Design, Synthesis, and Characterization of an Amphiphilic Lipoic Acid-Based Ru(III) Complex as a Versatile Tool for the Functionalization of Different Nanosystems
Abstract
1. Introduction
2. Results and Discussion
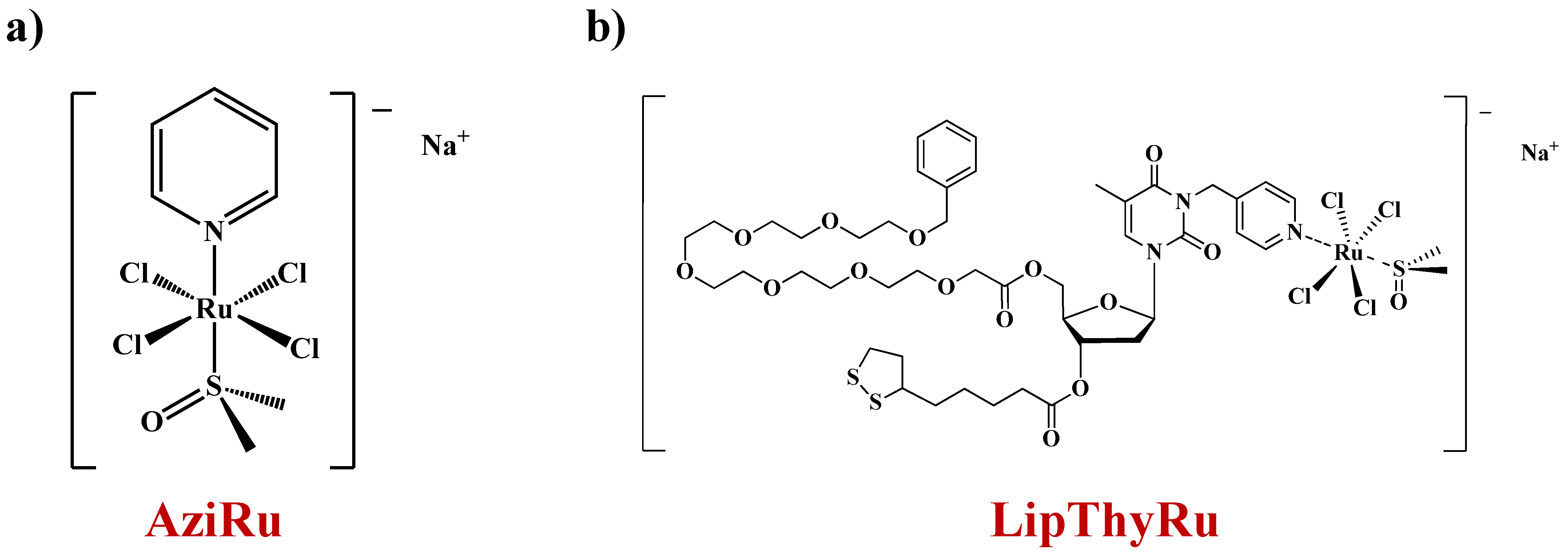
2.1. Design of LipThyRu
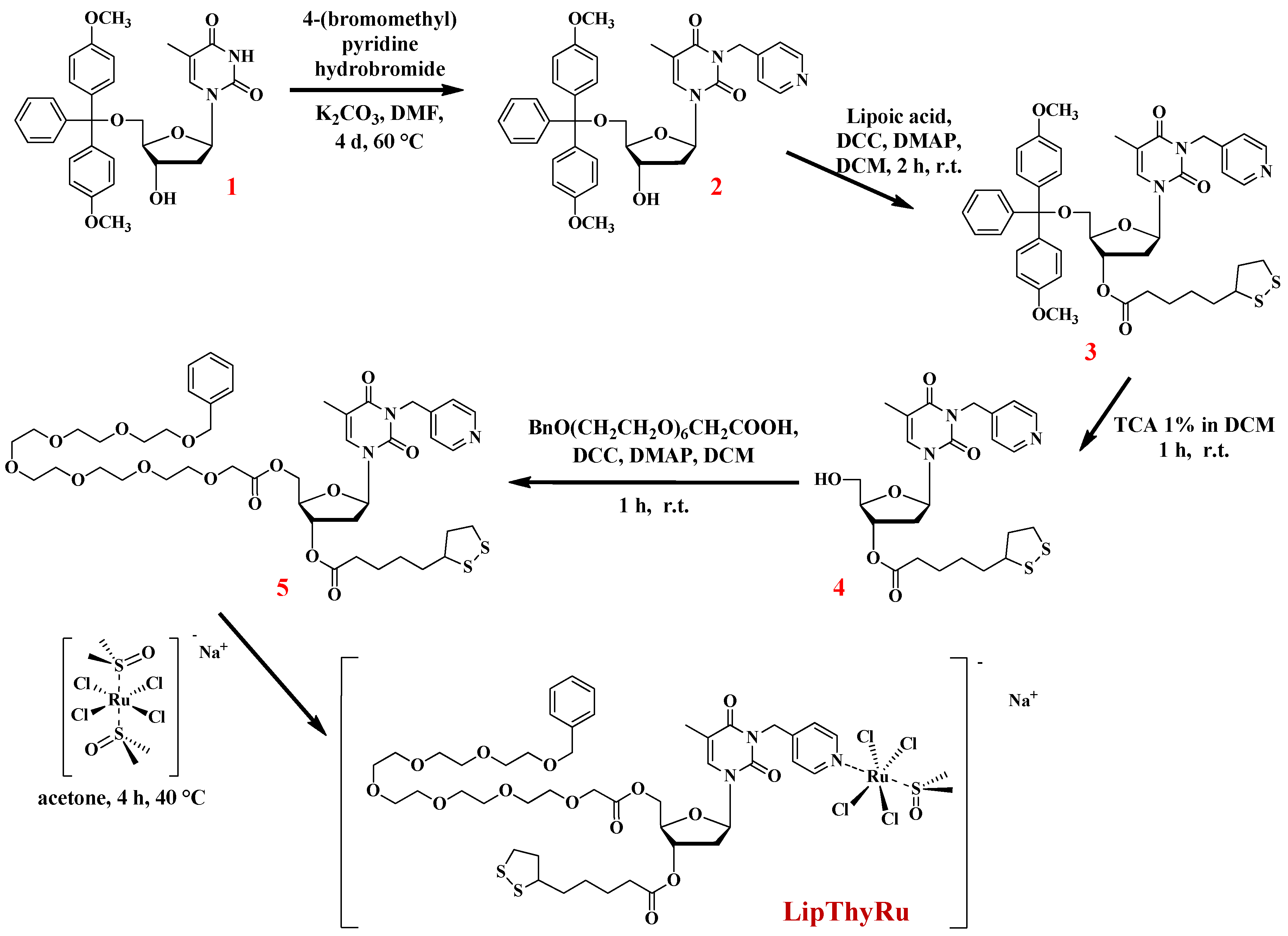
2.2. Synthesis and Characterization of LipThyRu
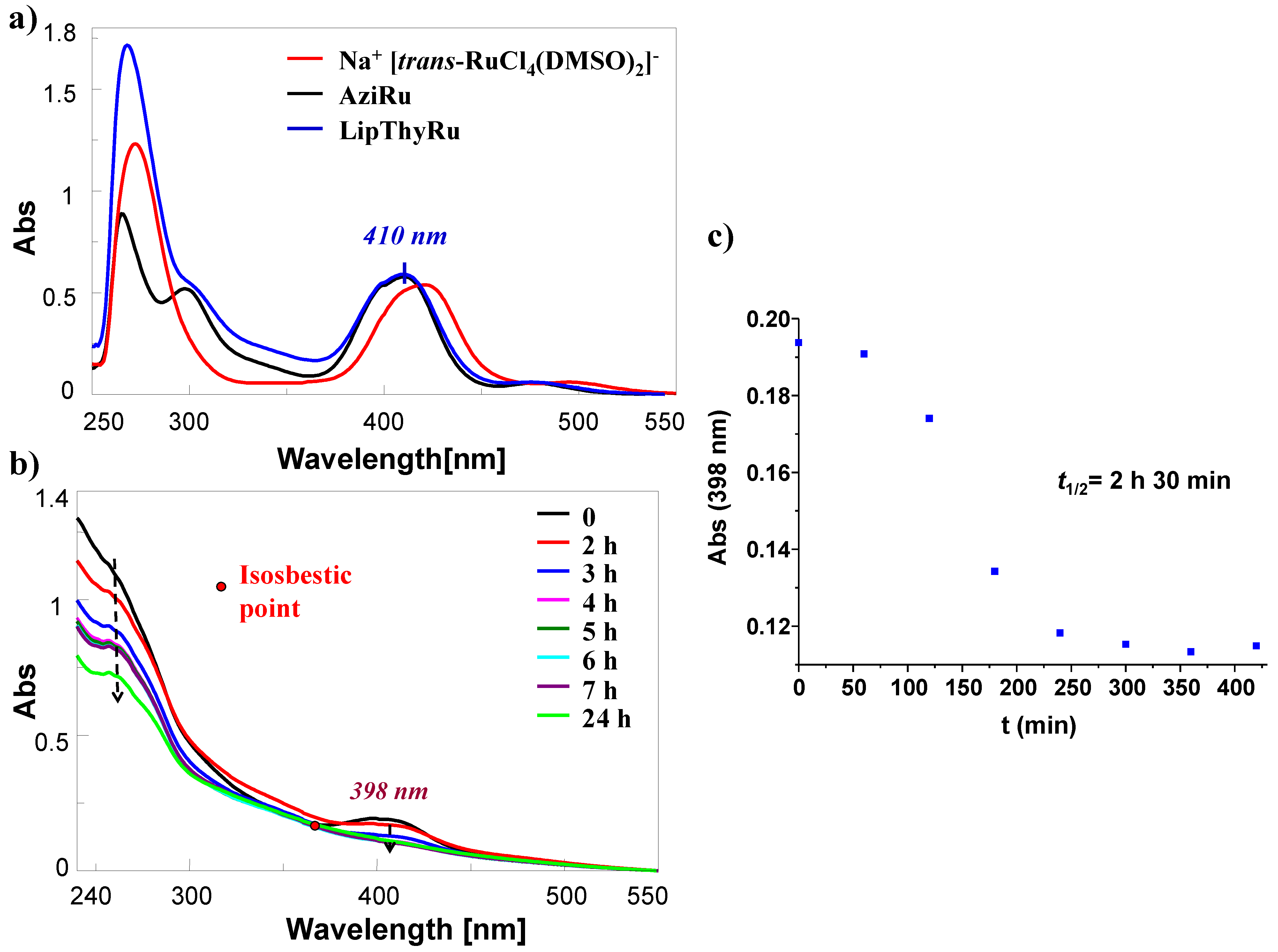
2.3. UV–Vis Studies of LipThyRu
3. Experimental Section
3.1. Materials and General Methods
3.2. Synthesis and Characterization of LipThyRu
3.2.1. Synthesis of 3-(4-pyridylmethyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-thymidine (2)
3.2.2. Synthesis of 3-(4-pyridylmethyl)-3′-O-lipoyl-5′-O-(4,4′-dimethoxytriphenylmethyl)-thymidine (3)
3.2.3. Synthesis of 3-(4-pyridylmethyl)-3′-O-lipoyl-thymidine (4)
3.2.4. Synthesis of 3-(4-pyridylmethyl)-3′-O-lipoyl-5′-O-(benzyloxy)hexaethylene glycol acetyl-thymidine (5)
3.2.5. Synthesis of LipThyRu
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs are unique: Opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef] [PubMed]
- Paprocka, R.; Wiese-Szadkowska, M.; Janciauskiene, S.; Kosmalski, T.; Kulik, M.; Helmin-Basa, A. Latest developments in metal complexes as anticancer agents. Coord. Chem. Rev. 2022, 452, 214307. [Google Scholar] [CrossRef]
- Lucaciu, R.L.; Hangan, A.C.; Sevastre, B.; Oprean, L.S. Metallo-drugs in cancer therapy: Past, present and future. Molecules 2022, 27, 6485. [Google Scholar] [CrossRef]
- Sales, T.A.; Prandi, I.G.; de Castro, A.A.; Leal, D.H.S.; da Cunha, E.F.F.; Kuca, K.; Ramalho, T.C. Recent developments in metal-based drugs and chelating agents for neurodegenerative diseases treatments. Int. J. Mol. Sci. 2019, 20, 1829. [Google Scholar] [CrossRef]
- Ma, D.L.; Wu, C.; Li, G.; Yung, T.L.; Leung, C.H. Transition metal complexes as imaging or therapeutic agents for neurodegenerative diseases. J. Mater. Chem. B 2020, 8, 4715–4725. [Google Scholar] [CrossRef]
- Huffman, S.E.; Yawson, G.K.; Fisher, S.S.; Bothwell, P.J.; Platt, D.C.; Jones, M.A.; Hamaker, C.G.; Webb, M.I. Ruthenium(III) complexes containing thiazole-based ligands that modulate amyloid-β aggregation. Metallomics 2020, 12, 491–503. [Google Scholar] [CrossRef]
- Rodríguez-Arce, E.; Saldías, M. Antioxidant properties of flavonoid metal complexes and their potential inclusion in the development of novel strategies for the treatment against neurodegenerative diseases. Biomed. Pharmacother. 2021, 143, 112236. [Google Scholar] [CrossRef]
