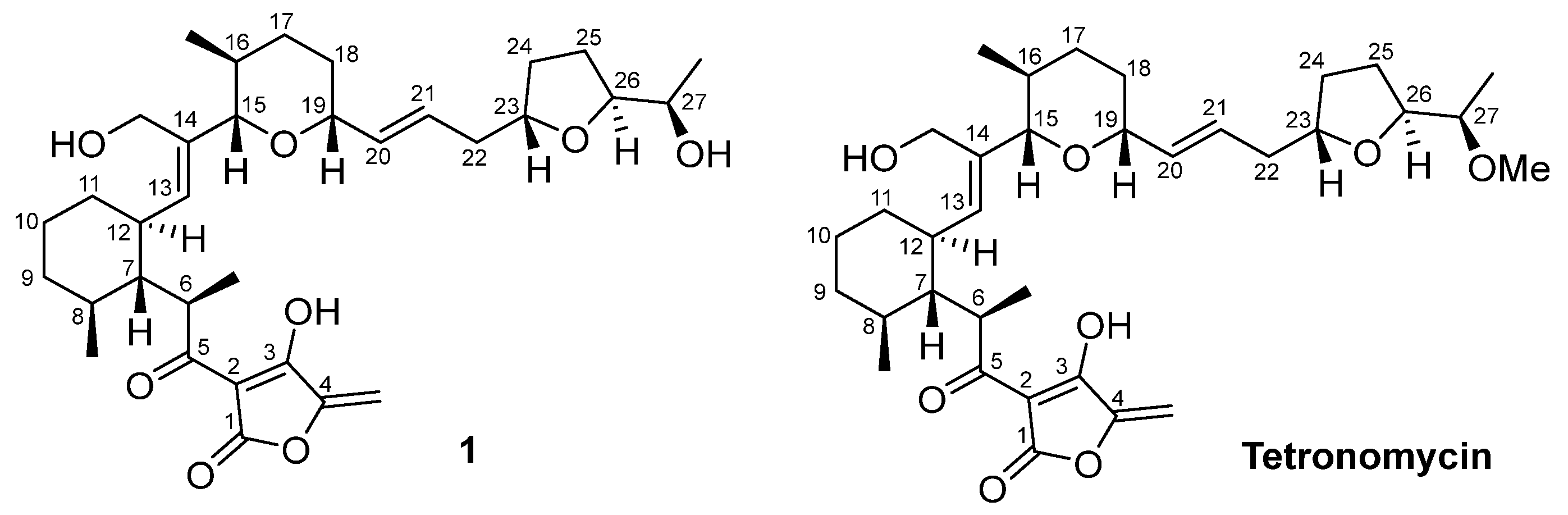

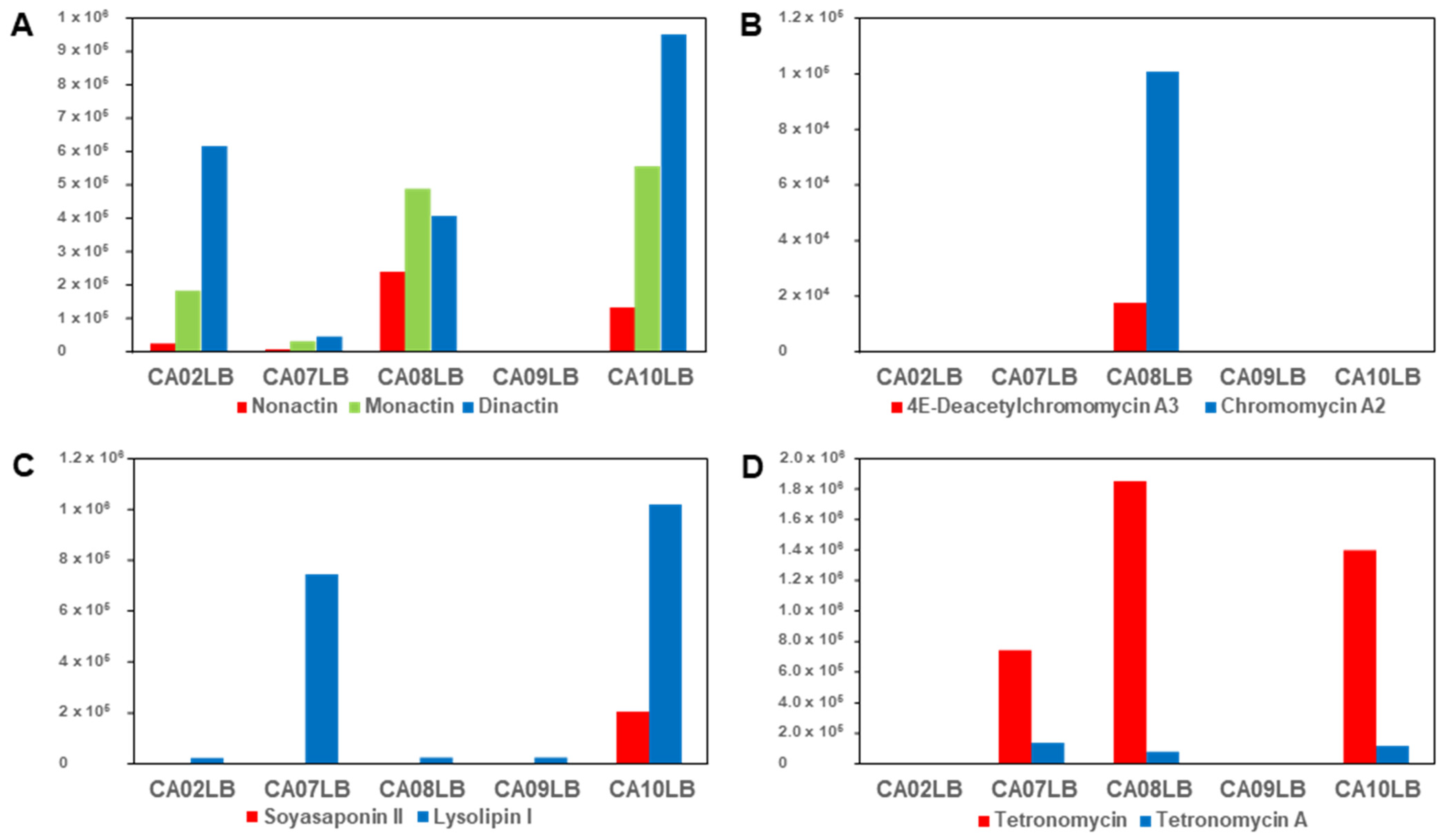
| Strain | Media | Compounds Confirmation |
|---|---|---|
| Streptomyces sp. A1099 | CA08LB | Nonactin, monactin, dinactin |
| Kitasatospora sp. A1174 | CA08LB | 4E-Deacetylchromomycin A3, chromomycin A2 |
| Streptomyces sp. A1301 | CA10LB | Soyasaponin II, lysolipin I |
| Streptomyces sp. A2461 | CA10LB | Tetronomycin A (1), tetronomycin and naphthomevalin |
| 1 | Tetronomycin | |||
|---|---|---|---|---|
| Pos. | 13C, Type 1 | 1H, Mult. (J = Hz) | 13C, Type 1 | 1H, Mult. (J = Hz) |
| 1 | 179.7, C | - | 180.5, C | - |
| 2 | n.d., C | - | n.d., C | - |
| 3 | 182.6, C | - | 182.3, C | - |
| 4 | 155.2, C | - | 155.6, C | - |
| 4-CH2 | 85.9, CH2 | 4.55, d (1.0); 4.93, d (1.0) | 85.9, CH2 | 4.54, d (1.0); 4.93, d (1.0) |
| 5 | 201.4, C | - | 201.3, C | - |
| 6 | 43.3, CH | 3.73, m | 43.3, CH | 3.81, m |
| 6-Me | 8.6, CH3 | 0.94, d (7.0) | 9.0, CH3 | 0.98, d (7.1) |
| 7 | 48.5, CH | 1.83, m | 48.5, CH | 1.81, m |
| 8 | 33.1, CH | 1.48, m | 33.1, CH | 1.45, m |
| 8-Me | 20.0, CH3 | 1.14, m | 20.1, CH3 | 1.14, m |
| 9 | 36.3, CH2 | 1.08, m; 1.64, m | 36.1, CH2 | 1.06, m; 1.64, m |
| 10 | 25.9, CH2 | 1.30, m; 1.59, m | 25.9, CH2 | 1.27, m; 1.58, m |
| 11 | 35.4, CH2 | 1.01, m, 1.44, m | 35.7, CH2 | 1.00, m, 1.45, m |
| 12 | 36.4, CH | 2.54, m | 36.5, CH | 2.55, m |
| 13 | 141.6, CH | 5.10, d (10.1) | 141.5, CH | 5.10, d (10.1) |
| 14 | 132.5, C | - | n.d., C | - |
| 14-CH2 | 56.5, CH2 | 3.83, m; 4.14, m | 56.5, CH2 | 3.84, m; 4.19, m |
