Modification of the 4-Hydroxyphenylacetate-3-hydroxylase Substrate Pocket to Increase Activity towards Resveratrol
Abstract
1. Introduction
2. Results and Discussion
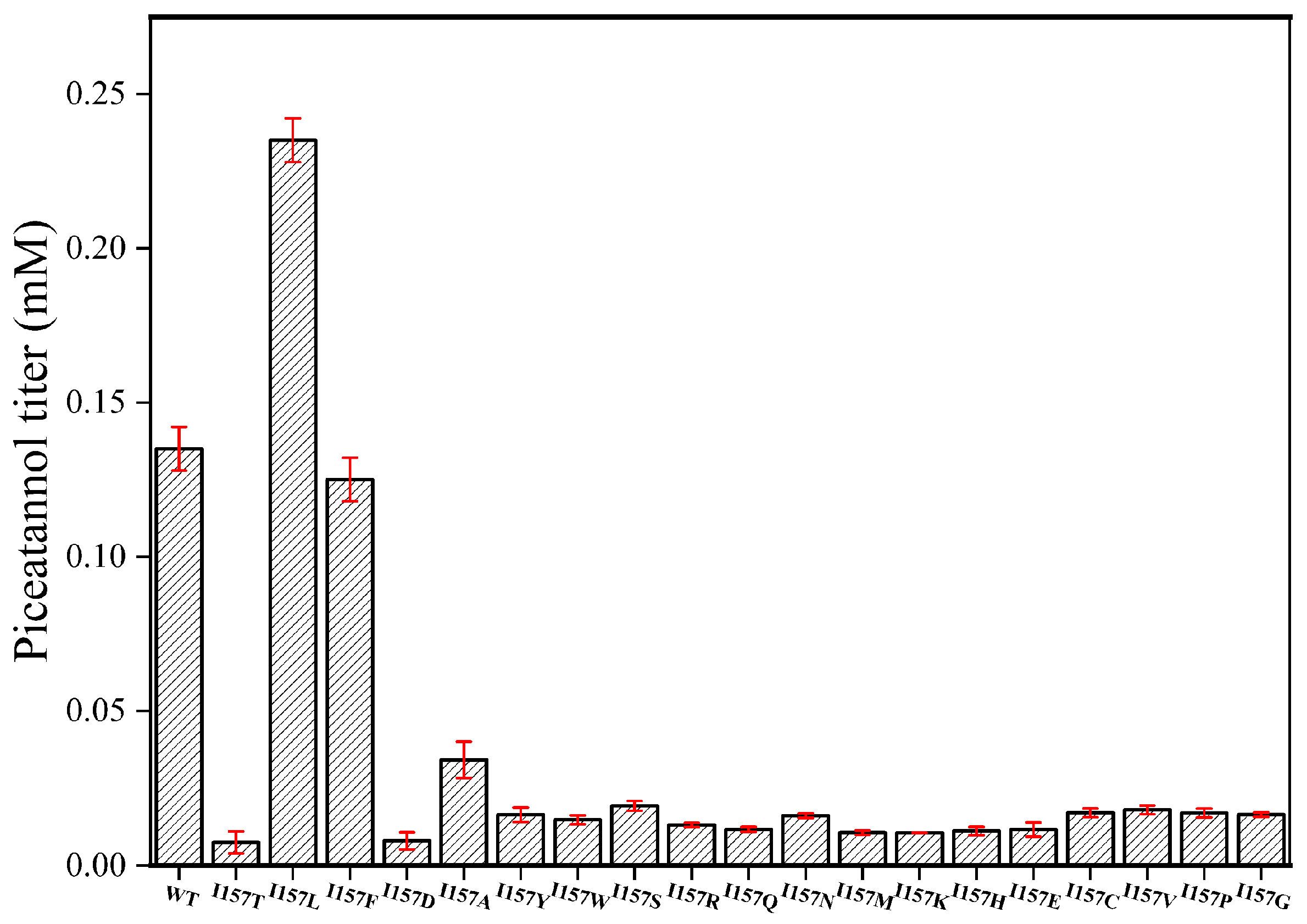
2.1. Key Catalytic Amino Acids in the Substrate Pocket
2.2. Determination of Enzymatic Parameters of Mutant Enzymes and Wild-Type (WT)
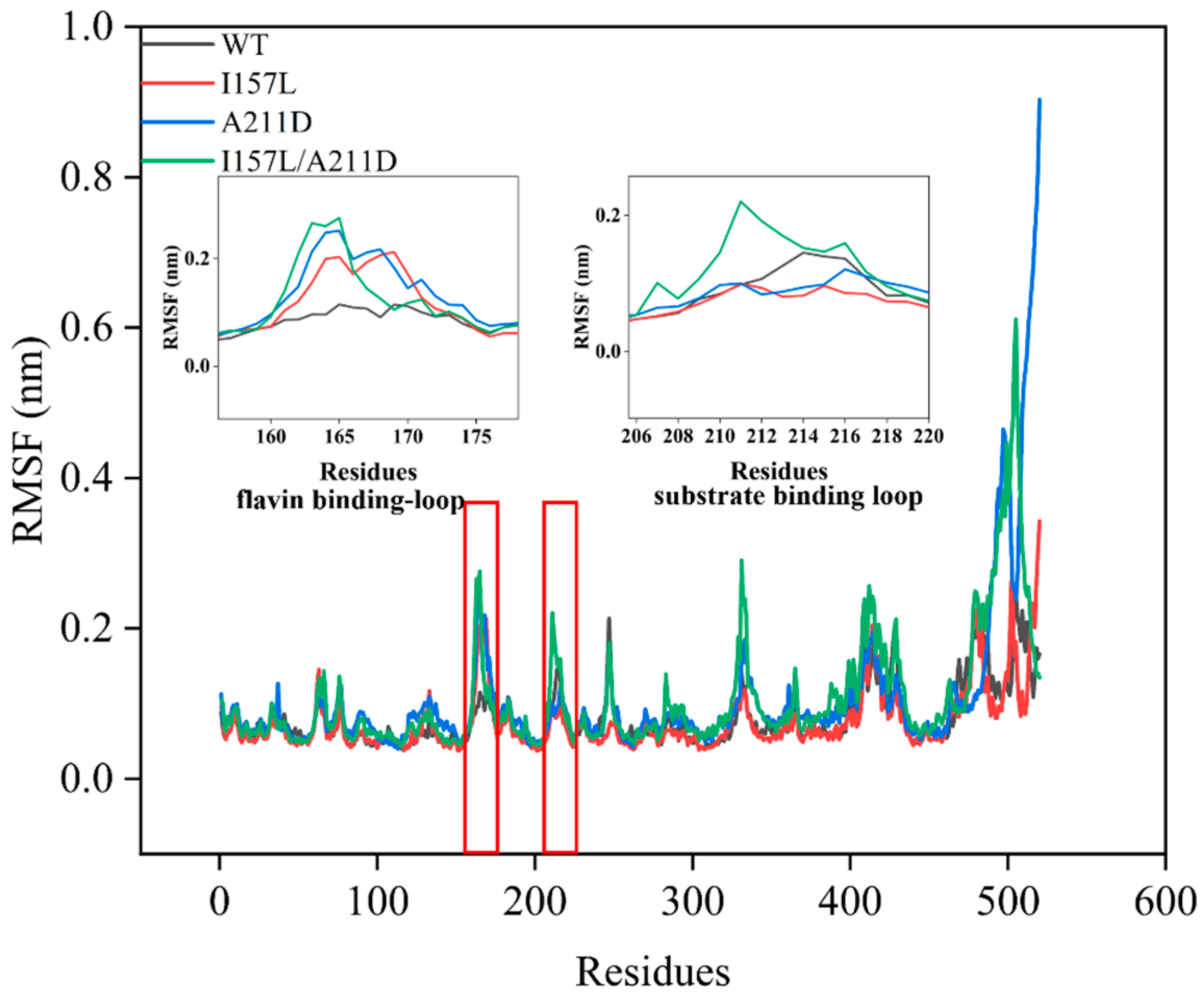
2.3. Mechanistic Insights into Enhanced Catalytic Efficiency of Mutants towards Resveratrol
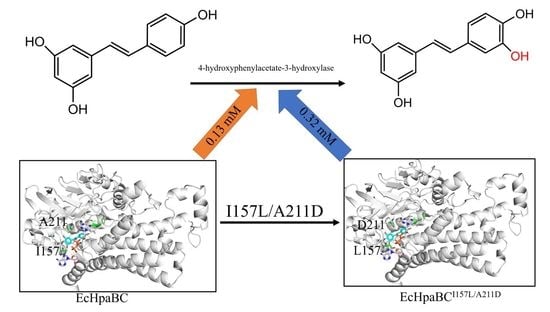
2.4. Preparation of Piceatannol Using Whole-Cell Catalysts
3. Materials and Methods
3.1. Strains and Materials
3.2. Plasmid Construction
3.3. Molecular Docking and Molecular Dynamics Simulations (MD)
3.4. Construction of Mutants
3.5. Preparation of the Whole-Cell Biocatalyst
3.6. Whole-Cell Biocatalytic Activity Assay
3.7. Enzyme Purification
3.8. Enzymatic Parameters of WT Enzyme and Mutant Enzymes
3.9. Optimization of the WT and I157L/A211D-Catalyzed Reactions
3.10. Production of Piceatannol from Resveratrol Using WT and I157L/A211D
3.11. High-Performance Liquid Chromatography (HPLC) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Lee, C.H.; Yang, H.; Park, J.H.Y.; Kim, J.-E.; Lee, K.W. Piceatannol, a metabolite of resveratrol, attenuates atopic dermatitis by targeting Janus kinase 1. Phytomedicine 2022, 99, 153981. [Google Scholar] [CrossRef] [PubMed]
- Ayan, I.C.; Guclu, E.; Vural, H.; Dursun, H.G. Piceatannol induces apoptotic cell death through activation of caspase-dependent pathway and upregulation of ROS-mediated mitochondrial dysfunction in pancreatic cancer cells. Mol. Biol. Rep. 2022, 49, 11947–11957. [Google Scholar] [CrossRef]
- Krishna, N.P.U.; Jalala, V.K.V.; Muraleedharan, K. Complexation behaviour of piceatannol ligand with Ti(IV) and Zr(IV) metal ions: A combined DFT and deep learning investigation. Struct. Chem. 2023. [Google Scholar] [CrossRef]