- Cao, K.; Zhu, Y.; Hou, Z.; Liu, M.; Yang, Y.; Hu, H.; Dai, Y.; Wang, Y.; Yuan, S.; Huang, G.; et al. α-synuclein as a target for metallo-anti-neurodegenerative agents. Angew. Chem. Int. Engl. 2023, 62, e202215360. [Google Scholar] [CrossRef]
- Gama Justi, F.V.; Matos, G.A.; De Sá Roriz Caminha, J.; Rodrigues Roque, C.; Carvalho, E.M.; Soares Campelo, M.W.; Belayev, L.; De França Lopes, L.G.; Oriá, R.B. The role of ruthenium compounds in neurologic diseases: A minireview. J. Pharmacol. Exp. Ther. 2022, 380, 47–53. [Google Scholar] [CrossRef]
- Coverdale, J.P.C.; Laroiya-McCarron, T.; Romero-Canelón, I. Designing ruthenium anticancer drugs: What have we learnt from the key drug candidates? Inorganics 2019, 7, 31. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.G. Ruthenium complexes as anticancer agents: A brief history and perspectives. Drug Des. Dev. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Skoczynska, A.; Lewinski, A.; Pokora, M.; Paneth, P.; Budzisz, E. An overview of the potential medicinal and pharmaceutical properties of Ru(II)/(III) complexes. Int. J. Mol. Sci. 2023, 24, 9512. [Google Scholar] [CrossRef] [PubMed]
- Rademaker-Lakhai, J.M.; Van Den Bongard, D.; Pluim, D.; Beijnen, J.H.; Schellens, J.H.M. A phase I and pharmacological study with imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a novel ruthenium anticancer agent. Clin. Cancer Res. 2004, 10, 3717–3727. [Google Scholar] [CrossRef]
- Leijen, S.; Burgers, S.A.; Baas, P.; Pluim, D.; Tibben, M.; Van Werkhoven, E.; Alessio, E.; Sava, G.; Beijnen, J.H.; Schellens, J.H.M. Phase I/II study with ruthenium compound NAMI-A and gemcitabine in patients with non-small cell lung cancer after first line therapy. Investig. New Drugs 2015, 33, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Jakupec, M.A.; Arion, V.B.; Kapitza, S.; Reisner, E.; Eichinger, A.; Pongratz, M.; Marian, B.; Keyserlingk, N.G.V.; Keppler, B.K. KP1019 (FFC14A) from bench to bedside: Preclinical and early clinical development- an overview. Int. J. Clin. Pharmacol. Ther. 2005, 43, 595–596. [Google Scholar] [CrossRef] [PubMed]
- Hartinger, C.G.; Zorbas-Seifried, S.; Jakupec, M.A.; Kynast, B.; Zorbas, H.; Keppler, B.K. From bench to bedside—Preclinical and early clinical development of the anticancer agent indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019 or FFC14A). J. Inorg. Biochem. 2006, 100, 891–904. [Google Scholar] [CrossRef]