| 15 | 91.7, CH | 3.19, m | 91.7, CH | 3.20, m |
| 16 | 34.5, CH | 1.40, m | 34.4, CH | 1.39, m |
| 16-Me | 18.2, CH3 | 0.58, d (6.6) | 18.3, CH3 | 0.58, d (6.8) |
| 17 | 32.6, CH2 | 1.23, m; 1.80, m | 32.7, CH2 | 1.22, m; 1.80, m |
| 18 | 32.1, CH2 | 1.48, m; 1.61, m | 32.0, CH2 | 1.46, m; 1.60, m |
| 19 | 80.0, CH | 3.80, m | 80.0, CH | 3.79, m |
| 20 | 132.2, CH | 5.57, dd (8.6, 15.1) | 132.8, CH | 5.54, dd (8.7, 15.6) |
| 21 | 135.4, CH | 6.19, m | n.d., CH | 6.14, m |
| 22 | 39.8, CH2 | 2.19, m; 2.39, m | 40.1, CH2 | 2.06, m; 2.38, m |
| 23 | 78.7, CH | 4.10, m | 78.9, CH | 4.11, m |
| 24 | 32.3, CH2 | 1.58, m; 2.12, m | 32.2, CH2 | 1.60, m; 2.12, m |
| 25 | 26.8, CH2 | 1.72, m; 1.92, m | 27.7, CH2 | 1.65, m; 1.96, m |
| 26 | 82.2, CH | 4.03, m | 80.7, CH | 4.15, m |
| 27 | 68.3, CH | 3.82, m | 78.8, CH | 3.37, dq (2.4, 6.4) |
| 27-Me | 16.5, CH3 | 0.97, d (6.3) | 11.1, CH3 | 0.95, d (6.4) |
| 27-OMe | - | - | 57.0, CH3 | 3.33, s |
| Sample | Media | Compound | Antimicrobial (µM) 1 | Cytotoxicity (µM) 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SA | CA | AF | A549 | MIA | PANC-1 | ||||||
| MIC90 | MBC90 | MIC90 | MFC90 | MIC90 | MFC90 | IC50 | IC50 | IC50 | |||
| A1099 | CA08LB | Nonactin | 49.2 | 64.9 | 38.1 | - | - | - | 10.1 | 2.3 | 2.9 |
| Monactin | 7.9 | - | 1.1 | 8.2 | - | - | 0.8 | 0.1 | 0.1 | ||
| Dinactin | 4.7 | - | 1.3 | 4.0 | - | - | 1.2 | 0.7 | 0.3 | ||
| A1174 | CA08LB | 4E-Deacetylchromomycin A3 | 2.9 | 13.4 | - | - | - | - | 1.7 | 1.9 | 3.0 |