- Majeed, M.; Nagabhushanam, K.; Bhat, B.; Ansari, M.; Pandey, A.; Bani, S.; Mundkur, L. The Anti-Obesity Potential of Cyperus rotundus Extract Containing Piceatannol, Scirpusin A and Scirpusin B from Rhizomes: Preclinical and Clinical Evaluations. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Tagai, N.; Ogino, Y.; Uozumi, H.; Kawakami, S.; Yamamoto, T.; Tanuma, S.-i.; Maruki-Uchida, H.; Mori, S.; Morita, M. Passion fruit seed extract protects beta-amyloid-induced neuronal cell death in a differentiated human neuroblastoma SH-SY5Y cell model. Food Sci. Nutr. 2022, 10, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Krambeck, K.; Santos, D.; Sousa Lobo, J.M.; Amaral, M.H. Benefits of skin application of piceatannol—A minireview. Australas. J. Dermatol. 2023, 64, 21–25. [Google Scholar] [CrossRef]
- Bavaresco, L.; Fregoni, M.; Trevisan, M.; Mattivi, F.; Vrhovsek, U.; Falchetti, R. The occurrence of the stilbene piceatannol in grapes. Vitis 2002, 41, 133–136. [Google Scholar]
- Heo, K.T.; Kang, S.-Y.; Jang, J.-H.; Hong, Y.-S. Sam5, a Coumarate 3-Hydroxylase from Saccharothrix espanaensis: New Insight into the Piceatannol Production as a Resveratrol 3′-Hydroxylase. Chemistryselect 2017, 2, 8785–8789. [Google Scholar] [CrossRef]
- Lin, Y.; Yan, Y. Biotechnological Production of Plant-Specific Hydroxylated Phenylpropanoids. Biotechnol. Bioeng. 2014, 111, 1895–1899. [Google Scholar] [CrossRef]
- Shrestha, A.; Pandey, R.P.; Sohng, J.K. Biosynthesis of resveratrol and piceatannol in engineered microbial strains: Achievements and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2959–2972. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Y.; Chen, D.; Tao, J.; Wang, H.; Liu, W. A Bacterial Cytochrome P450 Enzyme Catalyzes Multistep Oxidation Reactions in Pyrroindomycin Biosynthesis. Chin. J. Chem. 2023. [Google Scholar] [CrossRef]
- Pandey, B.P.; Lee, N.; Choi, K.Y.; Jung, E.; Jeong, D.H.; Kim, B.G. Screening of bacterial cytochrome P450s responsible for regiospecific hydroxylation of (iso)flavonoids. Enzym. Microb. Technol. 2011, 48, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kim, E.J.; Kim, B.G. Regioselective hydroxylation of trans-resveratrol via inhibition of tyrosinase from Streptomyces avermitilis MA4680. ACS Chem. Biol. 2012, 7, 1687–1692. [Google Scholar] [CrossRef]