- Hartinger, C.G.; Jakupec, M.A.; Zorbas-Seifried, S.; Groessl, M.; Egger, A.; Berger, W.; Zorbas, H.; Dyson, P.J.; Keppler, B.K. KP1019, a new redox-active anticancer agent—Preclinical development and results of a clinical phase I study in tumor patients. Chem. Biodivers. 2008, 5, 2140–2155. [Google Scholar] [CrossRef]
- Lentz, F.; Drescher, A.; Lindauer, A.; Henke, M.; Hilger, R.A.; Hartinger, C.G.; Scheulen, M.E.; Dittrich, C.; Keppler, B.K.; Jaehde, U. Pharmacokinetics of a novel anticancer ruthenium complex (KP1019, FFC14A) in a phase I dose-escalation study. Anticancer Drugs 2009, 20, 97–103. [Google Scholar] [CrossRef]
- Dickson, N.R.; Jones, S.F.; Burris, H.A.; Ramanathan, R.K.; Weiss, G.J.; Infante, J.R.; Bendell, J.C.; McCulloch, W.; Von Hoff, D.D. A phase I dose-escalation study of NKP-1339 in patients with advanced solid tumors refractory to treatment. J. Clin. Oncol. 2011, 29, 2607. [Google Scholar] [CrossRef]
- Thompson, D.S.; Weiss, G.J.; Jones, S.F.; Burris, H.A.; Ramanathan, R.K.; Infante, J.R.; Bendell, J.C.; Ogden, A.; Von Hoff, D.D. NKP-1339: Maximum tolerated dose defined for first-in-human GRP78 targeted agent. J. Clin. Oncol. 2012, 30, 3033. [Google Scholar] [CrossRef]
- Trondl, R.; Heffeter, P.; Kowol, C.R.; Jakupec, M.A.; Berger, W.; Keppler, B.K. NKP-1339, the first ruthenium-based anticancer drug on the edge to clinical application. Chem. Sci. 2014, 5, 2925–2932. [Google Scholar] [CrossRef]
- Burris, H.A.; Bakewell, S.; Bendell, J.C.; Infante, J.; Jones, S.F.; Spigel, D.R.; Weiss, G.J.; Ramanathan, R.K.; Ogden, A.; Von Hoff, D. Safety and activity of IT-139, a ruthenium-based compound, in patients with advanced solid tumours: A first-in-human, open-label, dose-escalation phase I study with expansion cohort. ESMO Open 2016, 1, e000154. [Google Scholar] [CrossRef] [PubMed]
- Ravera, M.; Baracco, S.; Cassino, C.; Zanello, P.; Osella, D. Appraisal of the redox behaviour of the antimetastatic ruthenium(III) complex [ImH][RuCl4(DMSO)(Im)], NAMI-A. Dalton Trans. 2004, 15, 2347–2351. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, L.; Liao, S.; Zheng, K.; Ji, L. A theoretical study on the hydrolysis process of the antimetastatic ruthenium(III) complex NAMI-A. J. Phys. Chem. B 2007, 111, 7862–7869. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Robertazzi, A.; Magistrato, A.; Ruggerone, P.; Carloni, P. The hydrolysis mechanism of the anticancer ruthenium drugs NAMI-A and ICR investigated by DFT-PCM calculations. J. Phys. Chem. B 2008, 112, 4401–4409. [Google Scholar] [CrossRef]