| Chromomycin A2 | 3.1 | 3.8 | - | - | - | - | 0.3 | 0.5 | 0.4 | ||
| A1301 | CA10LB | Lysolipin I | 0.01 | NT | 0.1 | NT | 0.9 | NT | 0.1 | 0.2 | 0.3 |
| Soyasaponin II | 2.8 | 2.4 | - | - | - | - | 2.1 | 3.1 | 2.1 | ||
| A2461 | CA10LB | Naphthomevalin | - | - | - | - | - | - | 3.7 | 6.4 | 9.0 |
| Positive Controls | Vancomycin hydrochloride | 0.6 | 3.5 | ||||||||
| Amphotericin B | 0.1 | 0.2 | 0.5 | 1.7 | |||||||
| Puromycin | 0.6 | 0.2 | 0.8 | ||||||||
| Compounds | Antimicrobial (µM) 1 | Cytotoxicity (µM) 2 | |||||
|---|---|---|---|---|---|---|---|
| SA25923 | MRSA33591 | A549 | MIA | PANC-1 | |||
| MIC90 | MBC90 | MIC90 | MBC90 | IC50 | IC50 | IC50 | |
| Tetronomycin A (1) | 2.2 | 9.2 | 3.9 | 11.8 | 16.5 | 13.6 | 11.1 |
| Tetronomycin | 0.8 | 1.7 | 0.9 | 2.1 | 8.1 | 6.6 | 4.2 |
| Vancomycin hydrochloride | 0.6 | 3.5 | 0.6 | 2.0 | |||
| Puromycin | 0.6 | 0.2 | 0.8 | ||||
| Components | Media (per L) | ||||
|---|---|---|---|---|---|
| CA02LB | CA07LB | CA08LB | CA09LB | CA10LB | |
| Lab-lemco, Oxoid LP0029 | - | - | - | 10 g | - |
| Cane molasses | - | - | 20 g | - | - |
| Cottonseed flour | - | - | 25 g | - | - |
| Glucose | - | - | 15 g | 20 g | - |
| Glycerol | - | 15 g | - | 3 g | - |
| Mannitol | 20 g | - | - | - | - |
| Oatmeal | - | 30 g | - | - | - |
| Soluble starch | - | - | 40 g | - | 20 g |
| Soybean meal | 20 g | - | - | - | 15 g |
| Yeast extract | - | 5 g | - | 4 g | - |
| CaCO3 | - | - | 8 g | - | - |
| KH2PO4 | - | 5 g | - | - | 3 g |
| Na2HPO4·12H2O | - | 5 g | - | - | 2 g |
| MgCl2·6H2O | - | 1 g | - | - | - |