- Deng, Y.; Faivre, B.; Back, O.; Lombard, M.; Pecqueur, L.; Fontecave, M. Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli. Chembiochem 2020, 21, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Hisano, T.; Iwasaki, W.; Ebihara, A.; Miki, K. Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity. Proteins-Struct. Funct. Bioinform. 2008, 70, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Chaiyen, P.; Suadee, C.; Wilairat, P. A novel two-protein component flavoprotein hydroxylase—p-hydroxyphenylacetate hydroxylase from Acinetobacter baumannii. Eur. J. Biochem. 2001, 268, 5550–5561. [Google Scholar] [CrossRef]
- Kim, S.H.; Hisano, T.; Takeda, K.; Iwasaki, W.; Ebihara, A.; Miki, K. Crystal structure of the oxygenase component (HpaB) of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8. J. Biol. Chem. 2007, 282, 33107–33117. [Google Scholar] [CrossRef]
- Yuenyao, A.; Petchyam, N.; Kamonsutthipaijit, N.; Chaiyen, P.; Pakotiprapha, D. Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands. Arch. Biochem. Biophys. 2018, 653, 24–38. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ortiz-Maldonado, M.; Entsch, B.; Ballou, D.P. Studies on the mechanism of p-hydroxyphenylacetate 3-hydroxylase from Pseudomonas aeruginosa: A system composed of a small flavin reductase and a large flavin-dependent oxygenase. Biochemistry 2010, 49, 372–385. [Google Scholar] [CrossRef]
- Arunachalam, U.; Massey, V.; Vaidyanathan, C.S. p-Hydroxyphenylacetate-3-hydroxylase. A two-protein component enzyme. J. Biol. Chem. 1992, 267, 25848–25855. [Google Scholar] [CrossRef]
- Yao, J.; He, Y.; Su, N.; Bharath, S.R.; Tao, Y.; Jin, J.-M.; Chen, W.; Song, H.; Tang, S.-Y. Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps. Nat. Commun. 2020, 11, 1515. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhou, D.; Lin, Y.; Wang, J.; Gao, S.; Kandavelu, P.; Zhang, H.; Zhang, R.; Wang, B.C.; Rose, J.; et al. Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli. Sci. Rep. 2019, 9, 7087. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-F.; Wang, C.-S.; Qiao, J.; Zhao, G.-R. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab. Eng. 2013, 19, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, H.; Song, W.; Chen, X.; Liu, J.; Luo, Q.; Liu, L. Engineering of the Conformational Dynamics of Lipase To Increase Enantioselectivity. ACS Catal. 2017, 7, 7593–7599. [Google Scholar] [CrossRef]