- Pashkunova-Martic, I.; Losantos, B.C.; Kandler, N.; Keppler, B. Studies of KP46 and KP1019 and the hydrolysis product of KP1019 in lipiodol emulsions: Preparation and initial characterizations as potential theragnostic agents. Curr. Drug Deliv. 2018, 15, 134–142. [Google Scholar] [CrossRef]
- Pal, M.; Nandi, U.; Mukherjee, D. Detailed account on activation mechanisms of ruthenium coordination complexes and their role as antineoplastic agents. Eur. J. Med. Chem. 2018, 150, 419–445. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Irace, C.; Paduano, L.; Montesarchio, D. Ru(III) complexes for anticancer therapy: The importance of being nucleolipidic. Eur. J. Org. Chem. 2017, 2017, 1100–1119. [Google Scholar] [CrossRef]
- Scintilla, S.; Brustolin, L.; Gambalunga, A.; Chiara, F.; Trevisan, A.; Nardon, C.; Fregona, D. Ru(III) anticancer agents with aromatic and non-aromatic dithiocarbamates as ligands: Loading into nanocarriers and preliminary biological studies. J. Inorg. Biochem. 2016, 165, 159–169. [Google Scholar] [CrossRef]
- D’Amora, A.; Cucciolito, M.E.; Iannitti, R.; Morelli, G.; Palumbo, R.; Ruffo, F.; Tesauro, D. Pyridine ruthenium (III) complexes entrapped in liposomes with enhanced cytotoxic properties in PC-3 prostate cancer cells. J. Drug Deliv. Sci. Technol. 2019, 51, 552–558. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, L.; Li, X.; Shi, Y.; Ding, R.; Teng, M.; Zhang, P.; Cao, C.; Stang, P.J. Self-assembled ruthenium (II) metallacycles and metallacages with imidazole-based ligands and their in vitro anticancer activity. Proc. Natl. Acad. Sci. USA 2019, 116, 4090–4098. [Google Scholar] [CrossRef] [PubMed]
- Brustolin, L.; Pettenuzzo, N.; Nardon, C.; Quarta, S.; Montagner, I.; Pontisso, P.; Rosato, A.; Conte, P.; Merigliano, S.; Fregona, D. Labelled micelles for the delivery of cytotoxic Cu(II) and Ru(III) compounds in the treatment of aggressive orphan cancers: Design and biological in vitro data. J. Inorg. Biochem. 2020, 213, 111259. [Google Scholar] [CrossRef] [PubMed]
- Orts-Arroyo, M.; Silvestre-Llora, A.; Castro, I.; Martínez-Lillo, J. Molecular self-assembly of an unusual dinuclear ruthenium(III) complex based on the nucleobase guanine. Crystals 2022, 12, 448. [Google Scholar] [CrossRef]
- Gallio, A.E.; Brustolin, L.; Pettenuzzo, N.; Fregona, D. Binuclear heteroleptic Ru(III) dithiocarbamate complexes: A step towards tunable antiproliferative agents. Inorganics 2022, 10, 37. [Google Scholar] [CrossRef]