| MgSO4·7H2O | - | - | - | - | 0.5 g |
| Trace salt sol 1 | - | - | - | - | 1 mL |
| pH | 7.5 | Natural | 7.2 | 7.0 | 7.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chin, E.-J.; Ching, K.-C.; Tan, Z.Y.; Wibowo, M.; Leong, C.-Y.; Yang, L.-K.; Ng, V.W.P.; Seow, D.C.S.; Kanagasundaram, Y.; Ng, S.-B. Natural Products from Singapore Soil-Derived Streptomycetaceae Family and Evaluation of Their Biological Activities. Molecules 2023, 28, 5832. https://doi.org/10.3390/molecules28155832
Chin E-J, Ching K-C, Tan ZY, Wibowo M, Leong C-Y, Yang L-K, Ng VWP, Seow DCS, Kanagasundaram Y, Ng S-B. Natural Products from Singapore Soil-Derived Streptomycetaceae Family and Evaluation of Their Biological Activities. Molecules. 2023; 28(15):5832. https://doi.org/10.3390/molecules28155832
Chicago/Turabian StyleChin, Elaine-Jinfeng, Kuan-Chieh Ching, Zann Y. Tan, Mario Wibowo, Chung-Yan Leong, Lay-Kien Yang, Veronica W. P. Ng, Deborah C. S. Seow, Yoganathan Kanagasundaram, and Siew-Bee Ng. 2023. "Natural Products from Singapore Soil-Derived Streptomycetaceae Family and Evaluation of Their Biological Activities" Molecules 28, no. 15: 5832. https://doi.org/10.3390/molecules28155832
APA StyleChin, E.-J., Ching, K.-C., Tan, Z. Y., Wibowo, M., Leong, C.-Y., Yang, L.-K., Ng, V. W. P., Seow, D. C. S., Kanagasundaram, Y., & Ng, S.-B. (2023). Natural Products from Singapore Soil-Derived Streptomycetaceae Family and Evaluation of Their Biological Activities. Molecules, 28(15), 5832. https://doi.org/10.3390/molecules28155832