- Liu, B.; Qu, G.; Li, J.K.; Fan, W.; Ma, J.A.; Xu, Y.; Nie, Y.; Sun, Z. Conformational Dynamics-Guided Loop Engineering of an Alcohol Dehydrogenase: Capture, Turnover and Enantioselective Transformation of Difficult-to-Reduce Ketones. Adv. Synth. Catal. 2019, 361, 3182–3190. [Google Scholar] [CrossRef]
- Wang, H.; Wang, S.; Wang, J.; Shen, X.; Feng, X.; Yuan, S.; Sun, X.; Yuan, Q. Engineering a Prokaryotic Non-P450 Hydroxylase for 3′-Hydroxylation of Flavonoids. ACS Synth. Biol. 2022, 11, 3865–3873. [Google Scholar] [CrossRef]
- Furuya, T.; Sai, M.; Kino, K. Biocatalytic synthesis of 3,4,5,3′,5′-pentahydroxy-trans-stilbene from piceatannol by two-component flavin-dependent monooxygenase HpaBC. Biosci. Biotechnol. Biochem. 2016, 80, 193–198. [Google Scholar] [CrossRef]
- Punnatin, P.; Chanchao, C.; Chunsrivirot, S. Molecular dynamics reveals insight into how N226P and H227Y mutations affect maltose binding in the active site of alpha-glucosidase II from European honeybee, Apis mellifera. PLoS ONE 2020, 15, e0229734. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Hu, S.; Zhao, W.R.; Huang, J.; Mei, J.Q.; Mei, L.H. Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase. Molecules 2020, 25, 690. [Google Scholar] [CrossRef]
- Furuya, T.; Sai, M.; Kino, K. Efficient monooxygenase-catalyzed piceatannol production: Application of cyclodextrins for reducing product inhibition. J. Biosci. Bioeng. 2018, 126, 478–481. [Google Scholar] [CrossRef]
- Zheng, F.; Tu, T.; Wang, X.; Wang, Y.; Ma, R.; Su, X.; Xie, X.; Yao, B.; Luo, H. Enhancing the catalytic activity of a novel GH5 cellulase GtCel5 from Gloeophyllum trabeum CBS 900.73 by site-directed mutagenesis on loop 6. Biotechnol. Biofuels 2018, 11, 76. [Google Scholar] [CrossRef]
- Han, S.-W.; Park, E.-S.; Dong, J.-Y.; Shin, J.-S. Active-Site Engineering of ω-Transaminase for Production of Unnatural Amino Acids Carrying a Side Chain Bulkier than an Ethyl Substituent. Appl. Environ. Microbiol. 2015, 81, 6994–7002. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, S.; Kim, D.; Yoon, S.H. A single amino acid substitution in aromatic hydroxylase (HpaB) of Escherichia coli alters substrate specificity of the structural isomers of hydroxyphenylacetate. BMC Microbiol. 