- Simeone, L.; Mangiapia, G.; Vitiello, G.; Irace, C.; Colonna, A.; Ortona, O.; Montesarchio, D.; Paduano, L. Cholesterol-based nucleolipid-ruthenium complex stabilized by lipid aggregates for antineoplastic therapy. Bioconjugate Chem. 2012, 23, 758–770. [Google Scholar] [CrossRef]
- Musumeci, D.; Rozza, L.; Merlino, A.; Paduano, L.; Marzo, T.; Massai, L.; Messori, L.; Montesarchio, D. Interaction of anticancer Ru(III) complexes with single stranded and duplex DNA model systems. Dalton Trans. 2015, 44, 13914–13925. [Google Scholar] [CrossRef]
- Simeone, L.; Mangiapia, G.; Irace, C.; Di Pascale, A.; Colonna, A.; Ortona, O.; De Napoli, L.; Montesarchio, D.; Paduano, L. Nucleolipid nanovectors as molecular carriers for potential applications in drug delivery. Mol. Biosyst. 2011, 7, 3075–3086. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Capuozzo, A.; Irace, C.; King, S.; Russo Krauss, I.; Paduano, L.; Montesarchio, D. “Dressing up” an old drug: An aminoacyl lipid for the functionalization of Ru(III)-based anticancer agents. ACS Biomater. Sci. Eng. 2018, 4, 163–174. [Google Scholar] [CrossRef]
- Montesarchio, D.; Mangiapia, G.; Vitiello, G.; Musumeci, D.; Irace, C.; Santamaria, R.; D’Errico, G.; Paduano, L. A new design for nucleolipid-based Ru(III) complexes as anticancer agents. Dalton Trans. 2013, 42, 16697–16708. [Google Scholar] [CrossRef]
- Riccardi, C.; Campanella, A.; Montesarchio, D.; Del Vecchio, P.; Oliva, R.; Paduano, L. Investigating the interaction of an anticancer nucleolipidic Ru (III) complex with human serum protein: A spectroscopic study. Molecules 2023, 28, 2800. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Trifuoggi, M.; Irace, C.; Paduano, L.; Montesarchio, D. Anticancer ruthenium (III) complexes and Ru(III) containing nanoformulations: An update on the mechanism of action and biological activity. Pharmaceuticals 2019, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, M.; Ferraro, M.G.; Raucci, F.; Riccardi, C.; Saviano, A.; Russo Krauss, I.; Trifuoggi, M.; Caraglia, M.; Paduano, L.; Montesarchio, D.; et al. Safety and efficacy evaluation in vivo of a cationic nucleolipid nanosystem for the nanodelivery of a ruthenium(III) complex with superior anticancer bioactivity. Cancers 2021, 13, 5164. [Google Scholar] [CrossRef]
- Ferraro, M.G.; Bocchetti, M.; Riccardi, C.; Trifuoggi, M.; Paduano, L.; Montesarchio, D.; Misso, G.; Santamaria, R.; Piccolo, M.; Irace, C. Triple negative breast cancer preclinical therapeutic management by a cationic ruthenium-based nucleolipid nanosystem. Int. J. Mol. Sci. 2023, 24, 6473. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Fàbrega, C.; Grijalvo, S.; Vitiello, G.; D’Errico, G.; Eritja, R.; Montesarchio, D. AS1411-decorated niosomes as effective nanocarriers for Ru(III)-based drugs in anticancer strategies. J. Mater. Chem. B 2018, 6, 5368–5384. [Google Scholar] [CrossRef] [PubMed]
- Thi, T.T.H.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The importance of poly(ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, S.; Yang, D.; Fang, Y.; Lin, X.; Jin, X.; Liu, Y.; Liu, X.; Su, K.; Shi, K. Influencing factors and strategies of enhancing nanoparticles into tumors in vivo. Acta Pharm. Sin. B 2021, 11, 2265–2285. [Google Scholar] [CrossRef]
- Turcu, I.; Zarafu, I.; Popa, M.; Chifiriuc, M.C.; Bleotu, C.; Culita, D.; Ghica, C.; Ionita, P. Lipoic acid gold nanoparticles functionalized with organic compounds as bioactive materials. Nanomaterials 2017, 7, 43. [Google Scholar] [CrossRef]
- Dzwonek, M.; Załubiniak, D.; Piatek, P.; Cichowicz, G.; Mȩczynska-Wielgosz, S.; Stȩpkowski, T.; Kruszewski, M.; Wiȩckowska, A.; Bilewicz, R. Towards potent but less toxic nanopharmaceuticals-lipoic acid bioconjugates of ultrasmall gold nanoparticles with an anticancer drug and addressing unit. RSC Adv. 2018, 8, 14947–14957. [Google Scholar] [CrossRef]
- Guzmán-Soto, I.; Omole, M.; Alarcon, E.I.; McTiernan, C.D. Lipoic acid capped silver nanoparticles: A facile route to covalent protein capping and oxidative stability within biological systems. RSC Adv. 2020, 10, 32953–32958. [Google Scholar] [CrossRef]
- Hajtuch, J.; Santos-Martinez, M.J.; Wojcik, M.; Tomczyk, E.; Jaskiewicz, M.; Kamysz, W.; Narajczyk, M.; Inkielewicz-Stepniak, I. Lipoic acid-coated silver nanoparticles: Biosafety potential on the vascular microenvironment and antibacterial properties. Front. Pharmacol. 2022, 12, 733743. [Google Scholar] [CrossRef]
- Hahm, J.I. Zinc oxide nanomaterials for biomedical fluorescence detection. J. Nanosci. Nanotechnol. 2014, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Trzciński, J.W.; Morillas-Becerril, L.; Scarpa, S.; Tannorella, M.; Muraca, F.; Rastrelli, F.; Castellani, C.; Fedrigo, M.; Angelini, A.; Tavano, R.; et al. Poly(lipoic acid)-based nanoparticles as self-organized, biocompatible, and corona-free nanovectors. Biomacromolecules 2021, 22, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, G.; Bang, E.K.; Molinard, G.; Tulumello, D.V.; Ward, S.; Kelley, S.O.; Roux, A.; Sakai, N.; Matile, S. Cellular uptake of substrate-initiated cell-penetrating poly(disulfide)s. J. Am. Chem. Soc. 2014, 136, 6069–6074. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Neises, B.; Steglich, W. Simple Method for the Esterification of Carboxylic Acids. Angew. Chem.-Int. Ed. Eng. 1978, 17, 522–524. [Google Scholar] [CrossRef]