2020, 20, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zheng, P.; Chen, P.; Wu, D. Engineering an α-L-rhamnosidase from Aspergillus niger for efficient conversion of rutin substrate. Biochem. Eng. J. 2022, 186, 108572. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, J.H.; Park, J.H.; Lee, N.; Kim, D.-H.; Jang, K.-S.; Park, I.L.H.; Kim, B.-G. Protein engineering of alpha 2,3/2,6-sialyltransferase to improve the yield and productivity of in vitro sialyllactose synthesis. Glycobiology 2014, 24, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, X.; Qin, Z.; Yu, S.; Chen, J.; Zhou, J. Combined engineering of L-sorbose dehydrogenase and fermentation optimization to increase 2-keto-L-gulonic acid production in Escherichia coli. Bioresour. Technol. 2023, 372, 128672. [Google Scholar] [CrossRef] [PubMed]










| Name | Km (mM) | Kcat (min−1) | Kcat/Km (min−1mM−1) |
|---|---|---|---|
| WT | 0.67 ± 0.12 | 0.81 ± 0.057 | 1.2 |
| I157L | 0.33 ± 0.056 | 0.77 ± 0.036 | 2.33 |
| A211D | 0.60 ± 0.030 | 1.89 ± 0.040 | 3.15 |
| I157L/A211D | 1.36 ± 0.3 | 7.79 ± 0.35 | 5.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Jin, Y.; Yang, K.; Hu, S.; Lv, C.; Huang, J.; Mei, J.; Zhao, W.; Mei, L. Modification of the 4-Hydroxyphenylacetate-3-hydroxylase Substrate Pocket to Increase Activity towards Resveratrol. Molecules 2023, 28, 5602. https://doi.org/10.3390/molecules28145602
Zhang Q, Jin Y, Yang K, Hu S, Lv C, Huang J, Mei J, Zhao W, Mei L. Modification of the 4-Hydroxyphenylacetate-3-hydroxylase Substrate Pocket to Increase Activity towards Resveratrol. Molecules. 2023; 28(14):5602. https://doi.org/10.3390/molecules28145602
Chicago/Turabian StyleZhang, Qianchao, Yuning Jin, Kai Yang, Sheng Hu, Changjiang Lv, Jun Huang, Jiaqi Mei, Weirui Zhao, and Lehe Mei. 2023. "Modification of the 4-Hydroxyphenylacetate-3-hydroxylase Substrate Pocket to Increase Activity towards Resveratrol" Molecules 28, no. 14: 5602. https://doi.org/10.3390/molecules28145602
APA StyleZhang, Q., Jin, Y., Yang, K., Hu, S., Lv, C., Huang, J., Mei, J., Zhao, W., & Mei, L. (2023). Modification of the 4-Hydroxyphenylacetate-3-hydroxylase Substrate Pocket to Increase Activity towards Resveratrol. Molecules, 28(14), 5602. https://doi.org/10.3390/molecules28145602