- Hassner, A.; Alexanian, V. Direct room temperature esterification of carboxylic acids. Tetrahedron Lett. 1978, 19, 4475–4478. [Google Scholar] [CrossRef]
- Alessio, E.; Balducci, G.; Calligaris, M.; Costa, G.; Attia, W.M.; Mestroni, G. Synthesis, molecular structure, and chemical behavior of hydrogen trans-bis(dimethyl sulfoxide)tetrachlororuthenate(III) and mer-trichlorotris(dimethyl sulfoxide)ruthenium(III): The first fully characterized chloride-dimethyl sulfoxide-ruthenium(III) comp. Inorg. Chem. 1991, 30, 609–618. [Google Scholar] [CrossRef]
- Alessio, E.; Balducci, G.; Lutman, A.; Mestroni, G.; Calligaris, M.; Attia, W.M. Synthesis and characterization of two new classes of ruthenium(III)-sulfoxide complexes with nitrogen donor ligands (L): Na[trans-RuCl4(R2SO)(L)] and mer, cis-RuCl3(R2SO)(R2SO)(L). The crystal structure of Na[trans-RuCl4(DMSO)(NH3)] · 2DMSO, Na[trans-RuCl. Inorg. Chim. Acta 1993, 203, 205–217. [Google Scholar] [CrossRef]
- Velders, A.H.; Bergamo, A.; Alessio, E.; Zangrando, E.; Haasnoot, J.G.; Casarsa, C.; Cocchietto, M.; Zorzet, S.; Sava, G. Synthesis and chemical-pharmacological characterization of the antimetastatic NAMI-A-yype Ru (III) complexes 5,7-dimethyl[1, 2, 4]triazolo[1, 5-a]pyrimidine). J. Med. Chem. 2004, 47, 1110–1121. [Google Scholar] [CrossRef]
- Riccardi, C.; Piccolo, M.; Ferraro, M.G.; Graziano, R.; Musumeci, D.; Trifuoggi, M.; Irace, C.; Montesarchio, D. Bioengineered lipophilic Ru (III) complexes as potential anticancer agents. Biomater. Adv. 2022, 139, 213016. [Google Scholar] [CrossRef]
- Webb, M.I.; Chard, R.A.; Al-Jobory, Y.M.; Jones, M.R.; Wong, E.W.Y.; Walsby, C.J. Pyridine analogs of the antimetastatic Ru(III) complex NAMI-A targeting non-covalent interactions with albumin. Inorg. Chem. 2012, 51, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Rosemeyer, H. Nucleolipids: Natural occurrence, synthesis, molecular recognition, and supramolecular assemblies as potential precursors of life and bioorganic materials. Chem. Biodivers. 2005, 2, 977–1063. [Google Scholar] [CrossRef] [PubMed]
- Berti, D.; Bombelli, F.B.; Fortini, M.; Baglioni, P. Amphiphilic self-assemblies decorated by nucleobases. J. Phys. Chem. B 2007, 111, 11734–11744. [Google Scholar] [CrossRef]
- Barthélemy, P. Nucleoside-based lipids at work: From supramolecular assemblies to biological applications. Comptes Rendus Chim. 2009, 12, 171–179. [Google Scholar] [CrossRef]
- Allain, V.; Bourgaux, C.; Couvreur, P. Self-assembled nucleolipids: From supramolecular structure to soft nucleic acid and drug delivery devices. Nucleic Acids Res. 2012, 40, 1891–1903. [Google Scholar] [CrossRef]
- Kashapov, R.; Gaynanova, G.; Gabdrakhmanov, D.; Kuznetsov, D.; Pavlov, R.; Petrov, K.; Zakharova, L.; Sinyashin, O. Self-assembly of amphiphilic compounds as a versatile tool for construction of nanoscale drug carriers. Int. J. Mol. Sci. 2020, 21, 6961. [Google Scholar] [CrossRef]
- Felton, C.; Karmakar, A.; Gartia, Y.; Ramidi, P.; Biris, A.S.; Ghosh, A. Magnetic nanoparticles as contrast agents in biomedical imaging: Recent advances in iron- and manganese-based magnetic nanoparticles. Drug Metab. Rev. 2014, 46, 142–154. [Google Scholar] [CrossRef]
- Caspani, S.; Magalhães, R.; Araújo, J.P.; Sousa, C.T. Magnetic nanomaterials as contrast agents for MRI. Materials 2020, 13, 2586. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riccardi, C.; Platella, C.; Musumeci, D.; Montesarchio, D. Design, Synthesis, and Characterization of an Amphiphilic Lipoic Acid-Based Ru(III) Complex as a Versatile Tool for the Functionalization of Different Nanosystems. Molecules 2023, 28, 5775. https://doi.org/10.3390/molecules28155775
Riccardi C, Platella C, Musumeci D, Montesarchio D. Design, Synthesis, and Characterization of an Amphiphilic Lipoic Acid-Based Ru(III) Complex as a Versatile Tool for the Functionalization of Different Nanosystems. Molecules. 2023; 28(15):5775. https://doi.org/10.3390/molecules28155775
Chicago/Turabian StyleRiccardi, Claudia, Chiara Platella, Domenica Musumeci, and Daniela Montesarchio. 2023. "Design, Synthesis, and Characterization of an Amphiphilic Lipoic Acid-Based Ru(III) Complex as a Versatile Tool for the Functionalization of Different Nanosystems" Molecules 28, no. 15: 5775. https://doi.org/10.3390/molecules28155775
APA StyleRiccardi, C., Platella, C., Musumeci, D., & Montesarchio, D. (2023). Design, Synthesis, and Characterization of an Amphiphilic Lipoic Acid-Based Ru(III) Complex as a Versatile Tool for the Functionalization of Different Nanosystems. Molecules, 28(15), 5775. https://doi.org/10.3390/molecules28155775









